Abstract
Polymorphism amongst the human leucocyte antigen (HLA) class II genes could influence antigen presentation and the ability to control human immunodeficiency virus (HIV)-1 by modulating the virus specific CD4 immune response. To examine the effect of such polymorphisms on disease progression, we studied a cohort of 46 HIV-1 infected long-term non-progressors (LTNPs), 87 intermediate progressors (IPs) and 26 rapid progressors. Kaplan–Meier survival analysis of all patients in the cohort on time to a CD4 count less than 350 cells/µl, showed a trend for a slower rate of CD4 decline in patients with, compared to those without, the DRB1*15-DQB1*06 haplotype (hazard ratio (HR) 0·69, 95% CI 0·46–1·01, P = 0·06). A similar effect was not observed with the DRB1*13-DQB1*06 haplotype (HR 1·18, 95% CI 0·75–1·88, P = 0·46), but was observed when DQB1*06 alleles were considered irrespective of their DR association (HR 0·74, 95% CI 0·52–1·05, P = 0·06). Major HLA-DQ6 alleles encode aspartate (Asp) at position 57 on the DQβ chain, a phenotype associated with protection from other immune disorders. We therefore examined the frequency of all DQβ57 Asp+ alleles, but could not detect a significant effect on the rate of CD4 decline. To examine whether the genotype associated with slower CD4 decline was over-represented in patients with a slow rate of disease progression, we conducted a categorical analysis of a subset of patients with an extended follow-up of 14+years. We found a higher proportion of LTNPs at 14+ years possessed the DRB1*15-DQB1*06 haplotype compared to IPs at 14+ years (38·46 versus 18·18%), though this difference did not reach statistical significance. When DQB1*06 alleles irrespective of their DR association were considered, the protective effect was greater (76·9% LTNPs versus 18·18% IPs, P = 0·04). Our results highlight the potential protective effect of HLA DQB1*06 alleles on the course of HIV disease.
Keywords: HIV-1, disease progression, HLA Class II, genes, haplotypes, polymorphisms, T cells: CD4+
Introduction
Control of human immunodeficiency virus (HIV)-1 infection involves the induction of virus-specific CD8 cytotoxic T cells (CTL), that, upon recognition of HIV-derived peptides presented by human leucocyte antigen (HLA) class I molecules, promote virus clearance via cytolysis and/or secretion of antiviral chemokines (see 1 and 2 for review). This model of infection control is emphasized by the absolute requirement for specific CD8 cells for virus clearance in the simian immunodeficiency virus/Macaque model.3,4 In addition, immunity to HIV-1 is dependent on CD4 T cells that recognize specific viral antigens in the context of HLA class II molecules.5,6 HIV-specific CD4 T cells orchestrate the virus-specific CD8 response, governing both its magnitude7–9 and quality in terms of proliferation and perforin expression.10 In addition to their helper role, virus specific CD4 cells may directly curtail HIV spread by the secretion of antiviral factors.11
In view of the known effects of HLA gene products on the immune response, there have been several attempts to correlate HLA gene polymorphisms with progression to disease following HIV infection12–14 in particular using cohorts representing the extremes of rapid progression and non-progression.15,16 In relation to class I HLA alleles, HLA-B14, -B27, -B57, and -B44 have been found to be associated with slower progression17–22 while the HLA-A29, -B22 and B-35 alleles were associated with rapid disease progression.17,23–25 In relation to class II HLA alleles, HLA-DR11 has been associated with rapid progression17 whilst three studies have highlighted the importance of the HLA-DRB1*13-DQB1*06 haplotype in slower progression.17,25,26 Indeed, in a large longitudinal study of 375 seroconverters aggregated from three cohorts, this was the only class II haplotype associated with acquired immune deficiency syndrome (AIDS)-free time.25 Inheritance of HLA-DRB1*13 alleles has also been linked with long-term survival among children with vertically transmitted HIV-1 infection.26 In addition, Malhotra et al.27 found that the DRB1*13-DQB1*06 haplotype was associated with an improved ability to suppress viral replication over time in response to early treatment in the acute phase of infection. Patients with this haplotype mounted a more vigorous HIV-specific CD4 response than those without, indicating that CD4 T-helper cells influenced by gene products associated with DRB1*13-DQB1*06 alleles may be important in immunity to HIV infection.27
In general, however, the reported effects of the HLA-DRB1*13-DQB1*06 haplotype have been weak (see 25). One possible explanation is that specific structural features that are shared by different HLA molecules and influence peptide-binding sites confer a protective effect. Under these circumstances there may be several HLA molecules with similar risk-associated structural features, but because they are encoded on different haplotypes, the association with risk remains obscure using haplotype analysis alone. A second possible explanation is that the effects of HLA class II gene structure and the ensuing CD4 helper cell response exert a protective effect only in subjects with a relatively benign disease course and are overcome by other viral, host or genetic factors in those with rapid disease progression. Such effects are therefore unlikely to be evident in studies that focus only on the extremes of rapid versus non-progression.
These issues were addressed by examining earlier reports of an association between HIV non-progression and the HLA-DRB1*13-DQB1*06 haplotype17,25–27 in our cohort of HIV+ patients with different rates of progression. As HLA-DQ6 alleles typically occur in linkage disequilibrium with HLA-DR15 and -13, the impact of the HLA-DRB1*15-DQB1*06 haplotype on disease progression was also assessed. We also examined the impact of one of the most studied structural features of HLA-DQ6, namely the presence of an aspartate (Asp) residue at position 57 on the DQβ chain, a region of the molecule known to influence the affinity and repertoire of peptide binding.28,29 Our results highlight the potential protective effect of HLA-DQB1*06 alleles and we were unable to detect an independent protective effect of β57Asp on different haplotypes on the course of HIV disease.
Materials and methods
Clinical features of cohort
Samples were taken from participants in a previously well-characterized HIV-infected cohort, established in 1995, of 165 long-term HIV-1-infected patients attending clinics in London, UK, who had been enrolled into a nested case–control study of the biological and behavioural correlates of non-progression in HIV-1 infection.30 The majority of subjects (95%) were white homosexual or bisexual men of European origin.30 Gentoype data were available on 159 patients: including 46 HIV-1 infected long-term non-progressors (LTNPs) defined as a CD4 count >500 cells/µl, at >10 years of infection, 87 intermediate progressors (IPs) defined as a CD4 count <500 cells/µl at >10 years of infection and 26 rapid progressors defined as development of AIDS within 5 years of infection. In order to study the potential impact of genotype on disease progression in patients with an extended follow-up, we identified 33 subjects (17 LTNPs and 16 IPs) who remained antiretroviral treatment naïve despite HIV infection of 14 or more years. Functional studies on these 33 subjects have shown that the HIV Gag-specific interleukin-2+ interferon-γ+ CD4 T-cell response, an important correlate of non-progression and low virus load was preserved in this LTNP group and conversely impaired in this group of IPs.7 Thirty of these 33 subjects were similar in race (Caucasian), gender (male) and behavioural characteristics (homosexual). Genotype analysis was known for 24 out of these 30 subjects (13 LTNPs and 11 IPs). The study was approved by the local Ethics Committee and written informed consent was obtained from all subjects.
DRB1 Gene amplification by polymerase chain reaction (PCR)
Primers employed for group-specific amplification and sequencing of DRB1 exon 2 were as detailed by Voorter et al.31 A multiplex mix of 5′ primers specific for all allelic groups was employed. In ambiguous cases, allele specific amplification was subsequently performed. The PCR reaction was set up in a volume of 50 µl containing 10 mm Tris (pH 8·3), 50 mm KCl, 1·5 mm MgCl2, 5% glycerol, 4 µg cresol red, 200 µm each dinuclotidetriphosphate (dNTP), 13·3 pmol of biotinylated primer, 26·6 pmol unlabelled primer, 200 ng of DNA and 1·4 U AmpliTaq DNA polymerase (Roche Ltd, East Sussex, UK). Cycle parameters used in a Perkin-Elmer 9600 thermal cycler were an initial denaturation step at 94° for 2 min prior to cycling, followed by 35 cycles at 94° for 30 s, 55° for 30 s, 72° for 45 s, followed by a final 5-min extension at 72°.
DQB1 amplification by PCR
Primers employed for amplification and sequencing of DQB1 exon 2 were as detailed by Voorter et al.31 DQB1 alleles were amplified employing the 5′ primer DQB96912 in combination with DQB96008b (DQB1*02/03/04) or DQB95011b (DQB1*05/06). PCR reactions were performed in a final volume of 50 µl. The PCR reaction was set up in a volume of 60 µl containing 10 mm Tris (pH 8·3), 50 mm KCl, 1·5 mm MgCl2, 5% glycerol, 1% dimethylsulphoxide, 6 µg cresol red, 200 µm each dNTP, 20 pmol of biotinylated primer, 40 pmol unlabelled primer, 500 ng of DNA and 2·0 U AmpliTaq DNA polymerase. Cycle parameters used in a Perkin-Elmer 9600 thermal cycler were an initial denaturation step at 94° for 2 min prior to cycling, followed by 10 cycles at 94° for 10 s, 65° for 60 s, and finally 20 cycles of 94° for 10 s, 61° for 50 s, and 72° for 30 s followed by a final 5-min extension at 72°.
Sequencing reactions and analysis
Ten µl of PCR reaction mixture was screened by agarose gel electrophoresis for the presence of PCR product of the correct size (bp). Positive PCR products were used as a template for sequencing. For DRB1 50 µl of each PCR product was subject to Solid phase sequencing reactions using the Pharmacia Autoload Solid Phase Sequencing Kit (Amersham Pharmacia Biotech UK Ltd, Amersham, UK) according to the manufacturer's protocol. Reaction products were separated on an ALFexpress automated sequencer. Generated sequencing data was processed automatically and evaluated manually, prior to HLA typing using the Pharmacia Sequityper software package. Sequencing ambiguities were resolved by allele specific amplification and sequencing or by DRB1/DQB1 SSP (Dynal Biotech UK, Wirral, UK) where appropriate.
Statistical analysis
Kaplan-Meier analysis was used to analyse the time to a CD4 count <350 cells/µl in individuals who had either one (heterozygous) or two alleles (homozygous) compared to neither; and Cox's proportional hazard regression models were used to estimate hazard ratios. Proportional hazards assumptions were tested and found to be not significant. Categorical analysis of LTNPs versus IPs at 14+ years was conducted. Association between possession of 1 (heterozygous) or 2 (homozygous) of HLA- DRB1*13-DQB1*06 or HLA- DRB1*15-DQB1*06 alleles or HLA-DQB1*06 alleles compared to none, was examined using Fisher's exact test. Since there had been previous reports of an association between the DRB1*13-DQB1*06 haplotype and non-progressors25–27 the association of this haplotype with non-progression was an apriori hypothesis, and accordingly, no correction was made for multiple comparisons.
Results
Possession of DRB1*15-DQB1*06 haplotype retards the rate of CD4 decline in HIV-1 infection
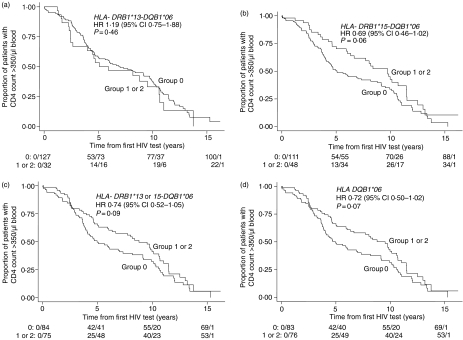
We first determined the effect of the DRB1*13-DQB1*06 haplotype on the rate of decline of CD4 T cells to below 350 cells/µl blood in all 159 patients in the cohort during the period from the first HIV test to more than 14 years of infection. Thirty-two patients at the time of their first HIV test had the the DRB1*13-DQB1*06 haplotype. No association between this haplotype and rate of CD4 decline was noted (Fig. 1a; hazard ratio (HR) = 1·19, 95% CI = 0·75–1·88, P = 0·46), for those with none versus one or two HLA-DR13/DQ6 haplotypes. We reasoned that the effect of the DRB1*13-DQB1*06 haplotype observed in other studies 17,25,26 might be related to the presence of genes encoding the HLA-DQ6 molecule, rather than HLA-DR alleles, and since HLA-DQ6 genes are typically in linkage disequilibrium with either HLA-DR13 or HLA–DR15 alleles, we next examined the frequency of the DRB1*15-DQB1*06 haplotype. Forty-eight subjects in the cohort presented with the DRB1*15-DQB1*06 haplotype. A strong trend for an association between the rate of CD4 T-cell decline and the DRB1*15-DQB1*06 haplotype in patients possessing one or more copies was observed (Fig. 1b, HR = 0·69, 95% CI 0·46–1·02, P = 0·06) and this did not change statistically when both the HLA-DQ6 encoding haplotypes were considered together (Fig. 1c, HR = 0·74, 95% CI 0·52–1·05, P = 0·09). Seventy-five subjects possessed the 13/6 or the 15/6 haplotypes. An additional five subjects had both the 13/6 and the 15/6 alleles, making the combined number of subjects with these alleles, 80. When we analysed the possession of HLA-DQ6 encoding alleles irrespective of the HLA-DR alleles (75 subjects in the cohort had DQ6 with DR13 and/or DR15, only one subject had DQ6 on a different DR allele), we found a similar level of protection as that noted with the DRB1*15-DQB1*06 haplotype (Fig. 1d, HR = 0·72, 95% CI 0·50–1·02, P = 0·07). Taken together these data indicate the potential importance of the DR15/DQ6 haplotype in delaying clinical progression in our cohort.
Figure 1.
Effect of HLA DQ6 alleles on rate of CD4 decline. Kaplan–Meier analysis of the rate of CD4 T-cell decline to <350 cells/µl from the time of the first HIV test to greater than 14 years of infection of 159 patients in the cohort is shown. Patients who possess one or two alleles of DQ6 (groups 1 or 2, respectively, d), or DQ6 linked to either HLA DRB1*13 only (a), HLA DRB1*15 only (c), or possess either the HLA DRB1*13-/-15-DQB1*06 haplotype (c), were compared to those who lacked these alleles (Group 0). Cox's proportional hazard regression models were used to estimate hazard ratios and two-tailed P-values represented. The rows below each survival curve show the number of events/number of individuals still at risk at 0, 5, 10, 15 years after the first HIV test.
DQ6-encoding alleles associate with a subset of non-progressors
We further hypothesized that the protective effect of HLA-DQ6-encoding-alleles may be more apparent by studying a subgroup of the 17 ‘best’ survivors, clinically stable and free of antiretroviral therapy despite a median duration of infection of 14·8 years (range 13–16·2 years). We therefore tested whether markers identified by the Kaplan–Meier analysis to be associated with a slower rate of CD4 decline were over-represented in this group of patients compared to a matched group of IPs who also had a median duration of infection of 14+ years (range 12·7–15·7 years) but whose CD4 count had declined below 350 cells/µl. Functional data on these IPs had previously revealed that their HIV-specific CD4 T-cell response was impaired compared to these LTNPs.7 We noted a higher prevalence of the DRB1*15-DQB1*06 and the DRB1*13-DQB1*06 haplotypes in this subset of LTNPs compared to the subset of IPs though group differences for either marker did not reach statistical significance (Table 1). However, in the combined analysis of DQB1*06 alleles linked to either DRB1*-15 or -13 alleles, the protective effect was stronger (76·9% LTNPs versus 27·27% IPs, P = 0·04).
Table 1.
Analysis of DQB1*06 alleles in long-term HIV-infected individuals at 14+years
| Parameters | LTNP n = 17 | IP n = 16 | P-value LTNP versus IP |
|---|---|---|---|
| Median duration of infection (years) | 14.8 (13–16.2) | 14.9 (12.7–15.7) | |
| Median virus load (copies/ml) (IQ range) | 13 510 (50–104 000) | 69 433 (1002–263 000) | 0.007 |
| Median CD4 count (×106/l) (IQ range) | 656.50 (497–1993) | 243 (85–530) | <0.0001 |
| Frequency of patients with HLA DRB1*15DQB1*06 haplotype | 5/13 (38.46%) | 2/11 (18.18%) | 0.3864 |
| Frequency of patients with HLA DRB1*13-DQB1*06 haplotype | 5/13 (38.46%) | 1/11 (9.09%) | 0.1660 |
| Frequency of patients with 1 or 2 HLA DQB1*06 alleles | 10/13 (76.9%) | 3/11 (27.27%) | 0.04 |
The genotype of a total of 13 LTNPs and 11 IPs at 14+ years was compared. The number and percentage of subjects who possessed one or two alleles of HLA DRB1 * 15- or DRB1 13-DQB1 *06 or DQB1 * 06 alleles on either DRB1 * 15- or DRB1 *13- alleles is summarized. Comparison of whether the frequency of one or two HLA alleles in the LTNP group was higher than that of the IP group was examined using Fischer's exact test and two-tailed P-values represented.
Aspartate at position 57 on the HLA-DQβ chain does not alter the rate of CD4 decline
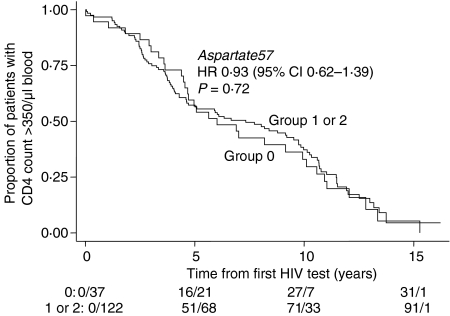
In structural terms, one of the most studied features of the protein products of the major alleles at the DQB1*06 locus is the presence of an Asp residue at position 57 on the DQβ chain (DQβ57Asp)28,29 which is, for example, associated with dominant protection from the autoimmune disease type 1 diabetes mellitus32 irrespective of haplotype or racial origin. We therefore examined associations of DQβ57Asp-encoding alleles with clinical groups and effects on the rate of CD4 decline. DQβ57Asp-encoding alleles were assigned as described.33 The majority of subjects in the cohort had one or two alleles encoding DQβ57 aspartate (122/159 = 76·7%) (Fig. 2). Kaplan–Meier survival and Cox-proportional hazards analysis showed no association with delay in CD4 decline and possession of 1 or more DQβ57Asp-encoding alleles (HR = 0·93, 95% CI 0·62–1·39, P = 0·72; Fig. 2). Similarly, we were unable to detect evidence of a strong protective effect of DQβ5Asp57 in our analysis of the subgroup of the ‘best’ survivors at 14+ years. The percentage of alleles with this residue did not differ statistically between LTNPs and IPs (84·6% LTNPs had one or two alleles encoding DQβ57 aspartate compared to 54·54% Ips, P = 0·18).
Figure 2.
Effect of DQβ57Asp alleles on rate of CD4 decline. Kaplan–Meier analysis of the rate of CD4 T-cell decline to <350 cells/µl from the time of the first HIV test to greater than 14 years of infection of 159 patients in the cohort is shown. Patients who possess one or two alleles of DQβ57Asp (groups 1 or 2, respectively) were compared to those who lacked these alleles (Group 0). Cox's proportional hazard regression models were used to estimate hazard ratios and two-tailed P-values represented. The rows below the survival curve show the number of events/number of individuals still at risk at 0, 5, 10, 15 years after the first HIV test.
Discussion
This study highlights the potential importance of the HLA-DRB1*15-DQB1*06 haplotype in the control of HIV infection and raises the possibility that previous observations reporting a similar protective effect of the HLA-DRB1*13-DQB1*06 haplotype 17,25,26 may reflect inheritance of DQ6 rather than DR13 alleles. The protective effect afforded by the HLA-DRB1*13-DQB1*06 haplotype was noted to be weaker when cohorts that included subjects representing the extremes of HIV progression and non-progression were studied;25 as emphasized by a recent study that looked at major histocompatibility complex (MHC) ancestral haplotypes and disease progression using a pooled cohort from several sites.34 Our data support and extend these earlier studies.
The strength of the protective effect exerted by MHC class II molecules and the ensuing specific CD4 response is likely to be governed by additional host factors. Thus the potential protective effect of DQB1*06 alleles was more apparent in the categorical analysis of the subgroup who included the extreme of non-progression at 14+ years, than in the survival analysis of all patients in the cohort. This is consistent with data from Malhotra et al.27 who showed that although patients with and without the DRB1*13-DQB1*06 haplotype had similar virus set-point after acquisition of HIV infection, patients with the haplotype had an improved ability to suppress viral replication over time in response to early treatment in the acute phase of infection. Taken together with our data, these results lead us to speculate that sustained levels of HIV-specific CD4 T-cell helper responses in patients inheriting the DRB1*15-DQB1*06 alleles may control small bursts of HIV-1 replication, and this may occur either through release of antiviral cytokines, and/or maintenance of help for cytotoxic responses. This is consistent with the general paradigm that CD4 T-cell help is critical in controlling virus infections, including HIV-1.5–9
It was important to examine whether the protective effect of the DR15/DQ6 haplotype could be mediated by the presence of an aspartate residue at position 57 on the DQβ chain. Although HLA-DQ6 appears to confer the strongest protection from autoimmune diabetes, other DQβ57Asp-encoding alleles also exert varying degrees of protection.32 Our survival analysis suggests that this particular structural feature may not, per se, confer protection. Rather, our data suggest that DQB1*06 alleles are protective. The protective effect of DQB1*06 alleles, when considered irrespective of their HLA-DR haplotype, was most evident in our categorical analysis of patients at 14+ years. Larger cohorts of patients with similar characteristics will be required to establish with confidence whether HLA-DQ6 haplotypes alone confer protection, or whether this effect requires particular structural features or extended haplotypes. The identification of HIV-encoded peptide epitopes that are selectively presented by HLA-DQ, but not by -DR molecules35 will facilitate functional studies that will help resolve this issue.
HLA molecule-associated delay in HIV-1 disease progression might operate through selective presentation of epitopes found more frequently in diverse HIV-1 proteins. This has been noted for the two most prominent protective HLA class I alleles, B*27 and B*57, that have been found to promote immunodominant CTL responses to conserved HIV-1 epitopes.36–38 The same HLA alleles were over-represented in canary pox HIV vaccine recipients with repeatedly detected CTL responses to certain viral proteins.39 Whether the same phenomenon would apply to MHC class II restricted responses is presently not known. It is known, however, that subjects with DRB1*13 alleles have a better prognosis in the context of other infections,40,41 including viruses.41,42 The pursuit of further comparable population studies combined with the demonstration of the functional significance of the putative markers will help refine and clarify the findings described in this report.
Acknowledgments
This work was supported by a grant from the Guy's, King's and St Thomas's Special Trustees Grant number: R990531.
References
- 1.Rowland-Jones S, Pinheiro S, Kaul R. New insights into host factors in HIV-1 pathogenesis. Cell. 2001;104:473–6. doi: 10.1016/s0092-8674(01)00235-5. [DOI] [PubMed] [Google Scholar]
- 2.Hogan C, Hammer S. Host determinants in HIV infection and disease. Ann Intern Med. 2001;134:761. doi: 10.7326/0003-4819-134-9_part_1-200105010-00013. [DOI] [PubMed] [Google Scholar]
- 3.Schmitz J, Kuroda M, Santra S, et al. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;283:857–60. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 4.Jin BX, Bauer DE, Tuttleton SE, et al. Dramatic rise in plasma viremia after CD8 T cell depletion in simian immunodeficiency virus–infected macaques. J Exp Med. 1999;189:991–8. doi: 10.1084/jem.189.6.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalams SA, Walker BD. The critical need for CD4 help in maintaining effective cytotoxic T lymphocyte responses. J Exp Med. 1998;188:2199. doi: 10.1084/jem.188.12.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altfeld M, Rosenberg ES. The role of CD4+ T-helper cells in the cytotoxic T-lymphocyte response to HIV-1. Current Opinion Immunol. 2000;12:375. doi: 10.1016/s0952-7915(00)00103-5. [DOI] [PubMed] [Google Scholar]
- 7.Boaz M, Waters A, Murad S, Easterbrook P, Vyakarnam A. Presence of HIV-1 Gag-specific IFN-γ+ IL2+ and CD28+ IL2+ CD4 T-cell responses is associated with non-progression in HIV-1 infection. J Immunol. 2002;169:6376–85. doi: 10.4049/jimmunol.169.11.6376. [DOI] [PubMed] [Google Scholar]
- 8.Kalams SA, Buchbinder SP, Rosenberg ES, et al. Association between virus specific cytotoxic T-lymphocyte and helper responses in human immunodeficiency virus type 1 infection. J Virol. 1999;73:6715–20. doi: 10.1128/jvi.73.8.6715-6720.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenberg E, Altfeld M, Poon S, Phillips M, et al. Immune control of HIV-1 after early treatment of acute infection. Nature. 2000;407:523–6. doi: 10.1038/35035103. [DOI] [PubMed] [Google Scholar]
- 10.Migueles S, Laborico A, Shupert W, et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nature Immunol. 2002;3:1061–9. doi: 10.1038/ni845. [DOI] [PubMed] [Google Scholar]
- 11.Rosenberg E, Billingsley J, Caliendo A, Boswell S, Sax P, Kalams S, Walker B. Vigorous HIV-1 specific CD4+ T-cell responses associated with control of viremia. Science. 1997;278:1447–50. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- 12.Roger M. Influence of host genes on HIV-1 disease progression. FASEB J. 1998;12:625. doi: 10.1096/fasebj.12.9.625. [DOI] [PubMed] [Google Scholar]
- 13.Al Jabri AA. HLA and in vitro susceptibility to HIV infection. Mol Immunol. 2002;38:959. doi: 10.1016/s0161-5890(02)00023-8. [DOI] [PubMed] [Google Scholar]
- 14.Keet IP, Klein MR, Just JJ, Kaslow RA. The role of host genetics in the natural history of HIV-1 infection: the needles in the haystack. AIDS. 1996;10(Suppl. A):S59–67. doi: 10.1097/00002030-199601001-00009. [DOI] [PubMed] [Google Scholar]
- 15.Rowland-Jones S. Long-term non-progression in HIV infection: clinico pathological issues. J Infect. 1999;38(2):67–70. doi: 10.1016/s0163-4453(99)90070-1. [DOI] [PubMed] [Google Scholar]
- 16.Kaslow RA, McNicholl JM. Genetic determinants of HIV-1 infection and its manifestations. Proc Assoc Am Physicians. 1999;111(4):299–307. doi: 10.1046/j.1525-1381.1999.99238.x. [DOI] [PubMed] [Google Scholar]
- 17.Hendel H, Caillat-Zucman S, Lebuanec H, et al. New class I and II HLA alleles strongly associated with opposite patterns of progression to AIDS. J Immunol. 1999;162:6942–6. [PubMed] [Google Scholar]
- 18.Kaslow RA, Carrington M, Apple R, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2(4):405–11. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 19.Migueles SA, Sabbaghian MS, Shupert WL, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci USA. 2000;97(6):2709–14. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Costello C, Tang J, Rivers C, Karita E, Meizen-Derr J, Allen S, Kaslow RA. HLA-B*5703 independently associated with slower HIV-1 disease progression in Rwandan women. AIDS. 1999;13(14):1990–1. doi: 10.1097/00002030-199910010-00031. [DOI] [PubMed] [Google Scholar]
- 21.Magierowska M, Theodorou I, Debre P, Sanson F, Autran B, Riviere Y, Charron D, Costagliola D. Combined genotypes of CCR5, CCR2, SDF1, and HLA genes can predict the long-term nonprogressor status in human immunodeficiency virus-1-infected individuals. Blood. 1999;93(3):936–41. [PubMed] [Google Scholar]
- 22.Flores-Villanueva PO, Yunis EJ, Delgado JC, et al. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc Natl Acad Sci U S A. 2001;98(9):5140–5. doi: 10.1073/pnas.071548198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Steel CM, Ludlam CA, Beatson D, Peutherer JF, Cuthbert RJ, Simmonds P, Morrison H, Jones M. HLA haplotype A1, B8 DR3 as a risk factor for HIV-related disease. Lancet. 1998;1(8596):1185–8. doi: 10.1016/s0140-6736(88)92009-0. [DOI] [PubMed] [Google Scholar]
- 24.Carrington M, Nelson GW, Martin MP, et al. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283(5408):1748–52. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 25.Keet PM, Tang J, Klein MR, et al. Consistent associations of HLA class I and II and transporter gene products with progression of human immunodeficiency virus type 1 infection in homosexual men. J Infect Dis. 1999;180:299–309. doi: 10.1086/314862. [DOI] [PubMed] [Google Scholar]
- 26.Chen Y, Winchester R, Korber B, et al. Influence of HLA alleles on the rate of progression of vertically transmitted HIV infection in children: association of several HLA-DR13 alleles with long-term survivorship and the potential association of HLA-A*2301 with rapid progression to AIDS. Long-Term Survivor Study. Hum Immunol. 1997;55(2):154–62. doi: 10.1016/s0198-8859(97)00092-x. [DOI] [PubMed] [Google Scholar]
- 27.Malhotra U, Holte S, Dutta S, et al. Role for HLA class II molecules in HIV-1 suppression and cellular immunity following antiretroviral treatment. J Clin Invest. 2001;107(4):505–17. doi: 10.1172/JCI11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kwok WW, Domeier ME, Johnson ML, Nepom GT, Koelle DM. HLA-DQB1 codon 57 is critical for peptide binding and recognition. J Exp Med. 1996;183:1253–8. doi: 10.1084/jem.183.3.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee KH, Wucherpfennig KW, Wiley DC. Structure of a human insulin peptide–HLA-DQ8 complex and susceptibility to type 1 diabetes. Nat Immunol. 2001;2:501–7. doi: 10.1038/88694. [DOI] [PubMed] [Google Scholar]
- 30.Easterbrook PJ, Rostron T, Ives N, Troop M, Gazzard BG. Chemokine receptor polymorphisms and HIV disease progression. J Infect Dis. 1999;180:1096–105. doi: 10.1086/314997. [DOI] [PubMed] [Google Scholar]
- 31.Voorter CE, Rozemuller EH, de Bruyn-Geraets D, van der Zwan AW, Tilanus MG, van den Berg-Loonen EM. Comparison of DRB sequence-based typing using different strategies. Tissue Antigens. 1997;49(5):471–6. doi: 10.1111/j.1399-0039.1997.tb02781.x. [DOI] [PubMed] [Google Scholar]
- 32.Todd JA, Acha-Orbea H, Bell JI, et al. A molecular basis for MHC class II-associated autoimmunity. Science. 1988;240:1003–9. doi: 10.1126/science.3368786. [DOI] [PubMed] [Google Scholar]
- 33.Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SG. IMGT/HLA and IMGT/MHC. sequence databases for the study of the major histocompatibility complex. Nucl Acids Res. 2003;31:311–4. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Flores-Villanueva PO, Hendel H, Caillat-Zucman S, et al. Associations of MHC ancestral haplotypes with resistance/susceptibility to AIDS disease development. J Immunol. 2003;170:1925–9. doi: 10.4049/jimmunol.170.4.1925. [DOI] [PubMed] [Google Scholar]
- 35.Pancre V, Georges B, Angyalosi G, et al. Novel promiscuous HLA-DQ HIV Nef peptide that induces IFN-gamma-producing memory CD4+ T cells. Clin Exp Immunol. 2002;129(3):429–37. doi: 10.1046/j.1365-2249.2002.01934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gillespie GM, Kaul R, Dong T, et al. Cross-reactive cytotoxic T lymphocytes against a HIV-1 p24 epitope in slow progressors with B*57. AIDS. 2002;16(7):961–72. doi: 10.1097/00002030-200205030-00002. [DOI] [PubMed] [Google Scholar]
- 37.Goulder PJ, Bunce M, Krausa P, et al. Novel, cross-restricted, conserved, and immunodominant cytotoxic T lymphocyte epitopes in slow progressors in HIV type 1 infection. AIDS Res Human Retroviruses. 1996;12(18):1691–8. doi: 10.1089/aid.1996.12.1691. [DOI] [PubMed] [Google Scholar]
- 38.Migueles SA, Sabbaghian MS, Shupert WL, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A. 2000;97(6):2709–14. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaslow RA, Rivers C, Tang J, Bender TJ, Goepfert PA, El Habib R, Weinhold K, Mulligan MJ. NIAID AIDS vaccine evaluation group. Polymorphisms in HLA class I genes associated with both favorable prognosis of human immunodeficiency virus (HIV) type 1 infection and positive cytotoxic T-lymphocyte responses to ALVAC-HIV recombinant canarypox vaccines. J Virol. 2001;75(18):8681–9. doi: 10.1128/JVI.75.18.8681-8689.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thursz MR, Kwiatkowski D, Allsopp CEM, Greenwood BM, Thomas HC, Hill AVS. Association between an MHC class II allele and clearance of hepatitis B virus in the Gambia. N Engl J Med. 1995;332:1065–9. doi: 10.1056/NEJM199504203321604. [DOI] [PubMed] [Google Scholar]
- 41.Apple RJ, Erlich HA, Klitz W, Manos MM, Becker TM, Wheeler CM. HLA DR–DQ associations with cervical carcinoma show papillomavirus-type specificity. Nat Genet. 1994;6:157–62. doi: 10.1038/ng0294-157. [DOI] [PubMed] [Google Scholar]
- 42.Hill AV, Allsopp CE, Kwiatkowski D, et al. Common west African HLA antigens are associated with protection from severe malaria. Nature. 1991;352:595–600. doi: 10.1038/352595a0. [DOI] [PubMed] [Google Scholar]