Abstract
Terminal deoxynucleotidyl transferase (TdT) is a template-independent DNA polymerase that catalyses the incorporation of deoxyribonucleotides into the 3′-hydroxyl end of DNA templates and is thought to increase junctional diversity of antigen receptor genes. TdT is expressed only on immature lymphocytes and acute lymphoblastic leukaemia cells and its transcriptional expression is tightly regulated. We had previously found that protein kinase C (PKC) activation down-regulates TdT expression. PKC-activation induces the synthesis of the Fos and Jun proteins, known as the major components of activation protein 1 (AP-1) transcriptional factor implicated in transcriptional control. Here we report the identification of several DNA–protein interactions within the TdT promoter region in non-TdT expressing human cells. Sequence analysis revealed the presence of a putative AP-1-like DNA-binding site, suggesting that AP-1 may play a relevant role in TdT transcriptional regulation. Using a different source of nuclear extracts and the AP-1–TdT motif as a probe we identified several DNA-protein retarded complexes in electrophoretic mobility shift assays. Super-band shifting analysis using an antibody against c-Jun protein confirmed that the main interaction is produced by a nuclear factor that belongs to the AP-1 family transcription factors. Our findings suggest that the TdT gene expression is down-regulated, at least in part, through AP-1-like transcription factors.
Introduction
Mature lymphocyte differentiation involves a complex combination of genetically preprogrammed events and responses to extracellular stimuli.1 This process occurs in a defined sequential order for both B and T lymphocytes and appears to drive cell migration, differentiation, gene rearrangement, cell-to-cell contacts, and positive and negative selection; all of which require the induction or down-regulation of distinct gene products in a tightly regulated specific sequential order. Even though many advances have been made toward the characterization of the intermediate stages of both B and T lymphocyte differentiation, at the present time our understanding of the molecular mechanisms directing lymphocytes through such events remain largely undefined.1
Regulated rearrangement of immunoglobulin and T-cell receptor (TcR) gene segments is an important event that occurs during lymphoid cell differentiation. Gene rearrangements are mediated by the V(D)J-recombinase complex, with multiple activities which are similar in both T and B lymphocytes.2 Low levels of V(D)J-recombinase are detected in early lymphoid cells; these levels then increase during rearrangement of lymphoid cell antigen receptors, and decrease again to undetectable levels in mature cells.3 TdT (terminal deoxynucleotidyl transferase: DNA deoxynucleotidyl exotransferase, EC: 2.7.7.31) is an extensively characterized, tissue-specific enzyme critical for immunoglobulin and TcR gene rearrangements.4,5 TdT is a 58 000 MW template-independent DNA polymerase which has been shown to account for the addition of non-germline-encoded ‘N’ nucleotides to double stranded DNA ends at the D/J, V/DJ, or V/J junctions during immunoglobulin and TCR gene rearrangements.6,7 The random insertion of ‘N’ nucleotides significantly increases the diversity of the immune repertoire.8 The TdT gene is expressed exclusively during very early stages of both B and T lymphocyte development, and is turned off by the time these cells reach maturity.9,10
Several compounds that increase intracellular cAMP levels induce TdT synthesis in transformed B-cell lines.11 Increased TdT expression has also been observed previously in normal, non-transformed thymocytes both in vivo and in vitro after treatment with thymosin12 and thymopoietin13,14 molecule that increase intracellular cGMP levels. However, the precise role that TdT plays in this particular cellular process is not yet fully understood.
The TdT gene is down-regulated by phorbol esters in normal thymocytes.10 Moreover, this regulatory response is also observed in human leukaemic cells of T and B lineages arrested at early stages of differentiation.15,16 This suggests that TdT expression is controlled, at least partially, by protein kinase C (PKC) activation. Furthermore, it has been reported that the PKC-dependent TdT gene expression is regulated at the transcriptional level.17–19 It is also known that PKC-activation induces the expression of the Fos/Jun heterodimer that is in turn responsible for the activation of transcription of different genes through the AP-1 pathways.20,21 Thus, a relevant aspect in understanding V(D)J recombinase regulation and function during lymphocyte differentiation is to identify mechanisms underlying the expression of its target genes.
Transcriptional regulation of the activation of early and late stages of lymphoid cell development, including turning on the TdT gene, is of special interest.22–24 The nucleotide sequence of the regulatory region of the human TdT gene, responsible for co-ordinated and tissue-specific expression, has been previously determined.14 The human TdT gene is regulated at the transcriptional level, it lacks a canonical TATA box, and GC-rich sequences characteristic of Sp1-binding sites; instead an initiator element (Inr) overlaps the transcription start site.25–28 The transcription initiation site and the core promoter region have previously been defined.22–24 In order, to carry out a detailed analysis of the TdT core promoter region, we decided to characterize the regulatory elements and/or nuclear factors involved in the regulation of TdT expression in human lymphoid cells. We found that in PKC-stimulated lymphoid cell lines Fos and Jun mRNA expression was up-regulated, which may correlate with an AP-1-like dependent induction of gene expression. Furthermore we identified by electrophoresis mobility shifting assays (EMSA), a transcription factor that interacts with an AP-1-like recognition sequence in TdT-non-expressing cell lines. We suggest that such an interaction may be responsible for the down-regulation of TdT gene expression.
Materials and methods
Cell lines and cells
The human lymphoblastoid T-cell line DND-41, an acute lymphoblastic leukaemic cell line, was provided by Dr T. W. Mak (Ontario Cancer Institute, University of Toronto). The pre-B-cell line HYON, an acute lymphoblastic leukaemic cell line, was generously supplied by Dr Michelle Letarte (Division of Immunology, The Hospital for Sick Children, Toronto). HL-60 promyelocytic leukaemic cell line was a gift from Dr Melvin Freedman laboratory (Division of Hematology, The Hospital for Sick Children, Toronto). HeLa cells were supplied by Dr A. García-Carrancá (Instituto de Investigaciones Biomédicas, Universidad Nacional Autónoma de México). Total RNA from human thymus and tonsils were kindly donated by Dr Amos Cohen from the Hospital for Sick Children, Toronto, from patients undergoing cardiac surgery following previous written agreement of the patients. Human thymocytes and tonsillar lymphocytes used in our experiments were obtained by Ficoll-Hypaque gradient centrifugation. Normal lymphoid cells and cell lines were maintained in Dulbecco's modified Eagle's minimal essential medium supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen, Carlsbad, CA), at a mean cell density of 2·5–5·0 × 106 cells/ml.
RNA extraction and analysis
Total RNA was extracted from 2·5 × 106 cells according to the method of Chomczynsky and Sacchi.29 After 2 hr incubation in the presence or absence of 12-tetradecanoylphorbol-13-acetate (TPA; 10 mm) and ionomycin (0·5 µm), total cell RNA was extracted and 10 µg of RNA was electrophoresed under denaturing conditions on a 1% agarose and 0·66 m formaldehyde gel, according to the method of Lehrch.30 To verify the integrity of RNA, gels were stained with 0·5 µg/ml of ethidium bromide. RNA was transferred onto nitrocellulose membranes, which were allowed to dry at room temperature and baked for 1 hr at 80° in a vacuum oven. The probes for c-fos, c-jun, SRF and C-Ha-ras were radiolabelled with [α-32P]-dCTP (Dupont, Boston, MA) and hybridized according to the method of Northern blot hybridization as described previously.31
Amplification of TdT promoter and oligonucleotides
Polymerase chain reaction (PCR) was performed by a modification of the method originally described.32 Two µg of human thymus DNA were diluted into 50 µl of a solution containing 10 mm dATP, dCTP, dGTP, and dTTP, 30 pmol of each primer, and 1·5 mm MgCl2. The mixture was heated at 94° for 90 s before 5 units of TaqI DNA polymerase were added. To amplify the 5′-regulatory region of TdT, human lymphocyte-derived genomic DNA was incubated for 90 s at 94° to denature the double stranded chain, and then 2 min at 67° to allow annealing of the primers to the template, and 3 min at 72° for primer extension for 35 cycles. We designed two oligonucleotides of the 5′-flanking region of the TdT gene with BamHI sequence ends of the sequence previously reported.14 The 5′ primer that corresponds to −600 to −580 is named primer 1 (primer 1: 5′-GGA-TCC-GGA-GCA-GTT-AGA-AGC-AAC-AGA-GC-3′). The 3′ primer, from −21 to −1 is named primer 2 (primer 2: 5′-GGA-TCC-GGG-AAG-AGG-CTG-CTG-CTG-CC-3′). The 600 bp DNA PCR-amplified product was cloned into the pBluescript vector and sequenced (data not shown). The 600 bp DNA fragment was digested with RsaI to generate three DNA subfragments.
Double-stranded DNA oligonucleotides for the AP-1 consensus (AP-1-Cons) and AP-1-TdT were synthesized and used in electrophoretic mobility shift assays (EMSA). The oligonucleotides for AP-1-Cons were sense 5′-CCT-TCG-TCA-GTC-AGC-GGG-A-3′ and antisense 3′-GGA-AGC-AGT-CAG-TCG-CCC-T-5′, while the oligonucleotides for AP-1-TdT were sense 5′-AAG-GGC-CTC-AGT-ACA-TTT-AG-3′ and antisense 3′-TTC-CCG-GAG-TCA-TGT-AAA-TC-5′, and corresponded to positions −345 to −339 upstream of the TdT transcriptional start site. The sequence of the mutated oligonucleotide for AP-1 (AP-1m) was similar to AP-1-Cons but the motif TCAGTCA was changed to TCAGTTG. The bold and underlined sequences in AP-1-Cons correspond to the sequence previously reported to interact with the AP-1 transcription complex,20 while the bold and underlined sequence in AP-1-TdT corresponds to a similar AP-1 sequence within the TdT promoter. Double-stranded DNA oligonucleotides for Sp1 were, sense 5-ATT-CGA-TCG-GGG-CGG-GGC-GAG-C-3′ and antisense 3′-TAA-GCT-AGC-CCC-GCC-CCG-CTC-G-5′, while the oligonucleotides for nuclear factor (NF)-κB were sense 5′-AGT-TGA-GGG-GAC-TTT-CCC-AGG-C-3′ and antisense 3′-TCA-ACT-CCC-CTG-AAA-GGG-TCC-G-5′. The bold and underlined sequences correspond to the target sequences for Sp1 and NF-κB transcriptional factors.
Nuclear extracts and EMSA
Nuclear extracts were obtained from TdT-expressing and -non-expressing cells and HeLa cells, and were prepared according to the method of Dignam et al.33 Briefly, the cultured cells were collected and resuspended in buffer A (20 mm HEPES pH 8·0, 10 mm KCl, 0·5 mm dithiothreitol (DTT), 1·5 mm MgCl2, 0·5 mm phenylmethylsulphonyl fluoride (PMSF), 0·25 mm ethylenediaminetetraacetic acid (EDTA), and 0·25 mm egtazic acid (EGTA)) at 3·5 × 106 cells/ml, and lysed with a glass homogenizer. The nuclei were pelleted and resuspended in 2 ml of buffer C (20 mm HEPES pH 8·0, 1·5 mm MgCl2, 0·5 mm DTT, 0·25 nm PMSF, 0·25 mm EDTA, 0·25 mm EGTA, 420 mm NaCl, and 50% glycerol) and homogenized. The suspension was pelleted and the supernatant divided into aliquots and stored at −70°. Protein determination was carried out by the Bradford method.34 EMSA was performed as reported previously,35 the oligonucleotides AP-1-Cons and AP-1-TdT were end-labelled with T4 DNA polynucleotide kinase using 30 µCi of [γ-32P]-dATP/100 ng of oligonucleotides. Protein extracts (5 µg) from HeLa cells were incubated for 20 min at room temperature with the labelled oligonucleotides (1 × 105 c.p.m.) in band shift buffer (10 mm Tris-HCl pH 7·5, 50 mm NaCl, 1 mm DTT, 1 mm EDTA, and 5% glycerol) containing 1 µg poly dI-dC as a non-specific competitor. The heterologous competitions were carried out with 10, 25, 50 and 100-fold molar excess of AP-1-Cons, mutated AP-1 m, Sp1, NF-κB probes, and self-competition with AP-1-TdT unlabelled probe. DNA–protein complexes were resolved in low-isotonic strength on non-denaturing 6% polyacrylamide gel electrophoresis (PAGE) containing 0·5× TBE. Gels were pre-electrophoresed for 30 min at 200 V, and the electrophoresis was carried out under buffer circulation at the same voltage for 3 hr at 4°. Gels were dried and subjected to autoradiogram at −70° with an intensifier screen. For competition experiments, 50-fold molar excess of unlabelled oligonucleotides was added 20 min before incorporating the labelled probe, after which the assays were performed as described above. For immune band shift assays, DNA–protein complexes were allowed to form prior to the addition of 3 µg of antic-Jun polyclonal antibody, or 1 µg of anti-IL-6 as irrelevant antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Samples were incubated with the antibodies for 4 hr at 4° prior to resolving the DNA–protein complexes in 6% PAGE.
Results
Induction of AP-1 mRNA in TdT-expressing cells
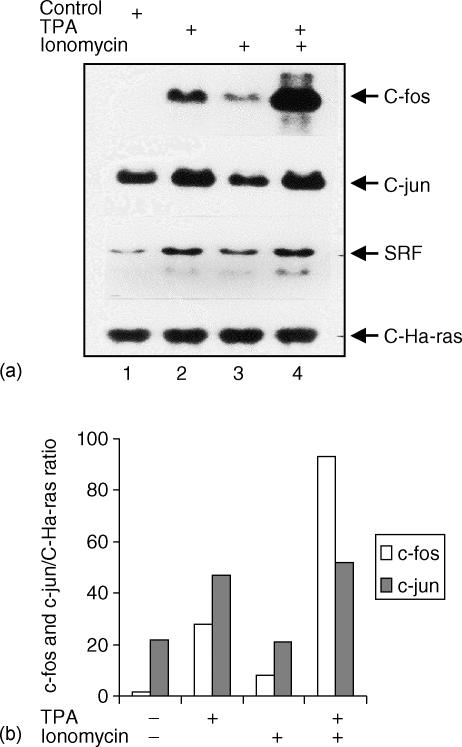
Signal transduction pathways, such as PKC-activation or Ca2+ mobilization regulate gene expression in human lymphoid cells.10,16,36,37 Some signals can be mimicked by pharmacological agents, such as TPA, which activates PKC or calcium ionophores (ionomycin) which increase intracellular Ca2+. These signals, alone or in combination, target the activation of transcription factors such as AP-1. Previous results from our laboratory showed that TdT mRNA expression is down-regulated in lymphoid cells following treatment with TPA.16 Therefore, we analysed the level of c-fos and c-jun mRNA expression in DND-41 cells, a TdT-expressing lymphoid T-cell line, after TPA and/or ionomycin treatment. Total RNA was blotted and hybridized in a Northern blot using c-fos, c-jun, SRF (serum response factor as a positive control) and c-Ha-ras (as a constitutive control) cDNA probes. As shown in Fig. 1, TPA induces a significant increase in SRF, c-fos (10-fold) and c-jun (2·8-fold) mRNA levels. Similarly, expression of these genes was greatly increased when cells were incubated with TPA/ionomycin (see Fig. 1, lower panel). On the other hand, ionomycin alone failed to induce substantial changes in mRNA levels. Importantly, no increases in transcriptional levels were observed when c-Ha-ras was used as the probe. These results suggest that PKC-activation alone significantly up-regulates c-fos and c-jun. Furthermore, PKC-activation and increased intracellular Ca2+ concentration have a synergistic effect on c-fos and c-jun expression in immature lymphoid cells. Based on our observations we asked whether a transcriptional factor of the AP-1 family could interact with the TdT regulatory region and play a role in regulating the expression of TdT during lymphoid maturation.
Figure 1.
Northern blot analysis of c-fos and c-jun mRNA in T-cells treated with TPA and ionomycin. The human lymphoblastoid T-cell line DND-41 was incubated for 2 hr in the absence (lane 1) or presence of phorbol ester TPA (lane 2), ionomycin (lane 3) and the combination of TPA and ionomycin (lane 4). Total RNA was extracted as described in Materials and Methods. Ten µg of RNA per lane were electrophoresed in 1% agarose gel, and transferred to nylon membranes according to Materials and Methods. The membranes were hybridized with the appropriate labelled 32P-labelled probe as indicated. The blot was exposed for autoradiography for 8 hr for c-fos, c-jun and c-Ha-ras probes and for 72 hr for the SRF (serum response factor) probe. As an internal control, the expression of the proto-oncogene C-Ha-ras was monitored. The blots (upper panel in (a)) were scanned using computer-assisted densitometry (Fluor-S-Multi-imager, Bio-Rad) and the data (c-fos and c-jun/C-Ha-ras mRNA signals) were plotted as the percentages of changes (lower panel in (b)). Data shown are representative of two independent experiments.
DNA–protein interactions within the TdT promoter regulatory region
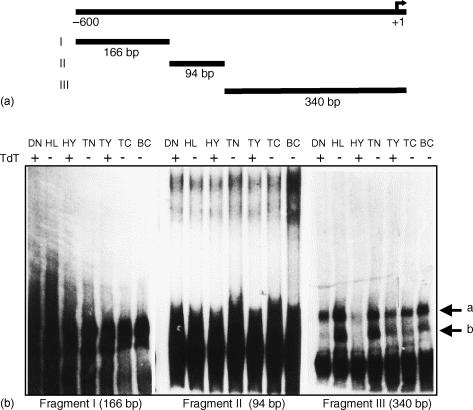
Next, we examined the transcriptional regulation of TdT in TdT expressing and non-expressing cells. To identify transcription factors involved in the negative regulation of TdT expression, we analysed the interaction between the TdT regulatory region with nuclear extracts from several sources of lymphoid cells: early T cells (DND-41, HYON, thymocytes (TdT expressing cells) and lymphoid cells (HL-60, a myeloid cell line), tonsil cells, and T and B lymphocytes (TdT-non-expressing cells). To map which DNA region of the TdT promoter interacts with nuclear factors, the 600 bp DNA fragment obtained by PCR (see Materials and methods) was partitioned into three subfragments with RsaI, and an EMSA was carried out for each fragment. As shown in Fig. 2, incubation of fragment III with nuclear extracts from TdT-non-expressing cells (HL-60, tonsil cells and peripheral T and B lymphocytes) identified several retarded DNA–protein complexes (Fig. 2b). No retarded complexes were identified with subfragment I, while there were additional retarded complexes with subfragment II. These additional complexes were similar in TdT-expressing and -non-expressing lymphoid cells and were not analysed further here. For subfragment III there is a single fast migrating retarded complex abundantly represented in nuclear extracts from TdT-nonexpressing cells, whereas TdT expressing cells had abundant slow complexes (Fig. 2b; arrows a and b). Importantly, as the size of the labelled probes were significantly different (fragments I: 166 bp, II: 94 bp and III: 340 bp), the retarded migration of the DNA–protein complexes could not be directly compared between the three distinct subfragments.
Figure 2.
EMSA analysis of the 5′-end regulatory region of human TdT gene in leukaemic and normal cells. (a) Diagram of regulatory region TdT with subfragments I (166 bp), II (94 bp), and III (340 bp). (b) Nuclear extracts of human leukaemic T-cells (DND-41 ‘DN’), myeloid cells (HL-60 ‘HL’), pre-B-cells (HYON ‘HY’), tonsil cells ‘TN’, thymocytes ‘TY’, T lymphocytes ‘TC’ and B lymphocytes ‘BC’ were analysed by EMSA using three subfragments of the 5′-end TdT promoter region obtained by PCR as described in Materials and Methods. The arrows (a) and (b) indicate the retarded complexes obtained with the fragment III. A representative of two independent experiments is shown.
The differential formation of the two complexes (arrows a and b) within the 340 bp DNA subfragment III indicates the presence of regulatory proteins (transcription factors) interacting with this DNA fragment in cells that do not express TdT. Furthermore our results show the presence of several DNA–protein complexes in TdT-non-expressing cells. Although these complexes were not present in TdT expressing cells (thymocytes), these cells are a heterogeneous population. Therefore only a few cells may be expressing the combination of transcriptional activators and repressors relevant for T-lymphocyte maturation, activation and differentiation. Therefore our finding is not opposed to the hypothesis that the DNA–protein complexes present in TdT-non-expressing cells could represent repressor factors. The presence of DNA–protein complexes in thymocyte subpopulations suggests the participation of repressor factors similar to those detected in other TdT-non-expressing cells. Moreover, this observation suggests that TdT gene expression requires distinct transcription factors induced specifically during cellular differentiation. These transcriptional factors are potential candidates to drive TdT transcriptional regulation during early and terminal differentiation of lymphoid cells.
An AP-1-like transcription factor binds to TdT regulatory region
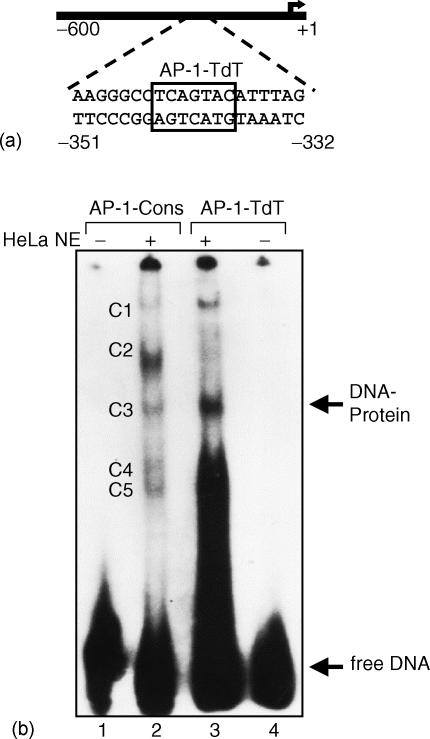
Human TdT mRNA is regulated by PKC activation10,16–19 through induction of the transcriptional factor AP-120,21 as shown on Fig. 1. The products of the nuclear proto-oncogenes c-jun and c-fos are components of the transcriptional activator protein AP-1, which represents one of the principal targets of signals elicited by growth factors or phorbol esters20,21,38,39,40–42 As AP-1 is a transcriptional regulator, deregulation of transcription in the cell has been considered as the mechanism of cell transformation by the viral oncogenes v-jun and v-fos.39 Because we had found that phorbol esters down-regulate TdT expression, we hypothesized that c-fos and c-jun could be responsible for such down-regulation. Computer-assisted DNA analysis revealed that subfragment III of the human TdT regulatory region contains a putative AP-1-like consensus sequence (here termed AP-1-TdT). To examine the possible role of AP-1-TdT in the negative regulation of TdT expression, we performed an EMSA with a 20 bp oligonucleotide probe containing the putative AP-1 recognition sequence (Fig. 3a; position −351 to −332). As a control we carried out an EMSA using the consensus sequence for AP-1 binding (here named AP-1-Cons). The radiolabelled AP-1-Cons and AP-1-TdT oligonucleotides were incubated with nuclear extracts from HeLa cells, which are rich source of AP-1 protein. We identified several retarded complexes (Fig. 3b; C1 to C5) and a specific retarded complex when nuclear extracts were incubated with AP-1-TdT. Some of these complexes had similar mobilities to those observed when using the AP-1-Cons sequence, while others had different mobilities (Fig. 3b; complex C3). However, we cannot rule out the possibility that the presence of several retarded complexes reflects the recruitment of other transcriptional factors or cofactors associated with the AP-1 complex throughout its autonomous trans-activation domain, as previously reported.43
Figure 3.
EMSA analysis of AP-1-like element of human TdT. (a) Localization of the putative AP-1-TdT-binding site. (b) The 20 bp DNA fragment of the 5′-end regulatory region of human TdT gene (−351 to −332) that contains the AP-1-like regulatory element was radiolabelled and incubated with nuclear extracts from HeLa cells (NE). An AP-1 consensus regulatory element was used as a probe (AP-1-Cons) in the absence (lane 1) or presence (lane 2) of nuclear extracts. AP-1-like regulatory element present in 5′-end regulatory region of TdT was used as a probe (AP-1-TdT) in presence (lane 3) or absence (lane 4) of nuclear extracts. The arrow shows the DNA–protein specific AP-1 retarded complex. Multiple retarded complexes are shown (C1 to C5). Notice the presence of the same complex (C3) with the two-independent probes. Data shown is representative of two independent experiments.
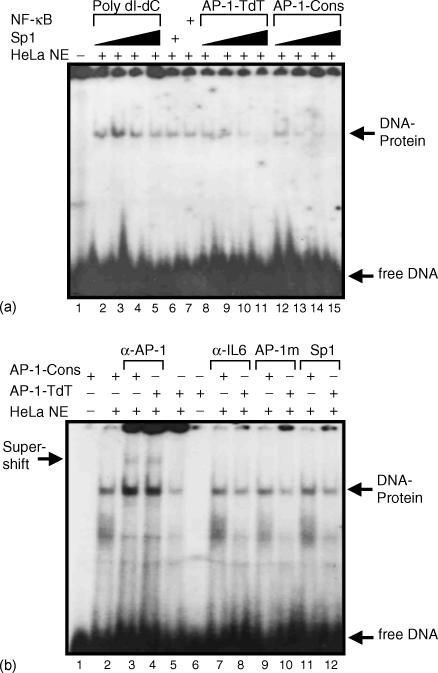
To verify the specificity of AP-1-like transcriptional factor binding to the TdT regulatory region we first performed competition assays varying the amounts of the non-specific competitor poly dI-dC with AP-1-TdT as the labelled probe (Fig. 4a; lanes 2–5), and for specific competition using the unlabelled AP-1-TdT probe at 10, 25, 50 and 100-fold molar excess (Fig. 4a; lanes 8–11). The specificity of the retarded complex was examined by means of Sp1 and NF-κB DNA-binding sequences as non-specific competitors at 100-fold molar excess (Fig. 4a; lanes 6 and 7). As expected, only specific competition with the AP-1-TdT probe reduced the formation of the DNA–protein complex, indicating that AP-1-TdT induced the formation of a DNA-protein migrating complex with a similar mobility as that generated by AP-1-Cons (Fig. 4a; lanes 8–15). In conclusion, we believe that AP-1 complex proteins interact with the TdT promoter.
Figure 4.
EMSA analysis of AP-1-TdT specific complex. (a) EMSA analysis. Radiolabelled AP-1-TdT was incubated with nuclear extracts (NE) of HeLa cells and increasing concentrations of non-specific competitor (1, 2, 4, and 8 µg of poly dI-dC) in the absence (lane 1) or presence (lanes 2–5) of nuclear extracts from HeLa cells. The nuclear extracts were preincubated with 100-fold molar excess of Sp1 and NF-κB heterologous probes before adding the labelled AP-1-TdT (lanes 6 and 7). Nuclear extracts were preincubated with 10, 25, 50 and 100-fold molar excess of autologous specific competitor AP-1-TdT and AP-1-Cons unlabelled probes (lanes 8–11 and 12–15, respectively). The arrows show the DNA-protein specific retarded complex and free DNA is indicated. (b) Super-shift assay analysis. Radiolabelled AP-1-Cons and AP-1-TdT were incubated in the absence (lanes 1 and 6) or presence (lanes 2–12) of HeLa cell nuclear extracts. The polyclonal antibody anti-jun was incubated with AP-1-Cons and AP-1-TdT probes, respectively (lanes 3 and 4) as well as an irrelevant antibody anti-interleukin-6 (lanes 7 and 8). The competition was performed using 100-fold molar excess of the unlabelled mutated probe AP-1 (AP-1 m, lanes 9 and 10), and also with 100-fold molar excess of the unlabelled probe Sp1 (lanes 11 and 12). The arrows indicate the formation of specific retarded complexes DNA-protein and the super-shift retarded complex is indicated. Data shown is representative of three independent experiments with two independent HeLa nuclear extract preparations.
AP-1-like site of the TdT belongs to AP-1 regulatory element family
To confirm the interaction between some AP-1 family members with AP-1-TdT, we carried out an EMSA with nuclear extracts from HeLa cells, using the AP-1-TdT labelled sequence as the probe and the AP-1-Cons as the cold competitor (Fig. 4a, lanes 12–15). AP-1-Cons as the source of cold competitor was incubated in increasing amounts with 10, 25, 50 and 100-fold molar excess in relation to the AP-1-TdT labelled probe. We found that the major complex competed efficiently with the unlabelled AP-1-Cons oligonucleotide, indicating that the bound protein belongs to the AP-1 family. To confirm this possibility, we evaluated the presence of the Jun protein in the AP-1-TdT retarded complex with an anti-jun polyclonal antibody. As shown in Fig. 4(b), we observed the formation of the retarded complex when we used the oligonucleotides AP-1-Cons and AP-1-TdT, and when these complexes were preincubated with the anti-jun polyclonal antibody a faint formation of a super-retarded complex was observed (Fig. 4b; lanes 3 and 4). There was no detection of any super-retarded complex when EMSA was performed with an irrelevant anti-interleukin-6 polyclonal antibody (Fig. 4b; lanes 7 and 8). Furthermore, to confirm the specificity of the DNA–protein complex we also performed a competition analysis with a mutated unlabelled probe of AP-1-Cons (Fig. 4b; lanes 9 and 10) and with the irrelevant unlabelled Sp1 probe in a 100-fold molar excess (Fig. 4b; lanes 11 and 12). No changes were seen on the formation of DNA–protein complexes following such competition. In conclusion, these results strongly support the notion that the transcription factor interacting with the AP-1-TdT binding sequence is a member of the AP-1 family of transcription factors.
Discussion
This work presents evidence that AP-1 family factors play a role in the regulation of TdT gene expression. Our results show that: (i) PKC-activation alone, without the need for an intracellular Ca2+ influx, induces c-fos and c-jun mRNA expression (Fig. 1); although Ca2+ alone induces c-fos expression, it has no effect on c-jun expression. (ii) We found an AP-1 binding site in the TdT regulatory region, further suggesting its role in TdT down-regulation (Fig. 3). (iii) EMSA assays indicated that an AP-1-like transcription factor binds to the TdT regulatory region (Fig. 4a); which was confirmed by super-shift with a polyclonal antibody directed to c-jun (Fig. 4b). Our findings indicate that transcription factors interacting at the TdT promoter region in TdT-nonexpressing cells may negatively influence TdT gene expression.
The data presented here do not allow the definition as to which particular member of the AP-1 family is required to repress TdT transcription through its promoter. However, our current model is that an AP-1 family member is essential for TdT gene regulation because we found that during the development of lymphoid cells (DND41) mRNA is expressed for two AP-1 family proteins c-fos and c-jun, proteins which are induced synergistically by PKC activation and increased levels of intracellular calcium (Fig. 1). We favour the idea that the most likely mediator for these effects is the Jun-D protein because it has previously been demonstrated to be capable of mediating repressive effects.44 Moreover, we found that the characterized AP-1-TdT binding site possesses a divergent sequence motif in comparison with the canonical AP-1 binding site. The relatively low sequence similarity is reminiscent of proteins that recognize the minor groove of DNA, such as HMG proteins45 and the TATA binding protein.46 This possibility is consistent with studies of murine Ets-1 suggesting that the AP-1–TdT binding protein binds to the major groove of DNA in a manner that permits the binding of other proteins.47
The Fos and Jun proteins belong to a class of transcription factors characterized by a leucine zipper dimerization domain and an amino-terminal adjacent DNA-binding domain rich in basic amino acids. In addition, c-Jun, Jun-B, Jun-D, and c-Fos, Fra, Fos-B are the major components of the AP-1 transcriptional regulator. Jun proteins can form homodimers and heterodimers more avidly with Fos protein, and they bind to the 5′-TGACTCA-3′ DNA sequence.20 Fos and Jun can also dimerize with members of the cAMP responsive element binding (CREB) protein family; these heterodimers have a preferential affinity for the CREB consensus sequence 5′-TGACGTCA-3′. Jun also interacts with the glucocorticoid receptors, modulating hormone dependent transcriptional regulation. Although the AP-1-family members are mainly considered to be activators of transcription, they can also have repressive functions. Thus, Jun-D has been reported to have a negative effect on cellular growth suggesting that the closely related factors c-Jun and Jun-D can function in opposite directions.44 Moreover, AP-1 factors have different transcriptional properties by means of specific activation and repression domains48,49 and/or differential post-translational modifications.50 The overall composition of the AP-1 complex may also be critical for its regulatory function because the DNA binding affinity of Jun members is greatly enhanced by the Fos protein. Binding of Jun homodimers or Fos–Jun heterodimers produces distinct degrees of DNA bending that could result in highly specific protein–protein interactions between AP-1 factors and other promoter-bound transcription complexes.51 Support for a contribution of c-fos to trans-activation of AP-1-dependent promoters came from studies using c-fos null cell lines, which provided the first direct evidence for an in vivo function of c-fos in the activation of a subset of AP-1-dependent promoters.52 Indeed, two independent groups have analyzed AP-1 binding activity and transcriptional regulation of several AP-1 target genes in fibroblasts lacking c-fos.52,53 These studies revealed normal AP-1 DNA-binding activity and similar levels of some known AP-1-dependent transcription. However, other AP-1 target genes were either down-regulated or up-regulated in response to growth factors.52,53 This evidence suggests that AP-1 sites by themselves can be divided into subtypes defined by their specificity for certain AP-1 family members or by their individual involvement in basal versus induced transcription. The present work demonstrates the formation of two major retarded complexes in cells that do not express TdT (Fig. 2). Based on these data, we propose that the nuclear factor that bind to the 5′-TCAGTAC-3′ sequence in AP-1-TdT is one of the AP-1-like regulatory factors.
An alternative possibility that may function in TdT gene regulation is that if an AP-1-like member, in conjunction with a corepressor, could recruit histone deacetylases directly thereby involving modification of the chromatin structure as a mechanism of TdT transcriptional repression in terminally differentiated lymphoid cells.54–56 Additionally, the kinetic studies of TdT gene demethylation and TdT transcription during thymus development have shown that changes in DNA methylation status are involved in the differential expression of TdT in the fetal and adult life of mice.57
In conclusion: (a) TdT expression occurs during lymphocyte development and transcriptional control must include mechanisms that restrict its expression to cells of the lymphoid lineage, specifically pre-B and pre-T cells; (b) TdT is not expressed by lymphoid mature cells; therefore, we believe that the AP-1-mediated repression regulates this control following PKC activation. It is well established that several genes are regulated by PKC-activation during human lymphoid maturation 10,16,20,21,36,37, and our results are in agreement with this notion. We cannot rule out the possibility that, in addition to the proximal regulatory elements, long-distance sequences, such as silencers, enhancers or locus control regions could also be contributing to basal promoter function and have tissue- and stage-specific effects on TdT expression. Recent studies have shown that interactions between TdT and PCNA (proliferating cell nuclear antigen) via its DNA polymerization domain have as a functional consequence the negative regulation of TdT activity.58
Acknowledgments
We thank Dr Yvonne Rosenstein and Dr José Moreno for critically reading the manuscript. This work was supported by a grant from Consejo Nacional de Ciencia y Tecnología (CONACYT) of México, no. 1018-M9111. Peralta-Zaragoza O. was supported by a fellowship number 117983 from CONACYT, México.
Abbreviations
- AP-1
activation protein 1
- CREB
cAMP responsive element binding
- EMSA
electrophoresis mobility shifting assay
- HMG
high mobility group protein
- PAGE
polyacrylamide gel electrophoresis
- PCR
polymerase chain reaction
- PKC
protein kinase C
- TdT
terminal deoxynucleotidyl transferase
- TPA
12-tetradecanoylphorbol-13-acetate
References
- 1.Ikuta K, Uchida N, Friedman J, Weissman IL. Lymphocyte development from stem cells. Annu Rev Immunol. 1992;10:759–83. doi: 10.1146/annurev.iy.10.040192.003551. [DOI] [PubMed] [Google Scholar]
- 2.Kallenbach S, Doyen N, Fanton d'Andon M, Rougeon F. Three lymphoid-specific factors account for all junctional diversity characteristic of somatic assembly of T-cell receptor and immunoglobulin genes. Proc Natl Acad Sci USA. 1992;89:2799–803. doi: 10.1073/pnas.89.7.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blackwell TK, Alt FW. Site-specific recombination between immunoglobulin D and JH segments that were introduced into the genome of a murine pre-B cell line. Cell. 1984;37:105–12. doi: 10.1016/0092-8674(84)90305-2. [DOI] [PubMed] [Google Scholar]
- 4.Bollum FJ. Terminal deoxynucleotidyl transferase as a hematopoietic cell marker. Blood. 1979;54:1203–15. [PubMed] [Google Scholar]
- 5.Coleman MS, Hutton JJ, Bollum FJ. Terminal riboadenylate transferase in human lymphocytes. Nature. 1974;248:407–9. doi: 10.1038/248407a0. [DOI] [PubMed] [Google Scholar]
- 6.Landau NR, Schatz DG, Rosa M, Baltimore D. Increased frequency of N-region insertion in a murine pre-B-cell line infected with a terminal deoxynucleotidyl transferase retroviral expression vector. Mol Cell Biol. 1987;7:3237–43. doi: 10.1128/mcb.7.9.3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bentolila LA, Wu GE, Nourrit F, Fanton d'Andon M, Rougeon F, Doyen N. Constitutive expression of terminal deoxynucleotidyl transferase in transgenic mice is sufficient for N region diversity to occur at any Ig locus throughout B cell differentiation. J Immunol. 1997;158:715–23. [PubMed] [Google Scholar]
- 8.Alt FW, Blackwell TK, Yancopoulos GD. Development of the primary antibody repertoire. Science. 1987;238:1079–87. doi: 10.1126/science.3317825. [DOI] [PubMed] [Google Scholar]
- 9.Hermans MH, Hartsuiker H, Opstelten D. An in situ study of B-lymphocytopoiesis in rat bone marrow. Topographical arrangement of terminal deoxynucleotidyl transferase-positive cells and pre-B cells. J Immunol. 1989;142:67–73. [PubMed] [Google Scholar]
- 10.Martinez-Valdez H, Cohen A. Coordinate regulation of mRNAs encoding adenosine deaminase, purine nucleoside phosphorylase, and terminal deoxynucleotidyltransferase by phorbol esters in human thymocytes. Proc Natl Acad Sci USA. 1988;85:6900–3. doi: 10.1073/pnas.85.18.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Siden EJ, Gifford A, Baltimore D. Cyclic AMP induces terminal deoxynucleotidyl transferase in immature B cell leukemia lines. J Immunol. 1985;135:1518–22. [PubMed] [Google Scholar]
- 12.Pazmino NH, Ihle JN, Goldstein AL. Induction in vivo and in vitro of terminal deoxynucleotidyl transferase by thymosin in bone marrow cells from athymic mice. J Exp Med. 1978;147:708–18. doi: 10.1084/jem.147.3.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Low TL, Hu SK, Goldstein AL. Complete amino acid sequence of bovine thymosin beta 4: a thymic hormone that induces terminal deoxynucleotidyl transferase activity in thymocyte populations. Proc Natl Acad Sci USA. 1981;78:1162–6. doi: 10.1073/pnas.78.2.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riley LK, Morrow JK, Danton MJ, Coleman MS. Human terminal deoxyribonucleotidyl transferase. molecular cloning and structural analysis of the gene and 5′ flanking region. Proc Natl Acad Sci USA. 1988;85:2489–93. doi: 10.1073/pnas.85.8.2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffbrand AV, Drexler HG, Ganeshaguru K, Piga A, Wickremasinghe RG. Biochemical aspects of acute leukemia. Clin Haematol. 1986;15:669–94. [PubMed] [Google Scholar]
- 16.Madrid-Marina V, Martinez-Valdez H, Cohen A. Phorbol esters induce changes in adenosine deaminase, purine nucleoside phosphorylase, and terminal deoxynucleotidyl transferase messenger RNA levels in human leukemic cell lines. Cancer Res. 1990;50:2891–4. [PubMed] [Google Scholar]
- 17.Trangas T, Coleman MS. Tissue-specific expression of human terminal deoxynucleotidyl transferase is regulated at the transcriptional level. Biochem Biophys Res Commun. 1989;164:750–7. doi: 10.1016/0006-291x(89)91523-4. [DOI] [PubMed] [Google Scholar]
- 18.Tillinghast JP, Russell JH, Fields LE, Loh DY. Protein kinase C regulation of terminal deoxynucleotidyl transferase. J Immunol. 1989;143:2378–83. [PubMed] [Google Scholar]
- 19.Koiwai O, Morita A, Yamada Y. Demonstration of promoter function for the 5′-flanking region of the human gene for terminal deoxynucleotidyl transferase. Biochem Biophys Res Commun. 1989;164:863–8. doi: 10.1016/0006-291x(89)91538-6. [DOI] [PubMed] [Google Scholar]
- 20.Ransone LJ, Verma IM. Nuclear proto-oncogenes fos and jun. Annu Rev Cell Biol. 1990;6:539–57. doi: 10.1146/annurev.cb.06.110190.002543. [DOI] [PubMed] [Google Scholar]
- 21.Engel JD. Meticulous AP-1 factors. Nature. 1994;367:516–7. doi: 10.1038/367516a0. [DOI] [PubMed] [Google Scholar]
- 22.Bhaumik D, Yang B, Trangas T, Bartlett JS, Coleman MS, Sorscher DH. Identification of a tripartite basal promoter which regulates human terminal deoxynucleotidyl transferase gene expression. J Biol Chem. 1994;269:15861–7. [PubMed] [Google Scholar]
- 23.Sorscher DH, Yang B, Bhaumik D, Trangas T, Philips AV, Chancellor KE, Coleman MS. Initiation of transcription at the human terminal deoxynucleotidyl transferase gene promoter. a novel role for the TATA binding protein. Biochemistry. 1994;33:11025–32. doi: 10.1021/bi00202a023. [DOI] [PubMed] [Google Scholar]
- 24.Garraway IP, Semple K, Smale ST. Transcription of the lymphocyte-specific terminal deoxynucleotidyl transferase gene requires a specific core promoter structure. Proc Natl Acad Sci USA. 1996;93:4336–41. doi: 10.1073/pnas.93.9.4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breathnach R, Chambon P. Organization and expression of eukaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–83. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- 26.Smale ST, Baltimore D. The ‘initiator’ as a transcription control element. Cell. 1989;57:103–13. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- 27.Hernandez N. TBP, a universal eukaryotic transcription factor? Genes Dev. 1993;7:1291–308. doi: 10.1101/gad.7.7b.1291. [DOI] [PubMed] [Google Scholar]
- 28.Smale ST, Jain A, Kaufmann J, Emami KH, Lo K, Garraway IP. The initiator element: a paradigm for core promoter heterogeneity within metazoan protein-coding genes. Cold Spring Harb Symp Quant Biol. 1998;63:21–31. doi: 10.1101/sqb.1998.63.21. [DOI] [PubMed] [Google Scholar]
- 29.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 30.Lehrach H, Diamond D, Wozney JM, Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977;16:4743–51. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- 31.Thomas PS. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci USA. 1980;77:5201–5. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–4. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 33.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucl Acids Res. 1983;11:1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell PJ, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–8. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 36.Peralta-Zaragoza O, Martínez-Valdez H, Madrid-Marina V. Regulation of gamma T-cell antigen receptor expression by intracellular calcium in acute lymphoblastic leukemia cell line DND41. Arch Med Res. 1996;27:305–10. [PubMed] [Google Scholar]
- 37.Martinez-Valdez H, Madrid-Marina V, Cohen A. Phorbol esters and cAMP differentially regulate the expression of CD4 and CD8 in human thymocytes. BMC Immunol. 2002;3:1–6. doi: 10.1186/1471-2172-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Curran T, Franza BR., Jr Fos and Jun: the AP-1 connection. Cell. 1988;55:395–7. doi: 10.1016/0092-8674(88)90024-4. [DOI] [PubMed] [Google Scholar]
- 39.Nishimura T, Vogt PK. The avian cellular homolog of the oncogene jun. Oncogene. 1988;3:659–63. [PubMed] [Google Scholar]
- 40.Chan AC, Desai DM, Weiss A. The role of protein tyrosine kinases and protein tyrosine phosphatases in T cell antigen receptor signal transduction. Annu Rev Immunol. 1994;12:555–92. doi: 10.1146/annurev.iy.12.040194.003011. [DOI] [PubMed] [Google Scholar]
- 41.Kobata T, Morimoto C. T cell receptor and its related molecules in signal transduction. Nippon Rinsho. 1999;57:273–7. [PubMed] [Google Scholar]
- 42.Lin J, Weiss A. T cell receptor signalling. J Cell Sci. 2001;114:243–4. doi: 10.1242/jcs.114.2.243. [DOI] [PubMed] [Google Scholar]
- 43.McBride K, Nemer M. The C-terminal domain of c-fos is required for activation of an AP-1 site specific for jun–fos heterodimers. Mol Cell Biol. 1998;18:5073–81. doi: 10.1128/mcb.18.9.5073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pfarr CM, Mechta F, Spyrou G, Lallemand D, Carillo S, Yaniv M. Mouse JunD negatively regulates fibroblast growth and antagonizes transformation by ras. Cell. 1994;76:747–60. doi: 10.1016/0092-8674(94)90513-4. [DOI] [PubMed] [Google Scholar]
- 45.van de Wetering M, Oosterwegel M, Dooijes D, Clevers H. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 1991;10:123–32. doi: 10.1002/j.1460-2075.1991.tb07928.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee DK, Horikoshi M, Roeder RG. Interaction of TFIID in the minor groove of the TATA element. Cell. 1991;67:1241–50. doi: 10.1016/0092-8674(91)90300-n. [DOI] [PubMed] [Google Scholar]
- 47.Nye JA, Petersen JM, Gunther CV, Jonsen MD, Graves BJ. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992;6:975–90. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- 48.Deng T, Karin M. JunB differs from c-Jun in its DNA-binding and dimerization domains, and represses c-Jun by formation of inactive heterodimers. Genes Dev. 1993;7:479–90. doi: 10.1101/gad.7.3.479. [DOI] [PubMed] [Google Scholar]
- 49.Suzuki T, Okuno H, Yoshida T, Endo T, Nishina H, Iba H. Difference in transcriptional regulatory function between c-Fos and Fra-2. Nucl Acids Res. 1991;19:5537–42. doi: 10.1093/nar/19.20.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karin M. The regulation of AP-1 activity by mitogen-activated protein kinases. J Biol Chem. 1995;270:16483–6. doi: 10.1074/jbc.270.28.16483. [DOI] [PubMed] [Google Scholar]
- 51.Nakabeppu Y, Ryder K, Nathans D. DNA binding activities of three murine Jun proteins: stimulation by Fos. Cell. 1988;55:907–15. doi: 10.1016/0092-8674(88)90146-8. [DOI] [PubMed] [Google Scholar]
- 52.Hu E, Mueller E, Oliviero S, Papaioannou VE, Johnson R, Spiegelman BM. Targeted disruption of the c-fos gene demonstrates c-fos-dependent and -independent pathways for gene expression stimulated by growth factors or oncogenes. EMBO J. 1994;13:3094–103. doi: 10.1002/j.1460-2075.1994.tb06608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brusselbach S, Mohle-Steinlein U, Wang ZQ, Schreiber M, Lucibello FC, Muller R, Wagner EF. Cell proliferation and cell cycle progression are not impaired in fibroblasts and ES cells lacking c-Fos. Oncogene. 1995;10:79–86. [PubMed] [Google Scholar]
- 54.Yang B, Gathy KN, Coleman MS. Mutational analysis of residues in the nucleotide binding domain of human terminal deoxynucleotidyl transferase. J Biol Chem. 1994;269:11859–68. [PubMed] [Google Scholar]
- 55.Bakin AV, Curran T. Role of DNA 5-methylcytosine transferase in cell transformation by fos. Science. 1999;283:387–90. doi: 10.1126/science.283.5400.387. [DOI] [PubMed] [Google Scholar]
- 56.Lee SK, Kim JH, Lee YC, Cheong J, Lee JW. Silencing mediator of retinoic acid and thyroid hormone receptors, as a novel transcriptional corepressor molecule of activating protein-1, nuclear factor-kappa-B, and serum response factor. J Biol Chem. 2000;275:12470–4. doi: 10.1074/jbc.275.17.12470. [DOI] [PubMed] [Google Scholar]
- 57.Nourrit F, Coquilleau I, D'Andon MF, Rougeon F, Doyen N. Methylation of the promoter region may be involved in tissue-specific expression of the mouse terminal deoxynucleotidyl transferase gene. J Mol Biol. 1999;292:217–27. doi: 10.1006/jmbi.1999.3079. [DOI] [PubMed] [Google Scholar]
- 58.Ibe S, Fujita K, Toyomoto T, et al. Terminal deoxynucleotidyl transferase is negatively regulated by direct interaction with proliferating cell nuclear antigen. Gene Cells. 2001;6:815–24. doi: 10.1046/j.1365-2443.2001.00460.x. [DOI] [PubMed] [Google Scholar]