Abstract
Streptococcal inhibitor of complement (SIC) is a 31 kDa extracellular protein produced by a few highly virulent strains of Streptococcus pyogenes (in particular the M1 strain). It has been shown additionally to inhibit four further components of the mucosal innate response—lysozyme, secretory leucocyte proteinase inhibitor, human α-defensin 1 and the cathelicidin LL-37 which are all bactericidal against Group A Streptococci (GAS). We now show that SIC also inhibits variably the antibacterial action of hBD-1, -2 and -3. By enzyme-linked immunosorbent assay (ELISA), SIC binds strongly to hBD-2 and hBD-3, but not at all to hBD-1. Investigation of the antimicrobial action of β-defensins hBD-1, -2 and -3 against GAS in two different buffer systems shows that both the killing efficiencies of all three defensins, and the binding of SIC to them, occurs more efficiently in 10 mm Tris buffer than in 10 mm phosphate. The lower ionic strength of the Tris buffer may underlie this effect. hBD-1 kills the M1 strain of GAS only in 10 mm Tris, but is able to kill an M6 (SIC negative) strain in 10 mm phosphate. The inhibition of hBD-3 by SIC is clearly of physiological relevance, that of hBD-2 is likely to be so, but the inhibition of hBD-1 occurs only at lower ionic strength than is likely to be encountered in vivo. Elastase digestion of SIC yields three major fragments of MW 3·843 kDa comprising residues 1–33 (fragment A); 10·369 kDa comprising residues 34–126 (fragment B); and MW 16·487 kDa, comprising residues 127–273 (fragment C). By ELISA, only fragment B binds to hBD-2 and hBD-3 and this may indicate the inhibitory portion of the SIC molecule.
Introduction
Streptococcal inhibitor of complement (SIC) is a 31 kDa extracellular protein produced by a few highly virulent strains of Streptococcus pyogenes (predominantly by the M1 strain), and was initially identified as an inhibitor of the membrane attack complex of complement.1 The mechanism for this inhibition was found subsequently to be prevention of the insertion of C567 into the cell membrane.2 SIC has since been reported to inhibit the antimicrobial properties of four more proteins of the innate immune system found on mucosal surfaces.3,4 The first study3 showed that SIC bound to secretory leucocyte proteinase inhibitor (SLPI) at high affinity, and totally inhibited the ability of SLPI to kill Group A Streptococci (GAS). Interestingly, SIC had no effect on the antiproteinase activity of SLPI located in the COOH-terminal domain of the protein, implying that its action is specifically with the NH2-terminal antimicrobial domain of SLPI. SIC was also shown to bind to lysozyme, but at lower affinity, and to inhibit the antimicrobial and catalytic functions of both human and hen egg lysozyme.3 The second study4 showed that SIC also bound to and inhibited the antibacterial action of the cathelicidin LL-37 and the human α-defensin hNP-1.
Other observations support the proposition that SIC is an important virulence factor. Lukomski and colleagues5 showed that mice inoculated intranasally with the sic-positive wild-type M1 strain had a significantly higher incidence of throat colonization within 4 days postinfection compared to those inoculated with a sic-negative M1 mutant strain. A recent study by Hoe and colleagues,6 using the same two strains, showed that the sic-negative M1 mutant strain adhered to, and was phagocytosed and killed more effectively by, human epithelial cells than the wild-type M1 strain, and that addition of SIC prevented adhesion of the mutant and naturally occurring sic-negative GAS to human epithelial cells. SIC protein was taken up quickly by human epithelial cells inducing cell flattening and loss of microvilli. They speculated that this binding interfered with GAS contact, internalization and killing, thus enhancing bacterial survival and dissemination by enabling the bacteria to remain longer in the extracellular environment. SIC has also been shown to be highly immunogenic7 and extremely variable, with over 300 variants having been recorded.8
However, there are many other components of the innate immune system found on mucosal surfaces (reviewed in Diamond et al.9) and the present study focuses on the interaction of SIC with three further antimicrobial peptides, namely, the human β-defensins hBD-1, hBD-2 and hBD-3. hBD-2 has recently been shown to kill M49 and M52 strains of GAS10 and hBD-3 has been shown to kill the same strain of M1 GAS used in this report together with a multiresistant strain of S. aureus.11 The action of hBD-1 against GAS has not hitherto been investigated.
The β-defensins are small, broad-spectrum, cationic antimicrobial peptides that are characterized by containing six cysteine residues connected by three disulphide bridges. They are expressed in a variety of different tissues, including those vulnerable to infection by GAS. For instance, hBD-1 is expressed constitutively in epithelial cells of the respiratory tract, salivary glands and kidney, and also in breast milk; hBD-2 is inducibly expressed in response to inflammatory stimuli in the epithelia of the skin, trachea and lung tissues, although GAS were reported to be relatively poor inducers of hBD-2 expression in keratinocytes compared to several other organisms (Pseudomonas aeruginosa, S. aureus, S. epidermidis and Escherichia coli);10 and hBD-3 is inducibly expressed in the trachea, oral epithelium and keratinocytes (reviewed in Schutte and McCray Jr12). The antibacterial action of hBD-1 and hBD-2 is sensitive to increasing concentration of salt,12 while that of hBD-3 is still functional at physiological salt concentration.11
In most reports, in vitro antimicrobial assays are performed in 10 mm sodium phosphate buffer, pH 7·2 or pH 7·4/1% growth medium, but in the recent study by Frick and colleagues4 into the interaction between SIC and hNP-1 and LL-37, the assays were performed in 10 mm Tris, pH 7·5/5 mm glucose. In this study we also compare the effects of the two different assay buffers on the antimicrobial activity of, and the interaction of SIC with, the three β-defensins and LL-37.
Materials and methods
Bacterial strains
Streptococcus pyogenes
Types M1 (SIC positive—NCTC 8198, ATCC 12344) and M6 (SIC negative—NCTC 8302) were obtained from the National Collection of Type Cultures, Central Public Health Laboratory, London. The sequence of the sic gene in the M1 strain is identical to that originally published (GenBank Accession no. X92968).1,2 M1 GAS were grown in Todd Hewitt broth/0·2% yeast extract (THBY) (Oxoid Ltd, Basingstoke, UK) and maintained in the short term on selective Columbia agar/5% horse blood agar plates (containing oxolinic acid 5 µg/ml and colistin sulphate 10 µg/ml; Sigma-Aldrich Co. Ltd, Gillingham, UK). M6 GAS were grown in brain heart infusion (BHI) (Oxoid Ltd) (as they were found to grow poorly in THBY and have low viability) and plated as above.
Purification of SIC
SIC was purified from 1·5 l overnight culture supernatants of strain M1 GAS, to which 10 mm EDTA had been added, as described.3
Purification and characterization of elastase-digested fragments of SIC
(a) Digestion and purification
In order to investigate which area of the SIC protein might interact with the β-defensins, 25 mg of SIC in phosphate buffered saline (PBS) was digested with 0·1% w/w human neutrophil elastase (Elastin Products Company, Inc., Owensville, MI, USA) for 1 h at 37°, and the reaction stopped by the addition of phenylmethanesulphonyl fluoride (PMSF) to 1 mm. Starting material was at an average concentration of 6 mg/ml. The digestion conditions were optimized for the generation of three major fragments of apparent molecular weights (by Tricine SDS-PAGE) 7·5 kDa (fragment A), 10 kDa (fragment B) and 16 kDa (fragment C) by performing a time–course experiment. The digested material was made to a final ionic strength of 0·1 m ammonium bicarbonate, pH 7 (start buffer), loaded onto a 10 ml Source 15Q column (Amersham Biosciences, Amersham, UK) equilibrated with start buffer and run on an FPLC system (Amersham Biosciences) at room temperature. The fractions were eluted with a 20-column-volume gradient of ammonium bicarbonate to 1 m. Fractions containing the three major digestion products were separately pooled, freeze dried and redissolved in 1 ml of 0·25 m ammonium bicarbonate immediately prior to loading onto a 94 × 1·5 cm (166 ml volume) Superdex 75 column (Amersham Biosciences) at 4° and run in the same buffer. Fractions containing the purified fragments were pooled and again freeze dried. In order to confirm purity, 2 µg aliquots of whole SIC and fragments A, B and C were run on a Tricine SDS-PAGE gel as above, and stained with Coomassie Blue.
(b) N-terminal sequencing
The purified fragments were run on a 16% Tricine SDS-PAGE gel and electroblotted onto an Immobilon membrane (Millipore (UK) Ltd, Watford, UK). N-terminal sequencing was carried out at the Protein and Nucleic Acid Chemistry Facility, Department of Biochemistry, University of Cambridge.
(c) Mass spectrometry
The size of fragment A was analysed using a Thermo Finnigan LCQ Classic ion-trap instrument (Thermo Finnigan, Hemel Hempstead, UK) fitted with a static nanoelectrospray source by Dr L. Packman in the Protein and Nucleic Acid Chemistry Facility, Department of Biochemistry, University of Cambridge. The monoisotopic mass was determined from the zoomscan of doubly and triply charged ions. These ions were analysed further by MS/MS fragmentation and the results submitted to a database search using Mascot (http://www.matrixscience.com) confirming the identity of the fragment. The size of fragments B and C was determined by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry on a Voyager DE-STR MALDI-TOF mass spectrometer equipped with a reflector analyser (PE Biosystems, Warrington, UK) at the Medical Research Council Centre, Cambridge
Enzyme linked immunosorbant assay (ELISA)
ELISA plates (Greiner Bio-One Ltd, Stonehouse, UK) were coated with recombinant hBD-1, hBD-2 or hBD-3 (PeproTech EC Ltd, London, UK) at 2 µg/ml in triplicate in 50 mm carbonate/bicarbonate buffer, pH 9·6. The plates were then blocked with 1% gelatine/PBS. SIC was added at 5 µg/ml diluted in PBS/0·05% Tween 20/0·1% gelatine (PBSTG) and bound protein detected with rabbit anti-SIC Ig fraction at 2 µg/ml,2 followed by alkaline phosphatase conjugated goat anti-rabbit Ig diluted 1/1200 (Sigma-Aldrich Co. Ltd). Both antibodies were diluted in PBSTG. A similar ELISA was performed in which the plate was blocked with 10 mm Tris-HCl, pH 7·5/1% gelatine, and all dilutions were in 10 mm Tris-HCl, pH 7·5/0·05% Tween 20/0·1% gelatine (TTG).
Alternatively, plates were coated with SIC at 1·5 µg/ml as above, then blocked in 10 mm Tris, pH 7·5/2·5% hydrolysed casein. hBD-1, -2 or -3 were added at 3 µg/ml in 10 mm Tris, pH 7·5/0·05% Tween 20/0·5% hydrolysed casein/(TTC) and bound peptide detected with goat anti-hBD-1 IgG fraction at 0·5 µg/ml (Santa Cruz Biotechnology, Inc., Autogen Bioclear, Calne, UK), affinity purified goat anti-hBD-2 at 0·5 µg/ml, or rabbit anti-hBD-3 IgG fraction at 1 µg/ml (Abcam Ltd, Cambridge, UK), followed by alkaline phosphatase conjugated monoclonal anti-goat Ig (Sigma Aldrich Co. Ltd) or alkaline phosphatase conjugated goat anti-rabbit Ig as above, all diluted in TTC. An identical ELISA was performed for hBD-3 only using PBS/gelatine buffers.
In other ELISAs, plates were coated with hBD-2 or hBD-3 as above, the plate blocked in Tris/gelatine (hBD-2) or PBS/gelatine (hBD-3) and whole SIC 5 µg/ml or the three elastase-digested fragments of SIC (at equimolar concentrations) added at 0·6 µg/ml (N-terminal fragment A), 1·68 µg/ml (mid-fragment B) or 2·67 µg/ml (C-terminal fragment C) diluted in TTG or PBSTG. Binding was detected as above, with the reagents being diluted in TTG or PBSTG.
Alternatively, plates were coated with SIC at 2 µg/ml, fragment A at 0·24 µg/ml, fragment B at 0·67 µg/ml or fragment C at 1 µg/ml, then blocked in Tris/casein. hBD-2 was added at 3 µg/ml in TTC and bound peptide detected with goat anti-hBD-2 as above.
In order to check that that the three elastase-digested fragments of SIC could be detected by the available antiserum, plates were coated with intact SIC and the three fragments at 2 µg/ml and bound protein detected as above, all dilutions being in PBSTG.
In all ELISAs control wells with either no ligand, no first antibody or with second antibody only were set up, together with matching sets of wells to which coating buffer only had been added as background controls. One hundred µl volumes were used throughout, except for the blocking step with 200 µl. Coating was at 4° overnight, subsequent incubations were for 1 h at 37°, and the final reagent was ‘Sigma Fast’ pNPP (4-nitrophenyl phosphate disodium salt hexahydrate) substrate/buffer system (Sigma Aldrich Co. Ltd) at room temperature. Absorbance was read at 405 nm (reference wavelength 490 nm) in a microplate reader (Bio-Rad 3550; Bio-Rad Laboratories Ltd, Hemel Hempstead, UK). All ELISAs have been performed at least three times and usually more.
Antibacterial assays
(a) Titration of the effect of hBD-1, hBD-2 and hBD-3 peptides on M1 and M6 Group A Streptococci
The effect of hBD-1, hBD-2 and hBD-3 peptides on the viability of the M1 and that of hBD-1 on M6 strains of GAS was investigated as described.3 Briefly, overnight cultures of GAS were grown at 37° in THBY (M1) or BHI (M6) and used to inoculate fresh broth at 1/100. Bacteria were grown to mid log-phase, spun down and washed twice in 10 mm sodium phosphate buffer, pH 7·2 and resuspended to a concentration of 2 × 105/ml in phosphate buffer/1% THBY/BSA 2 mg/ml (M1) or phosphate buffer/1% BHI/BSA 2 mg/ml (M6); 15 µl aliquots of bacteria were added to equal volume double dilutions of hBD-1 from 40 µm, 3 in 4 dilutions of hBD-2 from 6 µm, or double dilutions of hBD-3 from 10 µm in phosphate buffer/1% medium, or buffer/1% medium only, in duplicate, giving final concentrations of peptide from half the above concentrations, plus bacteria at 105/ml and BSA at 1 mg/ml, and incubated for 2 hr at 37°. Bacteria were then diluted and plated and percentage survival calculated compared to bacteria incubated in buffer alone.
Similar titrations of the β-defensins were performed with M1 GAS in which the bacteria were washed and resuspended in 10 mm Tris HCl, pH 7·5/5 mm glucose and the reagents were diluted in the same buffer, final concentrations being hBD-1 from 20 µm, hBD-2 from 2 µm and hBD-3 from 3 µm.
For comparison, titrations of the human cathelicidin LL-37 (kindly provided by Dr Pieter Hiemstra, University of Leiden, the Netherlands) were performed with M1 GAS from 10 µm in phosphate and from 5 µm in Tris buffer as above.
(b) Titration of the effect of SIC on the interaction of hBD-1, hBD-2 and hBD-3 with M1 Group A Streptococci
M1 GAS were grown and prepared as above in phosphate buffer. Doubling dilutions of SIC from 6 µm were combined with 2 µm hBD-2, or SIC from 19·5 µm with 6·5 µm hBD-3 (all in phosphate buffer/1% THBY) in a total volume of 15 µl (in duplicate), for 2 hr at 37°; 15 µl of M1 GAS at 2 × 105/ml were added and incubated for a further 2 hr at 37°. Final overall concentrations were SIC from 1·5 µm plus 0·5 µm hBD-2, or SIC from 4·875 µm plus 1·625 µm hBD-3, each with bacteria at 105/ml. Controls were bacteria in phosphate buffer/1% THBY, matching dilutions of SIC, or peptide alone, all containing BSA 1 mg/ml.
Similar assays were performed in which the bacteria and reagents were diluted in 10 mm Tris HCl, pH 7·5/5 mm glucose. The final concentrations were SIC from 12 µm plus 3 µm hBD-1, SIC from 0·6 µm plus 0·15 µm hBD-2 and SIC from 2·4 µm plus 0·6 µm hBD-3.
Calculation of ionic strength
The ionic strength of the two buffers used in the above antimicrobial assays was calculated using the Debye-Hückel equation as follows:
where Σ indicates ‘sum of all terms of the following description’; m = molality (assumed here to be ∼ = molarity); z = valence; and µ = ionic strength. Thus, the ionic strength at 37° of 10 mm sodium phosphate buffer pH 7·2 is 0·0244 and that of 10 mm Tris/HCl pH 7·5 is 0·0063.
Results
SIC binds strongly to hBD-2 and hBD-3, but not at all to hBD-1, as determined by ELISA
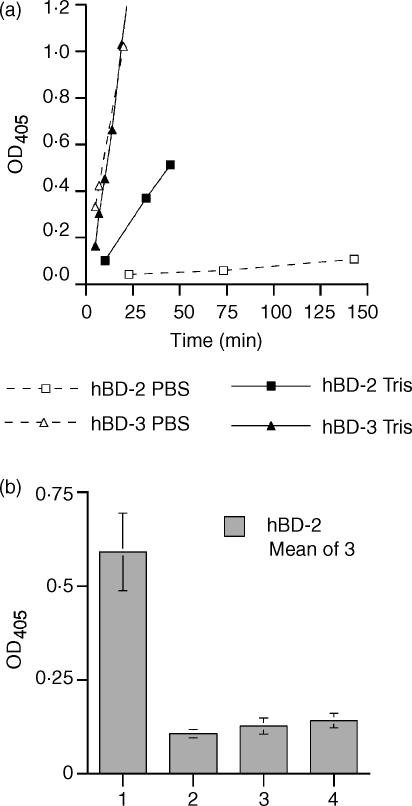
In the ELISA performed in PBS/gelatine, SIC bound very strongly to wells coated with hBD-3, an OD405 of >1 being achieved within about 20 min, a small amount of SIC bound to hBD-2 after a much longer incubation, but only minimal binding was observed between SIC and hBD-1. However, when the ELISA was performed in 10 mm Tris/gelatine, considerably better binding occurred between SIC and hBD-2, although this was still less than that between SIC and hBD-3, which was indistinguishable from that observed in PBS/gelatine. Again, no binding was observed between SIC and hBD-1 (data not shown). Figure 1a shows the rate of increase in the OD405 over time for SIC binding to hBD-2 and hBD-3 in both PBS and Tris/gelatine.
Figure 1.
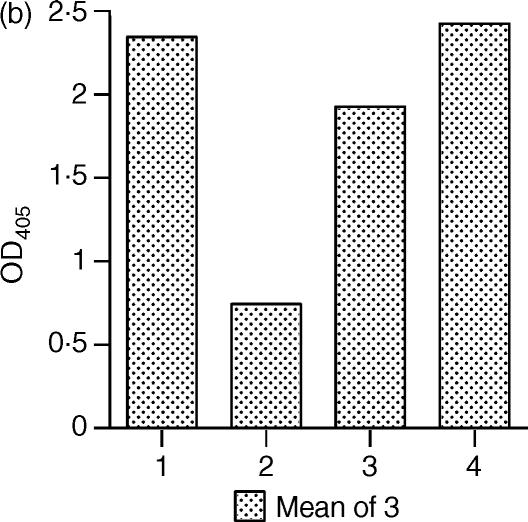
Binding of SIC to hBD-2 and hBD-3 in PBS or Tris buffer by ELISA. (a) Comparison of the binding efficiency between SIC and the β-defensins in different buffers, coating protein hBD-2 or hBD-3; ligand SIC; diluted in either PBS/Tween/0·1% gelatine or 10 mm Tris-HCl, pH 7·5/Tween/0·1% gelatine. Dotted lines and open symbols = PBS; solid lines and filled symbols = Tris. □ = hBD-2; ▵ = hBD-3. (b) Binding of hBD-2 to SIC in Tris buffer. Coating protein SIC; ligand hBD-2; dilution buffer 10 mm Tris-HCl, pH 7·5/Tween/0·5% hydrolysed casein. Bar 1 = SIC plus ligand plus first and second antibodies; bar 2 = SIC plus first and second antibodies; bar 3 = SIC plus second antibody; and bar 4 = SIC plus ligand plus second antibody. Results are mean ± SEM. In both panels, results are expressed as net OD405 after deduction of background values from matching uncoated wells.
When the ELISA was performed in the opposite orientation when the plate was coated with SIC and binding of the peptide was detected, hBD-2 bound strongly to SIC (Fig. 1b). It was necessary to use hydrolysed casein rather than gelatine in the buffers for this ELISA in order to reduce the background to low levels, as hBD-2 appeared to bind to gelatine. Again, no binding was observed between hBD-1 and SIC (data not shown). We were unable to obtain any interpretable results for hBD-3 and SIC in this orientation, in either buffer, due to the very high level of cross-reaction between the antibody and the SIC-coated plate, which we found impossible to eliminate despite trying a number of different blocking agents (data not shown). The epitope for this antibody has been mapped to residues 1–11 of the mature hBD-3 peptide (information from the datasheet provided by Abcam Ltd). Comparison of this sequence with that of SIC using the sequence alignment program pileup within the GCG-Package (Genetic Computer Group) did not reveal any obvious homology and the suppliers are unable to offer any suggestions, so the reason for this cross-reaction remains unclear.
hBD-2, hBD-3 and LL-37 kill the M1 strain of Group A Streptococci in both phosphate and Tris buffer but hBD-1 is effective only in Tris
In order to investigate or confirm the bactericidal activity of the human β-defensins 1, 2 and 3 and the cathelicidin LL-37 against an M1 strain of GAS, dilutions of hBD-1, hBD-2, hBD-3 or LL-37 were combined with bacteria at 105/ml in either phosphate or Tris buffer. hBD-2, -3 and LL-37 were bactericidal against M1 GAS in phosphate buffer, the concentrations required for 50% killing being 0·94, 1·03 and 1·82 µm, respectively. hBD-1 had no effect at all on M1 GAS in phosphate buffer even at the maximum concentration tested (20 µm). When similar titrations were performed in 10 mm Tris HCl/5 mm glucose the bactericidal activity of all four peptides was enhanced. Interestingly, hBD-1 now killed M1 GAS, 50% killing being achieved with 3·28 µm. hBD-2, -3 and LL-37 achieved 50% killing at approximately 7×, 2× and 3× lower concentrations than in phosphate buffer. The results are summarized in Table 1.
Table 1.
Concentration of antimicrobial peptide required for 50% killing of Group A Streptococci
| Peptide (µm) | M1 10 mm phosphate | M1 10 mm Tris | M6 10 mm phosphate |
|---|---|---|---|
| hBD-1 | N/K‡ | 3·28† | 10·28‡ |
| hBD-2 | 0·94* | 0·135† | N/T |
| hBD-3 | 1·03* | 0·47† | N/T |
| LL-37 | 1·82* | 0·66† | N/T |
N/K: no killing; N/T: not tested.
Mean of three experiments;
mean of two experiments;
mean of four experiments.
hBD-1 kills the M6 strain of Group A Streptococci in phosphate buffer
In order to investigate whether similar results would be obtained with a SIC negative strain of GAS, titration of hBD-1 was performed in phosphate buffer with the M6 strain. hBD-1 killed M6 GAS, 50% killing being achieved at 10·28 µm (Table 1).
SIC inhibits the bactericidal action of hBD-1, hBD-2 and hBD-3 against M1 GAS
Phosphate buffer
Dilutions of SIC from a threefold molar excess were preincubated with concentrations of hBD-2 or hBD-3 at which approximately 25% of the bacteria survive, and then incubated with M1 GAS at 105/ml. SIC inhibited both defensin peptides in a dose-dependent fashion (Fig. 2a), 50% survival of M1 GAS being restored by the addition of SIC at a molar SIC : hBD-2 ratio of 0·26 and a SIC : hBD-3 ratio of 1·12.
Figure 2.
SIC protects Group A Streptococci from killing by hBD-1, -2 and -3 in both (a) phosphate and (b) Tris buffer. Inhibition of the antibacterial action of the β-defensins by SIC. (a) Killing of M1 GAS by hBD-2 and hBD-3 in phosphate buffer is inhibited by SIC. Doubling dilutions of SIC from 6 µm were combined with 2 µm hBD-2 or SIC from 19·5 µm with 6·5 µm hBD-3, and incubated for 2 h at 37°. Bacteria at 2 × 105/ml were added, incubated for a further 2 h at 37°, then diluted and plated. Final overall concentrations were either SIC from 1·5 µm plus 0·5 µm hBD-2 or SIC from 4·875 µm plus 1·625 µm hBD-3, and bacteria at 105/ml. (b) Killing of M1 GAS by hBD-1, hBD-2 and hBD-3 in Tris buffer is inhibited by SIC. Doubling dilutions of SIC from 48 µm were combined with 12 µm hBD-1, SIC from 2·4 µm with 0·6 µm hBD-2 or SIC from 9·6 µm with 2·4 µm hBD-3, and incubated for 2 h at 37°. Bacteria at 2 × 105/ml were added, incubated for a further 2 h at 37°, then diluted and plated. Final overall concentrations were either SIC from 12 µm plus 3 µm hBD-1, SIC from 0·6 µm plus 0·15 µm hBD-2 or SIC from 2·4 µm plus 0·6 µm hBD-3, and bacteria at 105/ml. In both panels, results are expressed as percentage survival compared to bacteria incubated in the equivalent concentration of SIC alone ± SEM, against the number of molecules of defensin per molecule of SIC (hBD-1: three experiments; hBD-2: two experiments in each buffer; hBD-3: three experiments in phosphate, two experiments in Tris).
Tris buffer
Dilutions of SIC from a fourfold molar excess were preincubated with the β-defensins at concentrations at which approximately 20% of M1 GAS survive, and then incubated with bacteria as above. hBD-2 was inhibited with less variability in Tris buffer and hBD-3 much more efficiently (Fig. 2b), with 50% survival being achieved at SIC : defensin molar ratios of 0·3 and 0·42, respectively. In spite of the weaker antibacterial activity of hBD-1, SIC showed an inhibition of this effect at a comparable molar ratio of SIC : defensin of 0·49. The results are summarized in Table 2.
Table 2.
Concentration of SIC required, and molar ratios of SIC : defensin, for 50% survival of Group A Streptococci in the presence of hBD-1, -2 and -3
| 10 mm phosphate | 10 mm Tris | |||||
|---|---|---|---|---|---|---|
| Conc. peptide (µm) | Conc. SIC (µm) | Molar ratio SIC : defensin | Conc. peptide (µm) | Conc. SIC (µm) | Molar ratio SIC : defensin | |
| hBD-1 | N/T | N/T | N/T | 3 | 1·46 | 0·49 |
| hBD-2 | 0·5 | 0·13 | 0·26 | 0·15 | 0·045 | 0·3 |
| hBD-3 | 1·625 | 1·82 | 1·12 | 0·6 | 0·25 | 0·42 |
N/T: not tested.
Characterization of SIC fragments and their interaction with hBD-2 and hBD-3
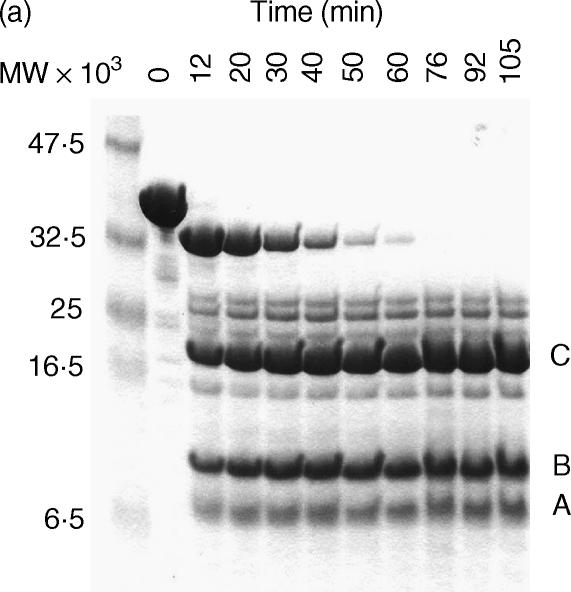
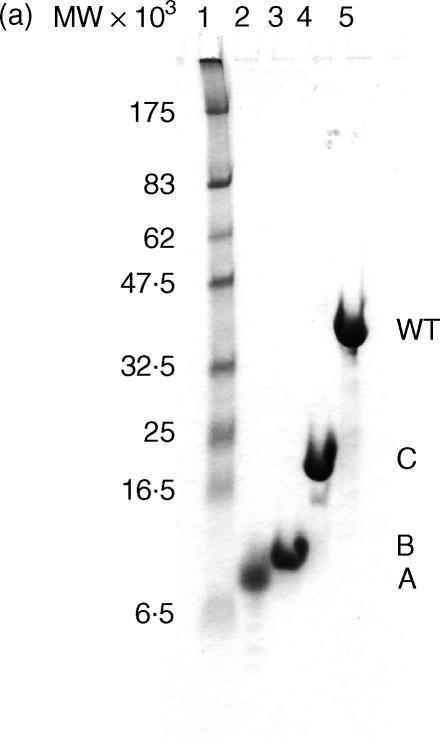
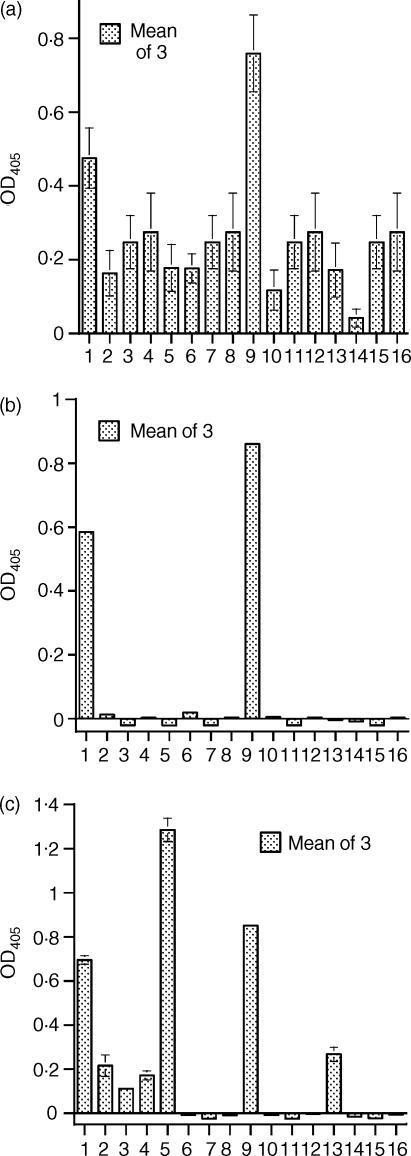
The time–course digestion of SIC with human sputum elastase generated three major fragments of apparent MW 7·5 kDa, 10 kDa and 16·5 kDa, respectively, by Tricine SDS-PAGE electrophoresis (together with several minor fragments) (Fig. 3a). The three major fragments were separated initially by ion exchange chromatography followed by gel filtration which resolved the fragments into sharp peaks (Figs 3b,c,d). Figure 4a shows a Tricine-SDS gel of the purified fragments, all of which could be detected by the polyclonal antibody in a direct ELISA (Fig. 4b). Mass spectrometry revealed that the molecular weights were 3·843 kDa (fragment A), i.e. substantially less than it appeared by Tricine-SDS PAGE analysis, 10·369 kDa (fragment B) and 16·487 kDa (fragment C) and these data, together with N-terminal sequencing results, showed that fragment A comprised amino acids 1–33, fragment B comprised amino acids 34–126 and fragment C amino acids 127–273, i.e. the remaining C-terminal part of the protein.
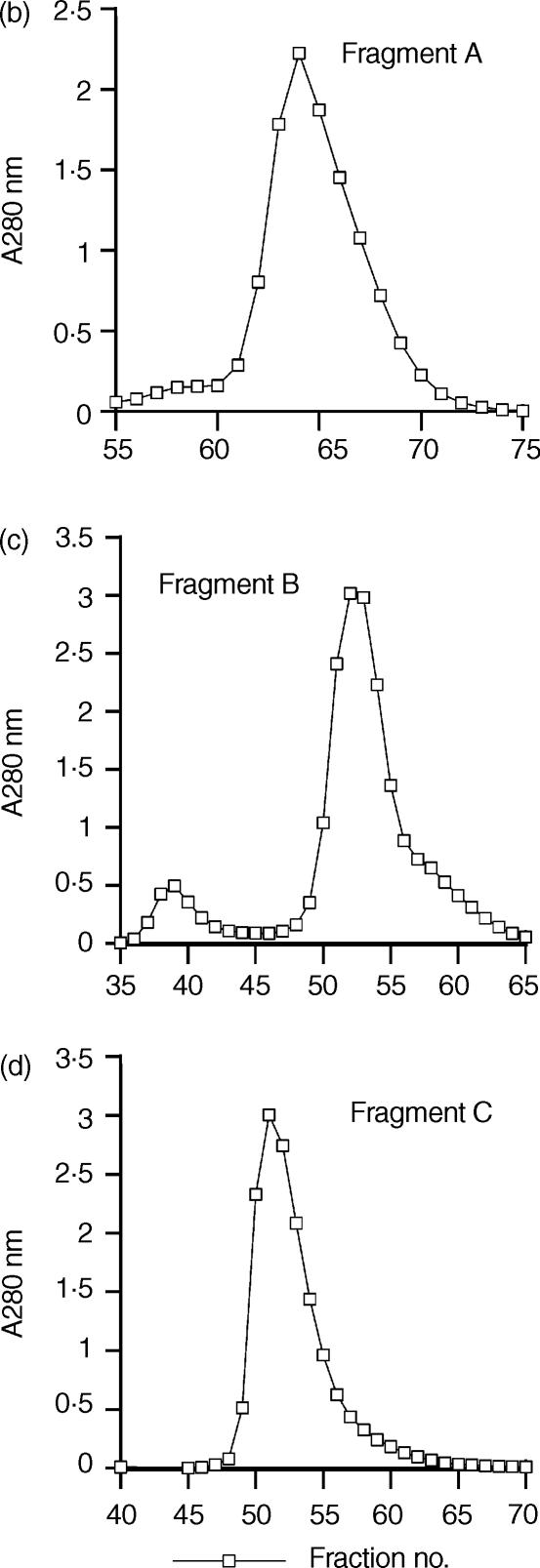
Figure 3.
Purification of elastase-digested fragments of SIC. (a) Time course digestion of SIC analysed on a Tricine-SDS gel. (b) Purification of Fragment A by gel filtration. (c) Purification of Fragment B by gel filtration. (d) Purification of Fragment C by gel filtration. (a) A time–course experiment was performed in which SIC was digested with 0·1% w/w human neutrophil elastase. Samples taken at approximately 10-min intervals were analysed on a 16% Tricine SDS-PAGE gel and Coomassie stained. Lane 1, molecular weight markers; lane 2, undigested SIC; lanes 3–11, digested SIC. The 50-min digest was chosen for purification of the three major fragments designated. (b) (c) and (d) Purification of Fragments A, B and C by gel filtration. Pooled fractions containing predominantly either fragments A, B or C, separated initially by ion-exchange chromatography on a Source Q column, were purified further on a Superdex 75 column and resolved into sharp peaks.
Figure 4.
Purified SIC fragments. (a) Tricine-SDS gel of purified SIC and fragments. (b) ELISA—detection of SIC and fragments by rabbit anti-SIC. Analysis of purified elastase-digested fragments of SIC. (a) SIC and purified elastase-digested fragments of SIC were run on a 16% Tricine SDS-PAGE gel and Coomassie stained. Lane 1, molecular weight markers; lane 2, fragment A, amino acids 1–33; lane 3, fragment B, amino acids 34–126; lane 4, fragment C, amino acids 127–273; lane 5, whole SIC. (b) An ELISA plate was coated with SIC or purified elastase-digested fragments of SIC, and bound protein detected. Bar 1 = intact SIC, bar 2 = fragment A, bar 3 = fragment B, bar 4 = fragment C. Results are expressed as net OD405 after deduction of background values from matching uncoated wells, ± SEM. Where no error bars are shown, the SEM is too small to be seen. Controls of coated wells plus second antibody are not shown, but all values were < 0·07.
Fragment B bound strongly to ELISA plates coated with hBD-2 and hBD-3 while fragments A and C did not bind at all (Figs 5a,b). Note that the hBD-2 ELISA was performed only in 10 mm Tris buffer in order optimize potential binding, and this probably accounts for the higher signal in the control wells compared with the hBD-3 ELISA which was performed in PBS. When the ELISA was performed in the opposite orientation and the plate coated with SIC and fragments, hBD-2 bound to all three fragments (Fig. 5c) A >B > C.
Figure 5.
ELISAs. Binding of SIC and SIC fragments to (a) hBD-2 and (b) hBD-3; (c) binding of hBD-2 to SIC and SIC fragments. In each panel, bars 1–4 = intact SIC, bars 5–8 = fragment A, bars 9–12 = fragment B, bars 13–16 = fragment C. Within each group of 4 bars, bar 1 = coating protein + ligand + both antibodies; bar 2 = coating protein + both antibodies; bar 3 = coating protein + second antibody; bar 4 = coating protein + ligand + second antibody. (a) coating peptide hBD-2, ligands SIC or fragments. Buffer = 10 mm Tris-HCl/Tween/gelatine. (b) coating protein hBD-3, ligands SIC or fragments. Buffer = PBS/Tween/gelatine. (c) coating protein SIC or fragments; ligand hBD-2. Buffer = 10 mm Tris-HCl/Tween/casein. Results are expressed as net OD405 after deduction of background values from matching uncoated wells, ± SEM. Where no error bars are shown, the SEM is too small to be seen.
Discussion
We have found that the β-defensins are a further group of antimicrobial peptides whose function can be inhibited by SIC; SIC inhibits the antibacterial activity of hBD-1, hBD-2 and hBD-3 against M1 GAS. The studies have been performed both in 10 mm phosphate buffer at pH 7·2 and in 10 mm Tris buffer at pH 7·5. The former is physiological; the latter has a substantially lower ionic strength which promotes charge interaction. The inhibition of hBD-2 by SIC is very similar in both buffers at a molar ratio of 1 : 3 SIC : defensin, while that of hBD-3 by SIC is more efficient in Tris buffer, i.e. 1 : 1 molar ratio in phosphate buffer but 1 : 2 in Tris buffer. SIC also inhibited hBD-1 at a molar ratio of 1 : 2 in Tris buffer. We have also reproduced the results obtained by Frick and colleagues for the inhibition of LL-37 by SIC in Tris buffer,4 but could not achieve reproducible significant inhibition in phosphate buffer (data not shown).
We have demonstrated that, by ELISA, SIC binds to hBD-3 strongly in both high (PBS) and low (10 mm Tris) ionic strength buffers, and that hBD-2 binds to SIC strongly at low ionic strength, particularly when SIC is bound to the plate. This observation may imply that when hBD-2 is bound to a plate this largely masks the site that binds to SIC. As hBD-3 is usually found as a dimer,13 it is less likely that all SIC binding sites would be hidden. The poor binding between SIC and hBD-1 by ELISA, whichever is bound to the plate, may also reflect differences between interaction in the fluid phase and that when a protein or small peptide is deposited on a plastic surface, as it was somewhat surprising that the functional inhibition of hBD-1 by SIC was of comparable efficiency to the other two defensins.
It would appear likely that a large component of the interaction between SIC and the β-defensins (and LL-37) is ionic, which would explain the more efficient inhibition of hBD-3 and LL-37 at lower ionic strength, that of 10 mm Tris being approximately four times lower than that of 10 mm phosphate. However, the interaction between SIC and the β-defensins may not be entirely ionic, as demonstrated by the identical inhibition results for hBD-2, and identical ELISA results for hBD-3, in both buffers.
Both hBD-1 and hBD-2 have been estimated conservatively to occur at a concentration of approximately 150 ng/ml in saliva (∼30 nm)14 and SIC is produced at approximately 5 µg/ml in mid-log phase culture (∼161 nm).4 If this is also the case in vivo, then there could easily be local concentrations of SIC high enough to inhibit these defensins. The concentration of hBD-3 in the throat has not yet been reported. The area of the throat involved in GAS pharyngitis is predominantly bathed in saliva, which is generally considered to be hypotonic and to have a pH of between 6 and 7 (reviewed in Humphrey & Williamson15]). We have measured the conductivity of unstimulated saliva to be approximately 7·35 milli-Siemens ± 1·15 (mS) (mean of four individuals ± SEM) compared to 4·15 mS for 10 mm phosphate, 1 mS for 10 mm Tris and 20 mS for PBS at 37°. The conductivity of phosphate buffer is not dissimilar to that of saliva, but we would hesitate to attribute biological significance to the reactions occurring only in the Tris buffer.
We have found that the β-defensin hBD-1 is bactericidal against the SIC-positive M1 strain of GAS only under optimal assay conditions (10 mm Tris HCl, pH 7·5) while it is able to kill the SIC-negative M6 strain in both Tris buffer and the more commonly employed phosphate buffer. The reason for this difference is not entirely apparent; secretion of SIC could be a contributing factor but probably not the sole reason. We have confirmed the published observations10,11 that the β-defensins 2 and 3 are able to kill Group A Streptococci at µM concentrations in phosphate buffer and have found that the killing is much more efficient in Tris buffer. This effect is particularly marked for hBD-2 (approximately sevenfold lower concentration required for 50% killing in Tris than in phosphate buffer), whereas for hBD-3 the difference was not so great (approximately 2·2-fold). We also compared the relative antibacterial efficiency of LL-37 in both buffers and found that an approximately 2·7-fold lower concentration of LL-37 was required for 50% killing in Tris than in phosphate buffer. These large differences in killing efficiency for hBD-1 and hBD-2 are due probably to the lower ionic strength of 10 mm Tris as they are known to be salt-sensitive,12 whereas the smaller differences observed for hBD-3 and LL-37, probably reflect their relative insensitivity to higher salt concentrations.11,16
The ELISA results for binding of the elastase digested fragments of SIC to hBD-2 and hBD-3 show that only fragment B, amino acids 34–126, binds when the plate is coated with the defensin peptides. Neither defensin binds either the N-terminal or C-terminal fragments of SIC. However, when the plate coated with SIC fragments hBD-2 binds strongly to all three, and most strongly to the N-terminal fragment A. This result correlates strongly with the calculated isoelectric point of the fragments, as fragment A is the most anionic (pI 3·76), and fragment C the least (pI 4·58). We suspect that this second ELISA is misleading and merely represents electrostatic attraction. However, in view of the functional inhibition results, and that the polyclonal antibody to SIC is able to detect all three fragments in a direct ELISA, we consider that the binding of fragment B to the defensins is likely to reflect events in the fluid phase and could well contain the inhibitory area of the SIC protein.
The SIC protein sequence contains a number of repeat regions: a short repeat region between residues 1–74, followed by three longer tandem repeats between residues 75–153. Fragment A contains several of the short repeats, and fragment B contains several short repeats together with all of the first, and most of the second, long repeat. Fragment C contains the last six residues of the second long repeat and all of the third, which is slightly shorter than the first two. The remainder of the protein does not contain any predictable potential structure/function motifs. Sequencing of the sic gene from numerous M1 GAS isolates has shown that long repeats 1 and 2 are the least variable areas of the gene in terms of amino acid sequence (reviewed in Reid et al.17), although several isolates have been shown to contain variable numbers of long repeat 2, and this could well reflect conservation of a functional area of the protein. None the less, identification of linear epitopes in SIC by human antisera, using a synthetic peptide library of overlapping 15-mers, showed that there does appear to be an epitope in the centre of each of the long repeats,7 which is presumably under the same selective pressure to avoid the adaptive immune system as the rest of the molecule. Further studies will be required to elucidate whether any of these internal structures are involved in the binding of SIC to the β-defensins and quantitative physicochemical studies of the binding between SIC and the β-defensins are in progress.
As observed previously,2 intact SIC runs at an apparent MW of >40 kDa on Tricine SDS-PAGE, although its MW is ∼31 kDa. This is thought to be due to the high number of prolines and lack of cysteines, resulting in a non-globular structure that could retard migration. It was also observed that a spontaneous breakdown product of SIC occurred after storage at −20°, in which the first 33 amino acids had been lost from the native protein2 and which we now know coincides with first elastase cutting site. This slightly smaller protein still ran higher than its true molecular weight on SDS-PAGE. Interestingly, fragment A runs at nearly twice its true MW as determined by Tricine SDS-PAGE, whereas the apparent MWs of fragments B and C are in accord with those obtained by mass spectrometry. This leads us to suspect that this section of the molecule is partly responsible for the anomalous migration of SIC on SDS-PAGE gels.
In conclusion, SIC would appear to be a multifunctional virulence factor of the M1 strain of Group A Streptococci which protects the bacteria from the antimicrobial action of several components of the innate immune system present on mucosal surfaces. The inhibition of hBD-3 by SIC is clearly of physiological relevance, that of hBD-2 is likely to be so, but the inhibition of hBD-1 occurs only at a lower ionic strength than is likely to be encountered in vivo.
Acknowledgments
We would like to thank the British Heart Foundation, Project Grant no. PG/03/084, who funded the latter stage of this study (BAF-K and DJS). We would also like to thank Dr Pieter S. Hiemstra and Ms Sandra Tjabringa, Department of Pulmonology, University of Leiden Medical Center, the Netherlands for the kind donation of synthetic LL-37 and Miss Heather Lindsay, Bacterial Infection Group, Centre for Veterinary Science, University of Cambridge for advice on growth conditions for Group A Streptococci.
References
- 1.Åkesson P, Sjöholm AG, Björck L. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J Biol Chem. 1996;271:1081–8. doi: 10.1074/jbc.271.2.1081. [DOI] [PubMed] [Google Scholar]
- 2.Fernie-King BA, Seilly DJ, Willers C, Würzner R, Davies A, Lachmann PJ. Streptococcal inhibitor of complement (SIC) inhibits the membrane attack complex by preventing uptake of C567 onto cell membranes. Immunol. 2001;103:390–8. doi: 10.1046/j.1365-2567.2001.01249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fernie-King BA, Seilly DJ, Davies A, Lachmann PJ. Streptococcal Inhibitor of Complement (SIC) inhibits two further components of the mucosal innate immune system: secretory leucokyte proteinase inhibitor (SLPI) and lysozyme. Infect Immun. 2002;70:4908–16. doi: 10.1128/IAI.70.9.4908-4916.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frick I-M, Åkesson P, Rasmussen M, Schmidtchen A, Björck L. SIC—a secreted protein of Streptococcus pyogenes that inactivates antibacterial peptides. J Biol Chem. 2003;278:16561–6. doi: 10.1074/jbc.M301995200. [DOI] [PubMed] [Google Scholar]
- 5.Lukomski S, Hoe NP, Abdi I, et al. Nonpolar inactivation of the hypervariable Streptococcal Inhibitor of Complement gene (sic) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonisation. Infect Immun. 2000;68:535–42. doi: 10.1128/iai.68.2.535-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoe NP, Ireland RM, DeLeo FR, et al. Insight into the molecular basis of pathogen abundance: Group A Streptococcus inhibitor of complement inhibits bacterial adherence and internalization into human cells. PNAS. 2002;99:7646–51. doi: 10.1073/pnas.112039899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoe NP, Kordari P, Cole R, et al. Human immune response to Streptococcal Inhibitor of Complement, a serotype M1 Group A Streptococcus extracellular protein involved in epidemics. J Infect Dis. 2000;182:1425–36. doi: 10.1086/315882. [DOI] [PubMed] [Google Scholar]
- 8.Hoe NP, Vuopio-Varkila J, Vaara M, et al. Distribution of Streptococcal Inhibitor of Complement variants in pharyngitis and invasive isolates in an epidemic of serotype M1 Group A Streptococcus infections. J Infect Dis. 2001;183:633–9. doi: 10.1086/318543. [DOI] [PubMed] [Google Scholar]
- 9.Diamond G, Legarda D, Ryan LK. The innate immune response of the respiratory epithelium. Immunol Rev. 2000;173:27–38. doi: 10.1034/j.1600-065x.2000.917304.x. [DOI] [PubMed] [Google Scholar]
- 10.Dinulos JGH, Mentele L, Fredericks P, Dale BA, Darmstadt G. Keratinocyte expression of human β-defensin 2 following bacterial infection: role in cutaneous host defense. Clin Diagn Lab Immunol. 2003;10:161–6. doi: 10.1128/CDLI.10.1.161-166.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harder J, Bartels J, Christophers E, Schröder J-M. Isolation and characterisation of human β-defensin 3, a novel human inducible peptide antibiotic. J Biol Chem. 2001;276:5707–13. doi: 10.1074/jbc.M008557200. [DOI] [PubMed] [Google Scholar]
- 12.Schutte BC, McCray PB., Jr β-defensins in lung host defense. Ann Rev Physiol. 2002;64:709–48. doi: 10.1146/annurev.physiol.64.081501.134340. [DOI] [PubMed] [Google Scholar]
- 13.Schibli DJ, Hunter HN, Aseyev V, et al. The solution structures of the human β-defensins lead to a better understanding of the potent bactericidal activity of hBD-3 against Staphylococcus aureus. J Biol Chem. 2002;277:8279–89. doi: 10.1074/jbc.M108830200. [DOI] [PubMed] [Google Scholar]
- 14.Mathews M, Jia HP, Guthmiller JM, Losh G, Graham S, Johnson GK, Tack BF, McCray PB., Jr Production of β-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect Immun. 1999;67:2740–5. doi: 10.1128/iai.67.6.2740-2745.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Humphrey SP, Williamson RT. A review of saliva: normal composition, flow, and function. J Prosthet Dent. 2001;85:162–9. doi: 10.1067/mpr.2001.113778. [DOI] [PubMed] [Google Scholar]
- 16.Travis SM, Anderson NN, Forsyth WR, et al. Bactericidal activity of mammalian cathelicidin-derived peptides. Inf Imm. 2000;68:2748–55. doi: 10.1128/iai.68.5.2748-2755.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reid SD, Hoe NP, Smoot LM, Musser JM. Group A Streptococcus: allelic variation, population genetics, and host–pathogen interactions. J Clin Invest. 2001;107:393–9. doi: 10.1172/JCI11972. [DOI] [PMC free article] [PubMed] [Google Scholar]