Abstract
Here, we describe the cloning, sequencing and characterization of an immunoglobulin A (IgA) Fc receptor from cattle (bFcαR). By screening a translated EST database with the protein sequence of the human IgA Fc receptor (CD89) we identified a putative bovine homologue. Subsequent polymerase chain reaction (PCR) amplification confirmed that the identified full-length cDNA was expressed in bovine cells. COS-1 cells transfected with a plasmid containing the cloned cDNA bound to beads coated with either bovine or human IgA, but not to beads coated with bovine IgG2 or human IgG. The bFcαR cDNA is 873 nucleotides long and is predicted to encode a 269 amino-acid transmembrane glycoprotein composed of two immunoglobulin-like extracellular domains, a transmembrane region and a short cytoplasmic tail devoid of known signalling motifs. Genetically, bFcαR is more closely related to CD89, bFcγ2R, NKp46, and the KIR and LILR gene families than to other FcRs. Moreover, the bFcαR gene maps to the bovine leucocyte receptor complex on chromosome 18. Identification of the bFcαR will aid in the understanding of IgA–FcαR interactions, and may facilitate the isolation of FcαR from other species.
Introduction
Phagocytes express specific receptors (FcRs) that bind the Fc regions of various immunoglobulin molecules. Ligation of FcRs can trigger numerous cellular effector functions, including phagocytosis, antibody-dependent cellular cytotoxicity, and the secretion of cytokines and other inflammatory mediators.1–3 FcRs therefore provide a crucial link between the humoral and cellular arms of the immune system.
Amongst FcRs, the three classes of immunoglobulin G (IgG) FcR (FcγRI, -II, and -III), present in humans and mice, are the best characterized.1–3 Although less well understood, cattle also possess homologues of FcγRI, -II and -III.4–6 In murine models, FcγRs have been shown to be important in protection against disease.7–10 More recently, numerous monoclonal and bispecific antibodies are being tested for their ability to exploit the powerful cellular triggering properties of FcγRs for therapeutic purposes.11
Numerous species have also been reported to possess immunoglobulin A (IgA) FcRs. However, to date, the human FcαRI (CD89) is the only IgA FcR that has been cloned from any species.12 CD89 has recently been shown to be as effective as the FcγRs in triggering cellular effector functions.13 Indeed, in some experiments, CD89 performed even better than FcγRs.14,15 Therefore, CD89 and related FcαRs may provide a powerful alternative or complementary way to effectively trigger immune phagocytes.
CD89 is expressed on the surface of neutrophils, monocytes and eosinophils. It consists of two extracellular immunoglobulin-like domains, a transmembrane region and a short cytoplasmic tail.12 Like many other FcRs, signal transduction via CD89 occurs through association with the FcR γ chain via oppositely charged residues within the transmembrane domains of the molecules.16
The FcR most closely related to CD89 is the bovine Fcγ2R (bFcγ2R), which binds bovine IgG2.17 CD89 and bFcγ2R represent a separate class of mammalian FcRs, more closely related to molecules such as the killer cell immunoglobulin-like receptors (KIR/CD158), NKp46, leucocyte-associated immunoglobulin-like receptor (LAIR)-1 and -2, GPVI and a family of proteins previously best known as immunoglobulin-like transcripts (ILTs), leucocyte immunoglobulin-like receptors (LIRs) or monocyte/macrophage immunoglobulin-related receptors (MIRs). This gene family has recently been renamed as the leucocyte immunoglobulin-like receptors (LILRs: see http://www.gene.ucl.ac.uk/nomenclature/genefamily/lilr.html). In humans, the KIR, NKp46, GPVI, LILR, LAIR and CD89 genes lie within the leucocyte receptor complex (LRC) on chromosome 19q13.4.18,19
Several reports have suggested that cattle may express an FcR homologous to CD89. Zhang et al. reported that bovine cells were able to bind human IgA via an unknown receptor.17 More recently, it was shown that recombinant bovine IgA could bind to CD89 on human neutrophils and trigger a respiratory burst.20 Therefore, we reasoned that the observed cross-reactivity between humans and cattle suggested the existence of a bovine IgA FcR, homologous to CD89. In this report we describe the cloning, sequencing and characterization of a bovine IgA FcR, which we believe to represent the bovine orthologue of CD89.
Materials and methods
Cell culture
COS-1 cells were maintained in Dulbecco's modified Eagle's minimal essential medium (DMEM; BioWhittaker Walkersville, MD) supplemented with 10% fetal bovine serum, 1 mm l-glutamine and 50 µg/ml gentamycin (Life Technologies, Paisley, UK).
Immunoglobulin preparations
Purified bovine IgA (bIgA) was purchased from Inter-Cell Technologies Inc. (Hopewell, NJ). The construction of relevant expression vectors, generation of immunoglobulin-secreting cell lines and purification of recombinant anti-NIP bovine IgA (rbIgA) has been described previously.20 Polymeric human IgA (hIgA) was purified at LIIPAT according to standard methodologies. Purified bovine IgG2 (bIgG2) was a kind gift from Dr C. J. Howard (Institute for Animal Health, Compton, UK). Purified human IgG was purchased from Rikshospitalet, Oslo.
Monoclonal antibodies (mAbs)
Sheep anti-bovine IgA-conjugated fluorescein isothiocyanate (FITC) was purchased from Serotec (Oxford, UK). Mouse anti-bovine κ-light chain and anti-bovine λ-light chain were purchased from VMRD (Pullman, WT). A hybridoma (AFRC IAH-CC42), secreting mIgG1 mAbs against bovine CD2, was obtained from ECACC (European Collection of Cell Cultures, Parton Down, UK). Goat anti-mouse immunoglobulin M (IgM)–, IgG1– and IgG2b–FITC conjugates were purchased from Southern Biotechnology (Birmingham, AL).
Isolation of bovine blood cells and preparation of bovine cDNA
Bovine mononuclear cells (MNCs) and polymorphonuclear neutrophils (PMNs) were purified from the peripheral blood of cattle by density centrifugation on lymphoprep (Nycomed, Oslo, Norway). PMNs were recovered from the red cell pellet by isotonic lysis. For isolation of specific cell subsets, the cells were first stained with anti-CD2 for T cells, anti-κ light chain/anti-λ light chain for B cells, or CC-G24 for PMNs and monocytes. Following incubation with an appropriate fluorochrome-labelled secondary Ab, the cells were sorted in a FACS Vantage cell sorter (Becton-Dickinson, Franklin Lakes, NJ) prior to RNA isolation. Total RNA was isolated using RNAWiz solution (Ambion, Austin, Tx), according to the manufacturer's instructions, and cDNA was subsequently synthesized using an oligo-dT primer.
Polymerase chain reaction (PCR) amplification, cloning and sequencing of bFcαR
The bFcαR cDNA was amplified using Pfu polymerase (Stragene, La Jolla, CA) from total bovine cDNA using forward (5′-CCATGGCCCCCAGACACATCACC-3′) and reverse (5′-GTCAGCTCCAGGAGTCTCCCTG-3′) primers which were designed according to the identified EST sequences. The PCR product obtained was cloned into the pCR-TOPO II vector (Invitrogen, Carlsbad, CA) prior to sequencing (Medigenomix, Martinsreid, Germany). The amplified cDNA was also cloned into the mammalian expression vector pCDNA3.1 (Invitrogen) for transfection studies.
Detection of bFcαR and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA by reverse transcription–PCR (RT–PCR)
Bovine cDNA, prepared as described previously, was used as template in a PCR reaction using primers specific for either bFcαR (see above for primer sequences) or bovine GAPDH (forward primer 5′-ATCACTGCCACCCAGAAGACT-3′; reverse primer 5′-TTGCCCTTCGAGTGACCGTAC-3′).
Transfections
COS-1 cells were transiently transfected with 1 µg of bFcαR cDNA constructs using the Fugene 6 transfection reagent (Boehringer Mannheim, Mannheim, Germany), according to the manufacturer's instructions. Cells to be used for immunoglobulin-binding assays were co-transfected with 0·05 µg of the pCMV-green fluorescent protein (GFP) plasmid in addition to the bFcαR constructs. Cells were incubated at 37° in a humidified CO2 atmosphere for 48 hr prior to harvesting.
Immunoglobulin-binding assays
Uncoated magnetic M-450 Dynabeads (Dynal, Oslo, Norway) were coated with NIP-BSA, bIgG2, hIgA or hIgG, according to the manufacturer's instructions. To prepare beads coated with rbIgA, NIP-BSA-coated beads were incubated with 100 µg/ml rbIgA for 1 hr at room temperature. These beads were washed three times in phosphate-buffered saline (PBS) prior to rosetting analysis. Beads coated with purified bIgA were prepared as follows: beads precoated with sheep anti-mouse IgG1 (Dynal) were incubated with a mixture of mouse anti-bovine light chain antibodies for 30 min at room temperature. The beads were then washed and incubated with 0·5 mg/ml purified bIgA for a further 30 min. The beads were again washed thoroughly with PBS prior to rosetting. In our rosetting procedure, transfected COS-1 cells were first enriched for those that had taken up DNA during the transfection procedure, as measured by co-expression of GFP. Binding assays were performed as follows: 0·5 × 105 GFP+ COS-1 cells (that had also been co-transfected with an FcR construct) were purified in a FACS Vantage cell sorter and mixed with immunoglobulin-coated Dynabeads, in a final volume of 50 µl per well, in V-bottomed microtitre plates. Following a 20-min incubation at room temperature, the plate was spun at 50 g for 1 min and incubated for an additional 45 min at room temperature. Cells and beads were then carefully resuspended and examined for the presence of rosettes in a Nikon Eclipse E800 microscope, combining ordinary light and fluorescence. Rosettes were defined as GFP+ cells binding four or more immunoglobulin-coated beads; at least 200 GFP+ COS-1 cells were counted for each determination.
Fluorescence-activated cell sorter (FACS) analysis
To examine the binding of small soluble IgA complexes in solution, transfected COS-1 cells were incubated with either rbIgA or hIgA previously heat-aggregated at 63° for 1 hr (HA-rbIgA or HA-hIgA). Briefly, cells were incubated with the HA-IgA preparations (20 µg/ml) for 1 hr at 4°, washed twice with FACS buffer, and then incubated for 30 min with either sheep anti-bIgA–FITC (Serotec) or goat anti-hIgA-FITC (Southern). Following a final wash, cells were analysed on a FACScan (Becton-Dickinson). Data acquisition was conducted using cellquest software (Becton-Dickinson), while analysis was performed using winmdi software (available from The Scripps Research Institute, La Jolla, CA).
Radiation hybrid (RH) mapping
The Roslin/Cambridge 3000Rad bovine/hamster (RH) cell panel21 (ResGen; Invitrogen) was screened with the following primers: 5′-CGTGGTGATGCCAGGGGAGA-3′ (forward), and 5′-TGGCCACTGTGCCAGCCATA-3′ (reverse). Thirty-five cycles of amplification were carried out, with an annealing temperature of 68° and in a 25-µl reaction volume, with 20 ng of template DNA, 6 pmol of each primer, 0·5 U of DyNAzyme II in 1× buffer (Finnzymes, Espoo, Finland) and 50 mm of each dNTP, using a hot-start protocol and four initial touchdown cycles (1° reduction for each cycle). Two investigators carried out scoring of PCR products independently. Two-point analysis, against bovine microsatellite markers typed previously on the panel, was performed using the rhmap 3·0 software package.22
Results
Isolation of a bFcαR cDNA
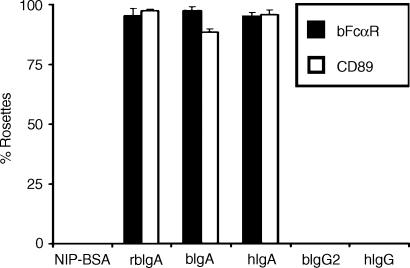
Screening of the translated EST database (which is composed of sequences from species other than humans or mice), using the human CD89 protein sequence, gave numerous matches, the best of which were predominantly bovine in origin. We identified several overlapping EST sequences that showed high homology to the entire CD89 coding region. Using this information, we designed PCR primers to amplify the predicted cDNA. PCR amplification with these primers, using bovine cDNA as template, gave only one clear band (of ≈ 900 bp) when the reaction was analysed by agarose-gel electrophoresis. The resultant PCR product was then cloned, in the correct orientation, into the mammalian expression vector, pCDNA3.1, ready for transfection. COS-1 cells were co-transfected with expression vectors encoding the putative bFcαR cDNA and GFP in order to select for cells taking up DNA during the transfection procedure. Therefore, GFP+ COS-1 cells were sorted and subjected to rosetting analysis with immunoglobulin-coated beads. Our results showed that COS-1 cells, transfected with the putative bFcαR cDNA, were able to efficiently bind beads coated with purified bovine IgA, recombinant bovine IgA, or human IgA (Fig. 1). The transfectants did not bind beads coated with bovine IgG2, human IgG, or NIP-BSA. These results confirmed that the isolated cDNA encoded a bovine IgA FcR.
Figure 1.
Functional characterization of a bovine immunoglobulin A (IgA) Fc receptor (bFcαR). COS-1 cells expressing either bFcαR or CD89 were generated and purified as described in the Materials and methods, prior to rosetting analysis with beads coated with NIP-BSA, recombinant bovine IgA (rbIgA), bovine IgA (bIgA), human IgA (hIgA), bovine IgG2 (bIgG2), or human IgG (hIgG), as indicated. Rosette formation was assessed microscopically. At least 200 cells were counted for each determination, and the number of cells binding four or more immunoglobulin-coated beads was recorded as percentage rosette formation. Results are expressed as the average + standard deviation of at least three separate experiments.
Nucleotide and protein sequence of bFcαR
The bFcαR cDNA was found to contain an 870 bp open-reading frame (GenBank accession number AY247821), encoding a 290 amino acid protein (Fig. 2). The first 21 amino acids were predicted to be an N-terminal leader sequence. The mature receptor would thus begin at Gln(Q)22 and be composed of a 204 amino acid extracellular region followed by a hydrophobic region of 19 amino acids (representing a putative transmembrane domain) and a 46 amino acid cytoplasmic tail devoid of known signalling motifs. The extracellular region was found to include four potential N-glycosylation sites (Asn21, Asn44, Asn118 and Asn161) and four conserved cysteines that form the characteristic disulphide bonds of the immunoglobulin domains. The conservation of the immunoglobulin domain structure suggested that Cys28 is linked to Cys77 to form domain 1 (EC1), and that Cys123 is paired with Cys170 to form domain 2 (EC2). The level of identity between bFcαR and CD89 at the amino acid level is: 56·2% for the complete protein; 58·2% for the extracellular domains alone; and 59·8% for the extracellular and transmembrane regions together.
Figure 2.
(a) Alignment of the deduced protein sequence of the bovine immunoglobulin A (IgA) Fc receptor (bFcαR) with those of CD89 and bFcγ2R. The GenBank accession numbers of nucleotide sequences of these three FcRs are AY247821, X54150 and Z37506, respectively. Identical residues are boxed in black, whereas conserved residues are in grey. The first amino acid of the mature bFcαR protein (Q-22) is indicated by ‘+1’ above the sequence. The four conserved extracellular cysteine residues that form the disulphide bonds of the two immunoglobulin domains are indicated (*). The four potential N-linked glycosylation sites are marked with a ‘+’. The putative transmembrane regions of all three FcRs are indicated by a dashed line above the sequence. (b) Close-up of the F–G loop regions of CD89, bFcαR and bFcγ2R. The residues of CD89 and bFcα2R, which have been shown to be important for binding to IgA and bIgG2, respectively, are indicated by arrows.
Expression pattern of bFcαR cDNA
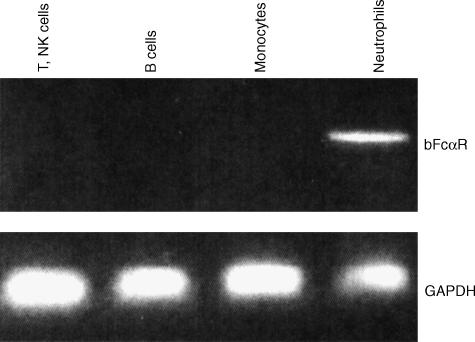
We examined the expression of the bFcαR cDNA in defined populations of circulating bovine leucocytes by RT–PCR. Using the mAbs available, we isolated CD2+ cells (mostly T cells and natural killer cells), κ- and λ light-chain positive cells (B cells), bFcγR2R+ granulocytes (neutrophils), and bFcγ2R+ mononuclear cells (monocytes). RT–PCR analysis of total RNA isolated from these cell fractions showed that the bFcαR cDNA was expressed only in neutrophils (Fig. 3).
Figure 3.
Expression pattern of bovine immunoglobulin A (IgA) Fc receptor (bFcαR) mRNA. Total mRNA was prepared and cDNA was synthesized from defined populations of bovine peripheral blood cells, as indicated. This cDNA was then used as a template in polymerase chain reaction (PCR) reactions with bFcαR or bovine glyceraldehyde 3-phosphate dehydrogenase (GAPDH)-specific primers. NK, natural killer.
Ligand-binding properties of the bFcαR
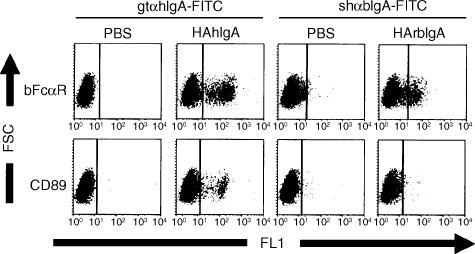
Rosetting analysis had previously shown that COS-1 cells transfected with the bFcαR cDNA were able to bind to beads coated with either bovine or human IgA (Fig. 1). However, with our immunoglobulin-coated bead-binding assay it is difficult to estimate the relative affinity of the bFcαR for these two ligands. Using a FACS-based IgA-binding assay we showed that the bFcαR was able to bind similar amounts of heat-aggregated bovine and human IgA. We also observed that (under the same conditions) CD89 bound less well to bovine IgA than to human IgA (Fig. 4). This may be explained by previous observations which indicated that bovine IgA has an affinity for CD89 of ≈2·5-fold lower than that of human IgA.20 This difference was not readily detected by our resetting method, presumably because levels of IgA, sufficient to achieve saturation binding, were used.
Figure 4.
Ligand specificity of the bovine immunoglobulin A (IgA) Fc receptor (bFcαR). (a) COS-1 cells, transfected with either bFcαR or CD89 (without green fluorescent protein), were incubated with heat-aggregated (HA) human IgA preparations (HAhIgA), as indicated. Following a second incubation with the relevant anti-IgA–fluorescein isothiocyanate (FITC) conjugates, the cells were analysed on a FACScan flow cytometer. Results are representative of at least three separate experiments. PBS, phosphate-buffered saline.
Chromosomal location of the bFcαR gene
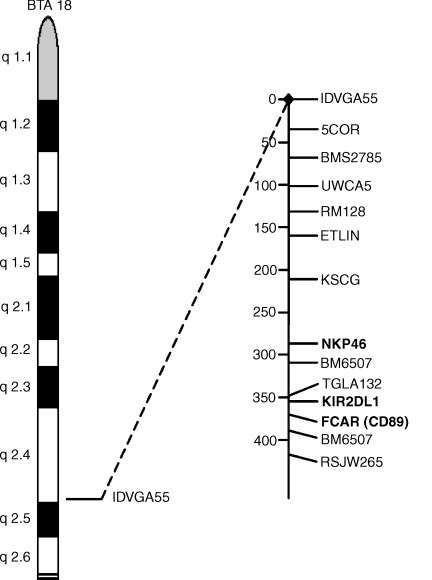
The chromosomal location of the bFcαR gene was determined by RH mapping (Fig. 5). Putative bFcαR exon-specific primers were designed by comparison to the gene structure of CD89. These primers were used to score each hybrid cell line for the presence of the bFcαR gene. Two-point analysis was carried out with resulting data compared against the RH map of genes, the chromosomal location of which was already known from linkage or physical mapping. The bFcαR gene was mapped to bovine chromosome 18, most closely linked to the microsatelite marker, TGLA132. Previously, bovine KIR2DL1 and NKp46 have also been mapped closely to this marker.23 These results suggested that bFcαR, KIR2DL1 and NKp46 are clustered in a bovine leucocyte receptor gene complex similar to the situation seen in humans, where the CD89, KIR, NKp46, LILR, GPVI and LAIR genes map to an analogous region of chromosome 19.19
Figure 5.
Chromosomal location of the bovine immunoglobulin A (IgA) Fc receptor (bFcαR) gene. The bFcαR gene was mapped to bovine chromosome 18, to a region close to KIR2DL1, by radiation hybrid mapping. Map distances (in centiRay) are shown.
Discussion
Here, we report the cloning, sequencing and functional characterization of an IgA Fc receptor from cattle. COS-1 cells, functionally expressing this receptor (that was designated bFcαR), were shown to specifically bind bovine IgA-coated beads. The bFcαR transfectants were also able to bind beads coated with human IgA, which was in agreement with previously published reports of a degree of cross-reactivity between IgA receptors from the two species.17,20
The bFcαR is highly homologous to human CD89 (FcαRI). Indeed, phylogenetic analysis of these two receptors and several related proteins confirmed the closeness of this relationship (Fig. 6). We therefore believe bFcαR to represent the bovine orthologue of CD89. In addition, bFcαR also shows high levels of homology with the other members of the gene family to which CD89 belongs. These include bFcα2R, NKp46, GPVI, paired immunoglobulin-like receptor (PIR)-A and -B, LAIR-1 and -2, and the KIR and LILR families of proteins. Indeed, the PIR proteins were first identified by screening a murine cDNA library with a probe from CD89.24 Moreover, our results also localize the bFcαR gene to bovine chromosome 18, to a region close to the KIR2DL1 and NKp46 genes, suggesting that cows possess a cluster of genes, encoding killer cell receptors and bFcαR, on chromosome 18, analogous to the LRC on human chromosome 19.18,19,23,25 The bFcγ2R gene has also been mapped to bovine chromosome 18, but its exact position is unknown.26
Figure 6.
Phylogram displaying amino acid similarity among the bovine immunoglobulin A (IgA) Fc receptor (bFcαR), CD89, bFcγ2R, bovine and human killer cell immunoglobulin-like receptors (KIRs), human leucocyte immunoglobulin-like receptors (LILRs) and selected closely related receptors. Prefices: b, bovine; h, human; m, mouse.
Although the existence of IgA FcRs has been reported in many mammals, bFcαR is the first CD89-like receptor to be cloned from a species other than the human. Owing to evolutionary conservation, it is probable that other mammals also possess a CD89-like FcαR, and work is currently underway in our laboratory to confirm this hypothesis. Our cloning of bFcαR, together with the recent description of bovine KIRs, suggests that receptors similar to these will be present in additional species.23,27 Recently, genomic Southern blotting has provided evidence that KIR genes may also be present in pigs, cats and dogs.25 The existence of an IgA FcR similar to bFcαR and CD89 should therefore be investigated in these species.
Structurally, bFcαR consists of two immunoglobulin-like extracellular domains: a transmembrane region containing a single positively charged residue; and a short cytoplasmic tail devoid of known signalling sequences. The presence of a positively charged residue in the transmembrane domain is a characteristic of a number of related receptors, including CD89, bFcγ2R, NKp46 and several KIRs. For many of these receptors it has been shown that they associate with specialized signalling molecules via these charged residues. For example, CD89 and NKp46 (like bFcαR) have an arginine residue within their transmembrane (TM) domains that is important for the association with the FcR γ chain, while several human KIRs associate with DAP-12 via a TM lysine.16,28,29 Both the FcR γ chain and DAP-12 contain activatory immunoreceptor tyrosine kinase activation motif (ITAM) motifs within their cytoplasmic domains, and cross-linking of these proteins via their associated receptors is generally considered to be a potent cellular activation signal. As cattle also possess an FcR γ chain, it therefore seems probable that it can associate with bFcαR, and that they form an activatory receptor complex on the surface of immune phagocytes.30
Owing to the fact that we did not have an antibody capable of recognizing bFcαR, we could not assess the expression pattern of the protein on the surface of bovine peripheral blood cells. Instead, we used RT–PCR to detect the expression of bFcαR mRNA in bovine cells. Unfortunately, antibodies specific for the range of cell-surface markers that can be used to isolate specific populations of cells in human and rodents are not yet available in cattle. We were therefore somewhat restricted in the type of cells we could investigate. In this study we examined bovine T cells (purified with an anti-bovine CD2 mAb), bovine B cells (purified with anti-bovine κ- and λ-light chain mAbs), bovine PMNs and monocytes (purified with anti-bFcγ2R mAb). By RT–PCR we showed that from the cell types analysed, only bovine PMNs expressed significant amounts of bFcαR mRNA (Fig. 4). This is somewhat similar to the situation in humans where CD89 expression is restricted to cells of the myeloid lineage.31 However, while human monocytes also express significant levels of CD89, bFcαR mRNA was not detected in bovine monocytes.
In terms of ligand binding, we showed that the bFcαR was able to bind both bovine and human IgA. Human IgA also bound well to transfectants, suggesting that bFcαR represents the IgA receptor previously identified, on bovine cells, by Zhang and colleagues.17 Our data also appeared to support an earlier observation which indicated that the affinity of CD89 for bovine IgA is somewhat lower than for its natural ligand.20 The CD89-binding site on IgA includes loop resides in both the Cα2 and Cα3 domains of the immunoglobulin.20,32 As the residues in these loops are completely conserved between human and bovine IgA, it is possible that these regions may also be involved in the binding of bovine IgA to the bFcαR.
The residues that form the IgA-binding site within CD89, and the bIgG2-binding site within bFcγ2R, have also been characterized, to some extent. Unlike other FcγRs, both CD89 and bFcγ2R bind their respective ligands via their membrane distal EC1 domains.33 Elegant site-directed mutational analysis has elucidated that the residues important for IgA binding lie predominantly in the F–G loop of EC1: histidine 85 and arginine 82 are essential for IgA binding, while arginine 87 also contributes.34 A later study, by the same authors, using a Biacore instrument, showed that two tyrosine residues at positions 35 and 81 are also essential.35 A similar mutagenesis study conducted with bFcγ2R showed that residues at similar positions were also essential for ligand binding by this receptor.36 As the EC1 F–G loop region of these molecules is clearly involved in ligand binding, it is important to examine the sequence of bFcαR in this region (Fig. 2b). At the start of the loop region, the bFcαR sequence closely resembles the corresponding region of CD89, with the arginine and histidine residues, important for the binding of hIgA to CD89, being absolutely conserved. Furthermore, tyrosine 35 is also conserved between these two receptors (Fig. 2a). As the residues within bovine and human IgA, implicated in receptor binding, appear to be conserved, it seems probable that Tyr35, Arg80 and His83 in bFcαR are also involved in the interaction with ligand. Interestingly, however, the residues towards the end of the bFcαR EC1 F–G loop are actually more similar to those in the corresponding positions within bFcγ2R (WSAPSE as compared to WSEPSE, respectively). Our experiments showed that bFcαR had no apparent affinity for bovine IgG2 (the ligand for bFcγ2R), so the relevance, if any, of the sequence similarity in this region is as yet unknown. Experiments to mutate the residues mentioned above, and to study their effect on the ability of bFcαR to bind IgA from both species, are currently underway.
The immunoglobulin system of cattle differs from that found in primates, rodents and rabbits.37 Primates and rabbits belong to the so-called Group I mammals in which virtually all IgG is transported in utero by a specific receptor, termed FcRn, in the placenta. Cattle belong to Group III mammals, which lack placental FcRn and thus transport no antibodies to their fetuses in utero. Instead, FcRn is expressed by acinar epithelial cells in the bovine mammary gland, which transports IgG1 into colostrum. In Group II mammals, such as rodents, both transport pathways operate. The relative levels of IgA in serum and secretions also vary among species. In comparison with most other mammals, cattle have relatively low levels of serum IgA. In addition, owing to the situation described above, the level of IgG1 in bovine colostrum is higher than that of IgA. This is in complete contrast to the situation in primates, rodents and rabbits, in which IgA is by far the most abundant immunoglobulin in colostrum. However, even though IgG1 predominates, probably owing to the efficiency of the transport mechanism, cattle still secrete significant amounts of colostral IgA (≈ 2 g/animal/day). IgA is also the major antibody in a number of other exocrine secretions, including tears and saliva. These data suggest that the role of IgA in bovine peripheral blood is perhaps somewhat different to its role in humans, but also that IgA antibodies may have a similar protective function at mucosal surfaces. Thus, mapping the expression pattern of bFcαR on various cell types and in tissues will no doubt help us to understand the immunological role of IgA in cattle.
In conclusion, we have isolated an IgA Fc receptor from cattle, which we have termed bFcαR. This receptor binds both bovine and human IgA, and we consider it to represent the bovine orthologue of human FcαRI/CD89. In the future, a detailed study of the expression pattern and function of bFcαR will lead to a greater understanding of the role of this FcR and its ligand in the immune repertoire of this agriculturally important species.
Acknowledgments
The authors would like to thank Katherine Hagelsteen and Linda Solfjell for excellent technical assistance. We also thank Dr Chris J. Howard for helpful discussions. This work was supported by The Research Council of Norway (HCM, AKS, PB) and the Wellcome Trust (RJP, JMW). JLW is grateful to the European Commission for funding the RH work through project QLRI-CT-2002-02744. HCM would also like to thank the Odd Fellows Medical Research Fund for additional financial support.
Abbreviations
- GAPDH
glycaraldehyde 3-phosphate dehydrogenase
- GFP
green fluorescent protein
- HA
heat aggregated
- IgA
immunoglobulin A
- IgG
immunoglobulinG
- ILT
immunoglobulin-like transcript
- ITAM
immunoreceptor tyrosine kinase activation motif
- KIR
killer cell immunoglobulin-like receptor
- LAIR
leucocyte-associated immunoglobulin-like receptor
- LILR
leucocyte immunoglobulin-like receptor
- LIR
leucocyte immunoglobulin-like receptor
- LRC
leucocyte receptor complex
- MIR
monocyte/macrophage immunoglobulin-related receptor
- MNC
mononuclear cell
- PIR
paired immunoglobulin-like receptor
- PMN
polymorphonuclear granulocyte (neutrophil)
References
- 1.Raghavan M, Bjorkman PJ. Fc receptors and their interactions with immunoglobulins. Annu Rev Cell Dev Biol. 1996;12:181–220. doi: 10.1146/annurev.cellbio.12.1.181. [DOI] [PubMed] [Google Scholar]
- 2.Ravetch JV. Fc receptors. Curr Opin Immunol. 1997;9:121–5. doi: 10.1016/s0952-7915(97)80168-9. [DOI] [PubMed] [Google Scholar]
- 3.Daeron M. Fc receptor biology. Annu Rev Immunol. 1997;15:203–34. doi: 10.1146/annurev.immunol.15.1.203. [DOI] [PubMed] [Google Scholar]
- 4.Yan Y, Li X, Wang A, Zhang G. Molecular cloning and identification of full-length cDNA encoding high affinity Fc receptor for bovine IgG (FcγRI) Vet Immunol Immunopathol. 2000;75:151–9. doi: 10.1016/s0165-2427(00)00197-5. [DOI] [PubMed] [Google Scholar]
- 5.Zhang G, Young JR, Tregaskes CR, Howard CJ. Cattle FcγRII: molecular cloning and ligand specificity. Immunogenetics. 1994;39:423–7. doi: 10.1007/BF00176160. [DOI] [PubMed] [Google Scholar]
- 6.Collins RA, Gelder KI, Howard CJ. Nucleotide sequence of cattle FcγRIII: its identification in γδ T cells. Immunogenetics. 1997;45:440–3. doi: 10.1007/s002510050228. [DOI] [PubMed] [Google Scholar]
- 7.Sylvestre DL, Ravetch JV. Fc receptors initiate the Arthus reaction: redefining the inflammatory cascade. Science. 1994;265:1095–8. doi: 10.1126/science.8066448. [DOI] [PubMed] [Google Scholar]
- 8.Clynes R, Ravetch JV. Cytotoxic antibodies trigger inflammation through Fc receptors. Immunity. 1995;3:21–6. doi: 10.1016/1074-7613(95)90155-8. [DOI] [PubMed] [Google Scholar]
- 9.Clynes R, Takechi Y, Moroi Y, Houghton A, Ravetch JV. Fc receptors are required in passive and active immunity to melanoma. Proc Natl Acad Sci USA. 1998;95:652–6. doi: 10.1073/pnas.95.2.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan R, Clynes R, Oh J, Ravetch JV, Scharff MD. Antibody-mediated modulation of Cryptococcus neoformans infection is dependent on distinct Fc receptor functions and IgG subclasses. J Exp Med. 1998;187:641–8. doi: 10.1084/jem.187.4.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Glennie MJ, Johnson PW. Clinical trials of antibody therapy. Immunol Today. 2000;21:403–10. doi: 10.1016/s0167-5699(00)01669-8. [DOI] [PubMed] [Google Scholar]
- 12.Maliszewski CR, March CJ, Schoenborn MA, Gimpel S, Shen L. Expression cloning of a human Fc receptor for IgA. J Exp Med. 1990;172:1665–72. doi: 10.1084/jem.172.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valerius T, Stockmeyer B, van Spriel AB, et al. FcαRI (CD89) as a novel trigger molecule for bispecific antibody therapy. Blood. 1997;90:4485–92. [PubMed] [Google Scholar]
- 14.Stockmeyer B, Dechant M, van Egmond M, et al. Triggering Fcα-receptor I (CD89) recruits neutrophils as effector cells for CD20-directed antibody therapy. J Immunol. 2000;165:5954–61. doi: 10.4049/jimmunol.165.10.5954. [DOI] [PubMed] [Google Scholar]
- 15.Stockmeyer B, Elsasser D, Dechant M, Repp R, Gramatzki M, Glennie MJ, van de Winkel JG, Valerius T. Mechanisms of G-CSF- or GM-CSF-stimulated tumor cell killing by Fc receptor-directed bispecific antibodies. J Immunol Methods. 2001;248:103–11. doi: 10.1016/s0022-1759(00)00346-x. [DOI] [PubMed] [Google Scholar]
- 16.Morton HC, van den Herik-Oudijk IE, Vossebeld P, Snijders A, Verhoeven AJ, Capel PJ, van de Winkel JG. Functional association between the human myeloid immunoglobulin A Fc receptor (CD89) and FcR γ chain. Molecular basis for CD89/FcR γ chain association. J Biol Chem. 1995;270:29781–7. doi: 10.1074/jbc.270.50.29781. [DOI] [PubMed] [Google Scholar]
- 17.Zhang G, Young JR, Tregaskes CA, Sopp P, Howard CJ. Identification of a novel class of mammalian Fcγ receptor. J Immunol. 1995;155:1534–41. [Google Scholar]
- 18.Wende H, Colonna M, Ziegler A, Volz A. Organization of the leukocyte receptor cluster (LRC) on human chromosome 19q13.4. Mamm Genome. 1999;10:154–60. doi: 10.1007/s003359900961. [DOI] [PubMed] [Google Scholar]
- 19.Martin AM, Kulski JK, Witt C, Pontarotti P, Christiansen FT. Leukocyte Ig-like receptor complex (LRC) in mice and men. Trends Immunol. 2002;23:81–8. doi: 10.1016/s1471-4906(01)02155-x. [DOI] [PubMed] [Google Scholar]
- 20.Pleass RJ, Dunlop JI, Anderson CM, Woof JM. Identification of residues in the CH2/CH3 domain interface of IgA essential for interaction with the human Fcα receptor (FcαR) CD89. J Biol Chem. 1999;274:23508–14. doi: 10.1074/jbc.274.33.23508. [DOI] [PubMed] [Google Scholar]
- 21.Williams JL, Eggen A, Ferretti L, et al. A bovine whole-genome radiation hybrid panel and outline map. Mamm Genome. 2002;13:469–74. doi: 10.1007/s00335-002-3001-x. [DOI] [PubMed] [Google Scholar]
- 22.Lange K, Boehnke M, Cox DR, Lunetta KL. Statistical methods for polyploid radiation hybrid mapping. Genome Res. 1995;5:136–50. doi: 10.1101/gr.5.2.136. [DOI] [PubMed] [Google Scholar]
- 23.Storset AK, Slettedal IO, Williams JL, Law A, Dissen E. Natural killer cell receptors in cattle: a bovine killer cell immunoglobulin-like receptor (KIR) multigene family contains members with divergent signaling motifs. Eur J Immunol. 2003;33:980–90. doi: 10.1002/eji.200323710. [DOI] [PubMed] [Google Scholar]
- 24.Kubagawa H, Burrows PD, Cooper MD. A novel pair of immunoglobulin-like receptors expressed by B cells and myeloid cells. Proc Natl Acad Sci USA. 1997;94:5261–6. doi: 10.1073/pnas.94.10.5261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volz A, Wende H, Laun K, Ziegler A. Genesis of the ILT/LIR/MIR clusters within the human leukocyte receptor complex. Immunol Rev. 2001;181:39–51. doi: 10.1034/j.1600-065x.2001.1810103.x. [DOI] [PubMed] [Google Scholar]
- 26.Klungland H, Vage DI, Lien S. Linkage mapping of the Fcγ2 receptor gene to bovine chromosome 18. Mamm Genome. 1997;8:300–1. doi: 10.1007/s003359900425. [DOI] [PubMed] [Google Scholar]
- 27.McQueen KL, Wilhelm BT, Harden KD, Mager DL. Evolution of NK receptors: a single Ly49 and multiple KIR genes in the cow. Eur J Immunol. 2002;32:810–7. doi: 10.1002/1521-4141(200203)32:3<810::AID-IMMU810>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 28.Pfefferkorn LC, Yeaman GR. Association of IgA-Fc receptors (FcαR) with FcεRI γ2 subunits in U937 cells. Aggregation induces the tyrosine phosphorylation of γ2. J Immunol. 1994;153:3228–36. [PubMed] [Google Scholar]
- 29.Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH. Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature. 1998;391:703–7. doi: 10.1038/35642. [DOI] [PubMed] [Google Scholar]
- 30.Morton HC, Storset AK, Brandtzaeg P. Cloning and sequencing of a cDNA encoding the bovine FcR γ chain. Vet Immunol Immunopathol. 2001;82:101–6. doi: 10.1016/s0165-2427(01)00352-x. [DOI] [PubMed] [Google Scholar]
- 31.Monteiro RC, Kubagawa H, Cooper MD. Cellular distribution, regulation, and biochemical nature of an Fcα receptor in humans. J Exp Med. 1990;171:597–613. doi: 10.1084/jem.171.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carayannopoulos L, Hexham JM, Capra JD. Localization of the binding site for the monocyte immunoglobulin (Ig) A-Fc receptor (CD89) to the domain boundary between Cα2 and Cα3 in human IgA1. J Exp Med. 1996;183:1579–86. doi: 10.1084/jem.183.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morton HC, van Zandbergen G, van Kooten C, Howard CJ, van de Winkel JG, Brandtzaeg P. Immunoglobulin-binding sites of human FcαRI (CD89) and bovine Fcγ2R are located in their membrane-distal extracellular domains. J Exp Med. 1999;189:1715–22. doi: 10.1084/jem.189.11.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wines BD, Hulett MD, Jamieson GP, Trist HM, Spratt JM, Hogarth PM. Identification of residues in the first domain of human Fcα receptor essential for interaction with IgA. J Immunol. 1999;162:2146–53. [PubMed] [Google Scholar]
- 35.Wines BD, Sardjono CT, Trist HH, Lay CS, Hogarth PM. The interaction of FcαRI with IgA and its implications for ligand binding by immunoreceptors of the leukocyte receptor cluster. J Immunol. 2001;166:1781–9. doi: 10.4049/jimmunol.166.3.1781. [DOI] [PubMed] [Google Scholar]
- 36.Morton HC, Howard CJ, Storset AK, Brandtzaeg P. Identification of residues within the extracellular domain 1 of bovine Fcγ2R essential for binding bovine IgG2. J Biol Chem. 2001;276:47794–800. doi: 10.1074/jbc.M104066200. [DOI] [PubMed] [Google Scholar]
- 37.Butler JE. Immunoglobulin diversity, B-cell and antibody repertoire development in large farm animals. Rev Sci Tech. 1998;17:43–70. doi: 10.20506/rst.17.1.1096. [DOI] [PubMed] [Google Scholar]