Abstract
Interleukin-13 (IL-13) is critical for the development of allergic asthma and is involved in the activation of eosinophils within the airways. IL-13 exerts its activity on target cells via the dimeric IL-13 receptor (IL-13R), which comprises the IL-13 receptor α1-chain (IL-13Rα1) as a specific component. The aim of this study was to investigate the expression of the IL-13Rα1-chain on primary human eosinophilic granulocytes. Furthermore, it addresses the regulatory influence of cytokines on the level of surface abundance of this receptor subunit. Expression of IL-13- and IL-4-receptor subunits in purified primary human eosinophils was monitored at the messenger RNA level by reverse transcription polymerase chain reaction and at the protein level by flow cytometry. For the analysis of IL-13Rα1 surface expression, a new monoclonal antibody, which was generated using genetic immunization, was employed. Different cytokines with established activity on eosinophils were studied with regard to their influence on IL-13Rα1 in vitro by flow cytometry. Whereas IL-13 and IL-4 had inhibitory effects on IL-13Rα1 expression on eosinophils, interferon-γ, tumour necrosis factor-α, and, to the largest extent, transforming growth factor-β, enhanced the expression of this receptor subunit. A positive regulatory response evoked by transforming growth factor-β and interferon-γ does not prevent inhibitory effects caused by IL-13. These findings suggest a regulatory cytokine network influencing the reactivity of eosinophils to IL-13.
Keywords: asthma, IL-13 receptor, surface expression, transforming growth factor-β
Introduction
Human interleukin-13 (IL-13) is a pleiotropic cytokine with a molecular weight of 12 000 which shows homology to IL-4.1–3 The human IL-13 gene is located on chromosome 5q31 in a cluster of genes encoding IL-3, IL-4, IL-5, IL-9 and granulocyte–macrophage colony-stimulating factor (GM-CSF). This chromosomal region has been linked with asthma.4 IL-13 is produced by T helper type 2 (Th2) lymphocytes and by a variety of other cell types, such as mast cells, basophils, eosinophils, dendritic cells, Th1 CD4+ T lymphocytes, Th0- and CD8+ T lymphocytes.5
IL-13 exhibits a broad range of activities in the regulation of inflammatory and immune responses, such as the induction of immunoglobulin class switching towards immunoglobulin E (IgE) in B cells and the activation of eosinophils within the airways. It was also found to be of critical importance for the development of allergic asthma.6,7 Allergic asthma is characterized by airway hyperresponsiveness, elevated serum IgE levels, mucus hypersecretion, subepithelial fibrosis and chronic pulmonary eosinophilia.8
The influence of IL-13 on eosinophils plays a pivotal role in asthma through regulation of recruitment, homing and activation. IL-13 is able to activate eosinophils and to prolong eosinophil survival.9 It also up-regulates vascular cell adhesion molecule 1 expression which leads to elevated eosinophil migration into inflamed lung tissue.10 Overexpression of IL-13 in the lungs of mice causes enhanced expression of IL-5 and eotaxin, two mediators strongly involved in lung eosinophilia.11,12 Interestingly, eosinophils are able to produce functional IL-13 in response to GM-CSF or IL-5.13
IL-13 mediates its effects via the IL-13 receptor (IL-13R). This receptor is a heterodimer composed of IL-13R α-chain 1 (IL-13Rα1) and IL-4R α-chain (IL-4Rα).14,15 IL-13Rα1 has a weak IL-13 binding capacity on its own but shows substantial IL-13 binding upon complexing with the IL-4Rα-chain.15,16 As a result of the heterodimeric structure (IL-13Rα1/IL-4Rα) of the IL-13R it can also be utilized by IL-4 which explains overlapping activities of IL-13 and IL-4.17
As a second IL-13 binding protein, the high-affinity IL-13R α-chain 2 (IL-13Rα2) has been described.18 Because of its short cytoplasmic tail IL-13Rα2 appears to have no signalling functions.18,19 It is supposed to act as a decoy receptor and might play a role in the regulation of IL-13 signal transduction. There is also evidence for a soluble form of IL-13Rα2.20
IL-13Rs are expressed on many different haematopoietic and non-haematopoietic cells.5 Within the airways, IL-13Rs are found on ciliated respiratory epithelial cells, smooth muscle cells in bronchial tissue, and submucosal glands in nasal mucosa. On eosinophils, the IL-13R has as yet been demonstrated only indirectly by functional studies.9,21
Recently, it has been shown that cytokine receptors involved in eosinophil functions are regulated by cytokines. IL-3, IL-5 and GM-CSF were found to reduce IL-5Rα expression and to increase IL-3Rα expression on eosinophils.22,23 There is also evidence that cytokines are involved in IL-13R regulation as detected by mRNA regulation studies in airway epithelial cells and macrophages of mice,24 but the mechanisms regulating IL-13R expression on eosinophils are still unknown. Various cytokines such as IL-3, IL-5, IL-13, interferon-γ (IFN-γ) and GM-CSF influence eosinophils, as determined by the expression of the eosinophil activation marker CD69.9,25 IFN-γ, as well as tumour necrosis factor-α (TNF-α), are able to up-regulate cell surface expression of CD95 on eosinophils, which leads to an increased susceptibility of eosinophils to CD95-mediated apoptosis.26 IL-4 has been reported to inhibit eosinophil survival and to induce apoptosis.27
These findings, and the role of eosinophilia in the asthmatic phenotype, prompted us to investigate the influence of cytokines on the expression of the IL-13R on eosinophils. We show for the first time that cytokines involved in eosinophil activation and lung inflammation alone and in combination exert positive and negative effects on the abundance of the IL-13R on eosinophils.
Materials and methods
Cytokines and antibodies
For eosinophil stimulation IL-3, IL-4, IL-5, IL-13, GM-CSF, IFN-γ (all from CellConcepts, Umkirch, Germany) and TNF-α (PBH, Hannover, Germany) at concentrations of 10 ng/ml and transforming growth factor-β1 (TGF-β1; CellConcepts) at concentrations of 1 ng/ml were used either alone or in combination.
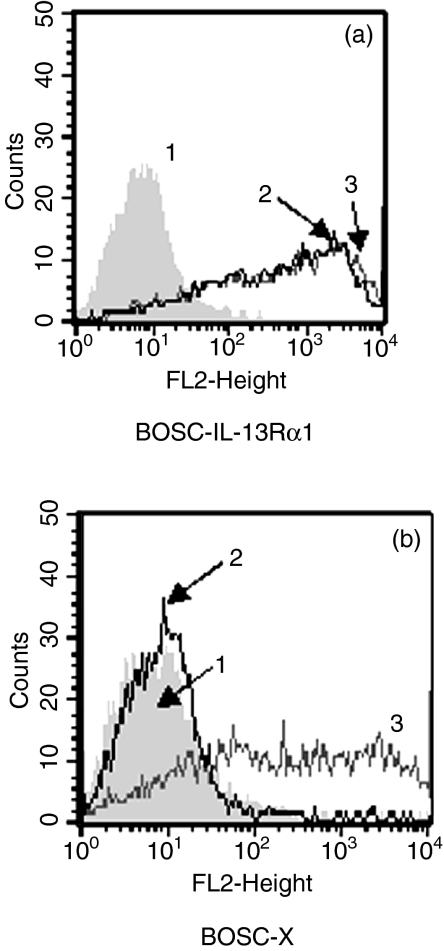
Using a cDNA of the IL-13Rα1 extracellular domain, the antibody AT-3D3 was generated using GENOVAC's technology, which enables the generation of antibodies that recognize the native protein (GENOVAC AG, Freiburg, Germany).28–30 Specificity testing of AT-3D3 was performed using a cell enzyme-linked immunosorbent assay. BOSC cells were transiently transfected with an expression vector encoding either the extracellular domain of IL-13Rα1 (BOSC-IL-13Rα1) or an irrelevant protein (BOSC-X). For specificity testing, IL-13Rα1-AT-3D3 supernatant was tested on both the IL-13Rα1 and the irrelevant protein transfectants (Fig. 1). A positive signal was only obtained with IL-13Rα1-transfected cells (curve 2). Furthermore, as described elsewhere (Krause et al. submitted for publication), AT-3D3 hybridoma culture supernatant was also tested for specificity by determining antibody reactivity to monocytes versus CD4 lymphocytes cytometrically and by receptor-binding studies on murine cells transfected with a human IL-13Rα1 derivative (Krause et al. submitted for publication). AT-3D3 hybridoma culture supernatant was applied for cytometric analysis of IL-13R expression on eosinophils. Mouse IgG1 and anti-mouse-fluorescein isothiocyanate (FITC) antibodies were obtained from Dako A/S (Hamburg, Germany).
Figure 1.
Specificity testing of AT-3D3 hybridoma culture supernatant. BOSC cells were transiently transfected with an expression vector encoding either (a) the extracellular domain of IL-13Rα1 (BOSC-IL-13Rα1) or (b) an irrelevant protein (BOSC-X). AT-3D3 was tested on both transfectants by flow cytometry (curve 2). As a control, cells were stained with negative (curve 1) and positive (curve 3) control antibodies.
Purification of eosinophils and mononuclear cells
Eosinophils were isolated from anti-coagulated (ethylenediaminetetraacetic acid) peripheral blood samples with magnetic CD16 microbeads (Miltenyi Biotech, Bergisch-Gladbach, Germany) as described before.31 Mononuclear cells were obtained during eosinophil isolation. Each experiment was performed 15 times, unless otherwise stated, with cells obtained from different healthy donors. Cells were counted using Kimura staining.32 The purity of eosinophils was consistently > 99%.
Cell culture and stimulation
Eosinophils (5 × 105 cells/ml) were cultured in medium (RPMI-1640; Gibco, Paisley, UK) containing 10% heat-inactivated fetal calf serum (Gibco, NY), 100 U/ml penicillin and 100 μg/ml streptomycin (Biochrom, Berlin, Germany), at 37° in a humidified CO2 (5%) atmosphere. For stimulation, different cytokines (either alone or in combination) were added and eosinophils were cultured for 20 hr. To investigate the influence of IL-13 on prestimulated eosinophils IL-13 was added after TGF-β/IFN-γ incubation for 20 hr followed by another 6-hr incubation.
RNA isolation and reverse transcription and polymerase chain reaction (RT-PCR) amplification
Total cellular RNA was extracted from 1 × 106 cells following lysis with PeqGold RNA Pure (Peqlab, Erlangen, Germany). The upper phase was transferred and the RNA was precipitated with isopropanol. Reverse transcription was performed starting from 5 μg RNA per sample using a Stratagene-Kit (Stratagene, La Jolla, CA). After heating up to 90°, distilled water was added to a final volume of 50 μl and the cDNA preparation was then stored at − 20°. PCR amplification was performed in a total volume of 25 μl (5 μl cDNA, 0·02 U/μl Taq polymerase, 1 μm primer mix, 50 μm dNTP) and overlaid with 20 μl mineral oil (Sigma, Deisenhofen, Germany). Amplification was performed over 35 cycles for IL-13Rα1 and common γ-chain and over 40 cycles for IL-13Rα2 and IL-4Rα. The annealing temperature was 60°, extension occurred at 72° and denaturation at 94°. One-fifth of the PCR product was separated by flat-bed electrophoresis in 1·6% agarose gels (USB, Cleveland, OH) and detected by ethidium bromide (USB) staining.
PCR products were quantified following agarose gel electrophoresis employing aida imaging software (Raytest, Straubenhardt, Germany). The average densitometry signals of five molecular weight marker bands, minus background, were calculated. The signals from PCR fragments were then normalized against average standard signals from the respective gels (arbitrarily set at 1) and expressed as ‘relative band intensities’.
The following primers were used: the IL-4Rα-primer: specific F-primer, 5′-TACTTGCGAGTGGAAGATGA-3′; specific R-primer, 5′-AGGGAGGGTTCTAGGTAGGT-3′; the common γ-chain primer: specific F-primer, 5′-CGCCATGTTGAAGCCATCAT-3′; specific R-primer, 5′-TCTGTGTGGCCTGTCTCCTG-3′; the IL-13Rα1-primer: specific F-primer, 5′-CTCCTTCCACAATGATGACC-3′; specific R-primer, 5′-GGAATTGCGCTTCTTACCTA-3′; the IL-13Rα2-primer: specific F-primer, 5′-GCTTGGCTATCGGATGCTTA-3′; specific R-primer, 5′-TTTCTGCCCAGGAACTTTGA-3′.
Flow cytometry
To analyse IL-13R expression, eosinophils were incubated with 10 μl unlabelled IL-13R antibody (AT-3D3 hybridoma supernatant) or mouse IgG1 antibody. Antibody detection was reached by incubation with FITC-conjugated secondary antibodies (anti-mouse-FITC). Flow cytometry was performed using FACSCalibur (Becton Dickinson, Heidelberg, Germany). Fluorescence intensity was measured on at least 1000 cells from each sample. Eosinophils were gated out on the basis of their forward and side scatter to exclude cell debris. Monocytes were defined by gating on the monocyte region of mononuclear cells. Dead cells were excluded by gating out propidium iodide (Sigma, Deisenhofen, Germany) -positive cells during assessment of IL-13R expression. Therefore, only viable cells were included in this study. Fluorescence data were obtained by determining the total fluorescence x (the fluorescence caused by the marked antibodies) and the nonspecific fluorescence y (the fluorescence caused by the isotype control). The specific mean fluorescence (SMF) was then calculated as SMF = x − y.
The indicated relative specific fluorescence (RSF) was derived from SMF × 100/y.
Statistics
Experiments were performed at least 15 times (unless otherwise stated) using cells from different healthy donors. Results were expressed as mean RSF ± SEM and analysed using the paired t-test. Results were considered statistically significant at a P-value < 0·05 (*) or highly significant at a P-value < 0·001 (**).
Results
Expression of IL-13Rα on eosinophils is much lower than on monocytes
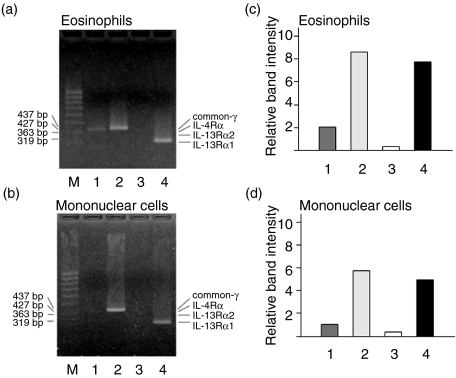
To obtain information about the expression of IL-13Rα1 on eosinophils, the abundance of its mRNA was investigated by RT-PCR using specific DNA primers. For comparison, the expressions of IL-4Rα, the common γ-chain and IL-13Rα2 were also determined (Fig. 2). IL-13Rα1 mRNA was expressed in eosinophils as well as in mononuclear cells. Both cell types also showed common γ-chain mRNA expression. IL-4Rα mRNA was expressed at low levels on both cell types and could only be detected by employment of increased DNA template concentrations or additional PCR cycles. IL-13Rα2 was not detectable in either cell type.
Figure 2.
Messenger RNA expression of IL-4R and IL-13R subunits on eosinophilic granulocytes (a and c) and mononuclear cells (b and d). Cells were separated from whole blood and subjected to RT-PCR amplification and agarose gel electrophoresis using specific primers for the IL-4Rα-chain (1), common γ-chain (2), IL-13Rα2-chain (3) and IL-13Rα1-chain (4). A molecular weight standard is shown in lane M in (a) and (b). Molecular weights in base pairs (bp) and positions of the specific products are indicated by arrows. Graphs in (c) and (d) show a densitometric relative quantitation of PCR products. ‘Relative band intensities’ express respective signal strengths in comparison with the average of signals from molecular weight standard bands.
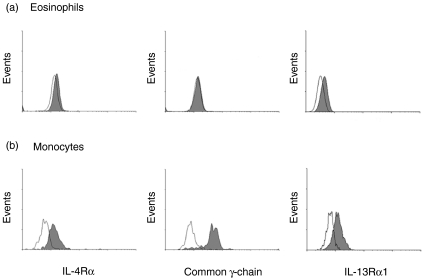
Analysis of low level IL-13R surface expression has as yet been hampered by the lack of antibodies with good specificity and affinity. Genetic immunization was used to develop a novel monoclonal antibody to the IL-13Rα1-chain (AT-3D3) to assess IL-13R surface expression on eosinophils and monocytes in a quantitative fashion by flow cytometry. In parallel, the expression of IL-4Rα and common γ-chain was measured (Fig. 3). IL-4Rα expression was weak on eosinophils. A strong IL-4Rα-chain expression was detectable on monocytes. Common γ-chain was clearly detectable on monocytes, whereas eosinophils showed only a marginal degree of common γ-chain expression. Eosinophils showed a weak, but unambiguous, IL-13R expression (18·99 ± 2·10; n = 15) which was, however, much lower compared to monocytes (116·56 ± 21·09; n = 7).
Figure 3.
Flow cytometric analysis of cell surface expression of IL-4Rα-chain, common γ-chain and IL-13Rα1-chain. Eosinophils (a) and monocytes (b) were separated from whole blood and stained with antibodies specific for either IL-4Rα-chain, common γ-chain or IL-13Rα1-chain and fluorescent secondary antibodies as indicated (filled curves). Staining with an unrelated primary antibody served as a negative control (open curves).
IL-13R expression on eosinophils is decreased by IL-13 and increased by TGF-β
Having shown IL-13Rα1 surface expression on eosinophils, we were interested in its possible regulation by external stimuli. To analyse the possible effects of cytokines, isolated peripheral blood eosinophils from human donors were incubated in medium alone or stimulated with cytokines for 20 hr (Fig. 4).
Figure 4.
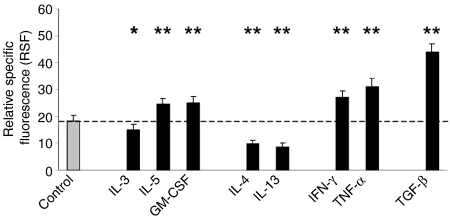
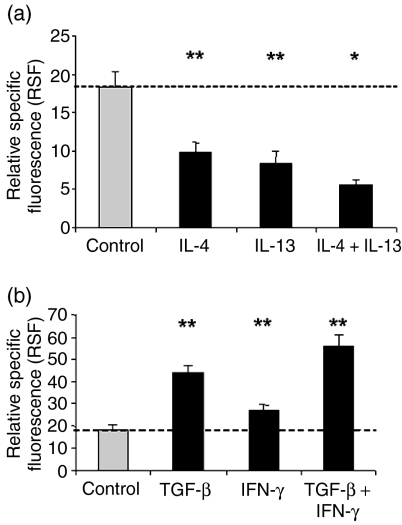
Cytometric analysis of IL-13Rα1 expression after 20 hr of incubation without stimulus or incubation with the indicated cytokines at the respective concentrations given in the methods (n = 15). Cells were stained with antibody AT-3D3 directed against the IL-13Rα1-chain. Data are expressed as relative specific fluorescence (RSF ± SEM). Asterisks indicate statistical significance of P < 0·05 (*) or P < 0·001 (**) compared to unstimulated control.
First the effects of the cytokines IL-3, IL-5 and GM-CSF were tested; these are known as potent activators of eosinophil function. All these cytokines caused a slight but significant change in IL-13R expression compared to unstimulated control (18·35 ± 2·02). A down-regulatory effect was observed for IL-3 (14·99 ± 2·02, P = 0·006), whereas stimulation with IL-5 and GM-CSF, respectively, up-regulated receptor expression (IL-5: 24·45 ± 2·30, P < 0·001; GM-CSF: 25·14 ± 2·38, P < 0·001).
Then, it was investigated whether the expression of the IL-13Rα1-chain on eosinophils is influenced by its own ligands IL-13 and IL-4. Interestingly, both IL-4 (9·87 ± 1·22, P < 0·001) and IL-13 (8·42 ± 1·56, P < 0·001) caused a highly significant decrease of IL-13Rα1 surface expression.
In contrast, TNF-α and IFN-γ which have been shown to up-regulate CD95 on eosinophils caused a highly significant increase of IL-13R expression (TNF-α: 31·12 ± 3·09, P < 0·001; IFN-γ: 27·13 ± 2·36, P < 0·001).
TGF-β was also included in the cytokine panel since there is evidence that TGF-β can inhibit human leucocyte antigen-DR expression on eosinophils.33 Surprisingly, TGF-β caused a highly significant increase of IL-13R expression, in fact the strongest effect observed in this investigation (43·84 ± 3·35, P < 0·001).
Systematic variation of the incubation times with cytokines showed that the cytokine-dependent effects on IL-13Rα1 expression were maximal within a period of 16–24 hr (data not shown).
Costimulatory effects of IL-13 and TGF-β
Both inhibitory and stimulatory effects on eosinophils by different cytokines were observed. It has to be assumed, however, that in vivo, eosinophils are not influenced by single cytokines but rather by their combination. We therefore analysed several combinations which might enhance the effects observed by individual stimuli. IL-13 and TGF-β, which had shown the strongest inhibitory or stimulatory effects, respectively, on IL-13Rα1 expression were therefore combined with cytokines that also showed tendencies in the same direction of regulation and the expression levels were again measured after 20 hr of incubation by flow cytometry.
Costimulation of eosinophils with IL-13 and IL-4 further reduced IL-13R expression (5·62 ± 0·61; n = 8) (Fig. 5a). Interestingly, the stimulatory effect of TGF-β (43·84 ± 3·35) on IL-13R expression was further enhanced in the presence of IFN-γ (56·09 ± 4·65, P < 0·001) (Fig. 5b). Co-incubation of eosinophils with this combination plus additional cytokines did not evoke any further additive effects (data not shown).
Figure 5.
Cytometric analysis of IL-13Rα1 expression on eosinophilic granulocytes in response to individual (n = 15) or simultaneous stimulation with inhibitory (IL-4 and IL-13; n = 8) (a) and stimulatory (TGF-β and IFN-γ; n = 15) (b) cytokines. Cells were treated with medium alone (control) or with the indicated individual or combined cytokines for 20 hr and stained with antibody AT-3D3. Data are expressed as relative specific fluorescence (RSF ± SEM). Asterisks indicate statistical significance of P < 0·05 (*) or P < 0·001 (**) compared to unstimulated control.
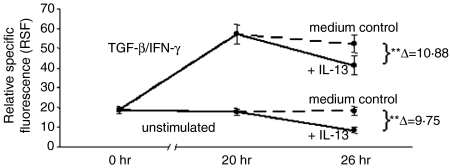
TGF-β/IFN-γ-induced up-regulation of IL-13R expression on eosinophils does not prevent IL-13R down-regulation by IL-13
Furthermore, the possible mutual effects of positive and negative regulators of IL-13R expression on eosinophils were investigated. To this end, cells were either left untreated or prestimulated with TGF-β/IFN-γ to obtain maximum receptor expression (compare Fig. 5b). After a 20-hr period of incubation, IL-13 was added to both cell populations and receptor expression was again monitored 6 hr later. A highly significant down-regulation in receptor expression was observed (P < 0·001). However, the decrease was similar in both cases, irrespective of prior TGF-β/IFN-γ stimulation (untreated cells: Δ = 9·75 versus prestimulated cells: Δ = 10·88; n = 12) (Fig. 6). This result shows that positive and negative mechanisms of cytokine-induced IL-13Rα1 expression regulation operate independently of each other.
Figure 6.
Influence of IL-13 on the cytokine-induced surface expression of IL-13Rα1. Eosinophils were pretreated for 20 hr with medium only or a combination of TGF-β/IFN-γ and then subjected to a subsequent 6-hr incubation with IL-13 (n = 12). Fluorescence was determined at the time-points indicated and expressed as in Figs 4 and 5. Asterisks indicate statistical significance of P < 0·001 (**).
Discussion
This study is the first to examine the expression of the IL-13Rα1 subunit on primary eosinophilic granulocytes directly. Furthermore, we studied the influence of different cytokines on the regulation of this receptor chain. The detection and quantification of the low baseline surface expression and stimulation-dependent alterations of this parameter were rendered feasible by the availability of a novel monoclonal antibody, which was generated by genetic immunization and allows the detection of native antigens.
Interestingly, we were able to show that cytokines which are potent activators of eosinophils, namely IL-3, IL-5 and GM-CSF, have only small effects on the surface expression of the IL-13Rα1. In contrast, IL-13 and IL-4 caused a strong reduction in IL-13Rα1 expression. The similar effects of these two cytokines are conceivable since the IL-13R and IL-4R share IL-4Rα as a common subunit.17
We provide evidence that IL-13 can down-regulate its own receptor. Down-regulation of cytokine receptors by their natural ligands has been widely reported and seems to be an important mechanism in the regulation and maintenance of eosinophil homeostasis. There are several possibilities how receptor down-regulation might occur, such as receptor endocytosis after ligand binding, receptor release from the cell surface via proteolytic cleavage or transcriptional regulation. An example for receptor down-regulation on eosinophils through a shedding mechanism is the IL-5R. There is strong evidence that a reduction of membrane IL-5Rα-chain after incubation with IL-5, at least in part, is caused by proteolytic cleavage by matrix metalloproteinases.34 A well-known example for the regulation of cytokine receptor expression by its own ligand on the level of transcription is the IL-2R on T cells. Its synthesis increases in response to IL-2 upon T-cell activation.35 Since in this study we observed a latency of 16–24 hr for the cytokine effects on IL-13R cell surface expression, a slow mechanism such as transcriptional control is most probably underlying this phenomenon.
We are also in the process of analysing cytokine-dependent expression regulation of the IL-4Rα subunit and have obtained evidence that it also becomes down-regulated in response to IL-4/IL-13 (D. Myrtek, unpublished results).
Comparing receptor expression data obtained by RT-PCR and flow cytometry revealed partly diverging results. Such differences are probably explicable by distinct relative stabilities of related mRNAs and receptor proteins. The greater explanatory power for the purpose of this study clearly lies in flow cytometry analyses, since this method determines the steady-state level of surface-presented functional receptor under different conditions.
It seems noteworthy that TGF-β showed the most pronounced stimulatory effect on IL-13Rα1 expression of all cytokines tested. TGF-β plays an important role in the regulation of chronic inflammation and fibrosis and it can reduce eosinophil viability.36–38 As IL-13 is able to activate eosinophils and prolong eosinophil survival via the IL-13R, our findings on TGF-β-induced IL-13R expression are unexpected at first sight. Obviously, TGF-β acts on eosinophils in a complex fashion. On the one hand it inhibits eosinophil activation as assessed by human leucocyte antigen-DR33 and CD69 expression (unpublished data), but on the other hand it possibly enables eosinophil activation indirectly via IL-13R expression. Possible explanations for this bimodal activity of TGF-β are dose-dependent cellular responses and an as yet not characterized inference with other molecules involved in signal transduction towards IL-13R expression. Evidence that TGF-β is able to exert distinct effects on cells in a dose-dependent fashion comes from studies on smooth muscle cells, fibroblasts and chondrocytes. At low concentrations of TGF-β it can stimulate proliferation whereas higher amounts are less stimulatory. This bimodal activity is apparently accomplished through modulation of the platelet-derived growth factor/platelet-derived growth factor receptor system, which in turn influences cell activation.39
Our study revealed that TGF-β elicited not only a stimulatory effect on its own, but further enhanced the expression of IL-13Rα1 when combined with IFN-γ. It is possible that one of these cytokines enhances the sensitivity for the other cytokine which then leads to the observed receptor increase. The role of IFN-γ in the development and maintenance of lung eosinophilia is poorly understood. Although animal studies revealed that IFN-γ can prevent the recruitment of eosinophils into the airways,40,41 IFN-γ is able to prolong eosinophil survival and to induce the expression of the eosinophil activation marker CD69.25,42 Our results suggest that IFN-γ as well as TGF-β may increase the sensitivity of eosinophils to IL-13 by up-regulation of the specific IL-13R.
Interestingly, cytokine-dependent up- and down-regulation of IL-13Rα1 expression, respectively, appear to be subject to independent triggers. Although initial up-regulation of IL-13R surface abundance by TGF-β/IFN-γ was reduced by IL-13, the degree of decrease was similar to that seen with unstimulated control cells.
Our study about interleukin-13Rα1-expression regulation suggests a scenario that multiple cytokines influence eosinophils towards IL-13 sensitivity through up- and down-regulation of the IL-13Rα1 subunit. We support the concept of IL-13 as the key factor of chronic asthmatic inflammation. Knowledge of this mechanism might help to understand the relevance of IL-13 within the cytokine network, and might help future therapeutical concepts against asthma.
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft (DFG LU623, FR854) and Bundesministerium für Wirtschaft und Technologie (KF 0238801KULO).
Abbreviations
- FITC
fluorescein isothiocyanate
- GM-CSF
granulocyte–macrophage colony-stimulating factor
- IFN-γ
interferon-γ
- IgG
immunoglobulin G
- IL
interleukin
- IL-3Rα
IL-3 receptor α-chain
- IL-4Rα
IL-4 receptor α-chain
- IL-5Rα
IL-5 receptor α-chain
- IL-13R
IL-13 receptor
- IL-13Rα1
IL-13 receptor α-chain 1
- IL-13Rα2
IL-13 receptor α-chain 2
- RSF
relative specific fluorescence
- SEM
standard error of mean
- SMF
specific mean fluorescence
- TGF-β
transforming growth factor-β
- TNF-α
tumour necrosis factor-α
References
- Minty A, Chalon P, Derocq JM, et al. Interleukin-13 is a new human lymphokine regulating inflammatory and immune responses. Nature. 1993;362:248–50. doi: 10.1038/362248a0. [DOI] [PubMed] [Google Scholar]
- Zurawski G, de Vries JE. Interleukin 13, an interleukin 4-like cytokine that acts on monocytes and B cells, but not on T cells. Immunol Today. 1994;15:19–26. doi: 10.1016/0167-5699(94)90021-3. [DOI] [PubMed] [Google Scholar]
- de Vries JE. Molecular and biological characteristics of interleukin-13. Chem Immunol. 1996;63:204–18. [PubMed] [Google Scholar]
- Marsh DG, Neely JD, Breazeale DR, et al. Linkage analysis of IL4 and other chromosome 5q31.1 markers and total serum immunoglobulin E concentrations. Science. 1994;264:1152–6. doi: 10.1126/science.8178175. [DOI] [PubMed] [Google Scholar]
- Hershey GK. IL-13 receptors and signaling pathways: an evolving web. J Allergy Clin Immunol. 2003;111:677–90. doi: 10.1067/mai.2003.1333. [DOI] [PubMed] [Google Scholar]
- Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, Donaldson DD. Interleukin-13: central mediator of allergic asthma. Science. 1998;282:2258–61. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- Grunig G, Warnock M, Wakil AE, et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282:2261–3. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills-Karp M, Chiaramonte M. Interleukin-13 in asthma. Curr Opin Pulm Med. 2003;9:21–7. doi: 10.1097/00063198-200301000-00004. [DOI] [PubMed] [Google Scholar]
- Luttmann W, Knoechel B, Foerster M, Matthys H, Virchow JC, Kroegel C., Jr Activation of human eosinophils by IL-13. Induction of CD69 surface antigen, its relationship to messenger RNA expression, and promotion of cellular viability. J Immunol. 1996;157:1678–83. [PubMed] [Google Scholar]
- Bochner BS, Klunk DA, Sterbinsky SA, Coffman RL, Schleimer RP. IL-13 selectively induces vascular cell adhesion molecule-1 expression in human endothelial cells. J Immunol. 1995;154:799–803. [PubMed] [Google Scholar]
- Zhu Z, Homer RJ, Wang Z, Chen Q, Geba GP, Wang J, Zhang Y, Elias JA. Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J Clin Invest. 1999;103:779–88. doi: 10.1172/JCI5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope SM, Brandt EB, Mishra A, Hogan SP, Zimmermann N, Matthaei KI, Foster PS, Rothenberg ME. IL-13 induces eosinophil recruitment into the lung by an IL-5- and eotaxin-dependent mechanism. J Allergy Clin Immunol. 2001;108:594–01. doi: 10.1067/mai.2001.118600. [DOI] [PubMed] [Google Scholar]
- Schmid-Grendelmeier P, Altznauer F, Fischer B, et al. Eosinophils express functional IL-13 in eosinophilic inflammatory diseases. J Immunol. 2002;169:1021–7. doi: 10.4049/jimmunol.169.2.1021. [DOI] [PubMed] [Google Scholar]
- Lin JX, Migone TS, Tsang M, et al. The role of shared receptor motifs and common Stat proteins in the generation of cytokine pleiotropy and redundancy by IL-2, IL-4, IL-7, IL-13, and IL-15. Immunity. 1995;2:331–9. doi: 10.1016/1074-7613(95)90141-8. [DOI] [PubMed] [Google Scholar]
- Aman MJ, Tayebi N, Obiri NI, Puri RK, Modi WS, Leonard WJ. cDNA cloning and characterization of the human interleukin 13 receptor alpha chain. J Biol Chem. 1996;271:29265–70. doi: 10.1074/jbc.271.46.29265. [DOI] [PubMed] [Google Scholar]
- Gauchat JF, Schlagenhauf E, Feng NP, et al. A novel 4-kb interleukin-13 receptor alpha mRNA expressed in human B, T, and endothelial cells encoding an alternate type-II interleukin-4/interleukin-13 receptor. Eur J Immunol. 1997;27:971–8. doi: 10.1002/eji.1830270425. [DOI] [PubMed] [Google Scholar]
- Callard RE, Matthews DJ, Hibbert L. IL-4 and IL-13 receptors: are they one and the same? Immunol Today. 1996;17:108–10. doi: 10.1016/0167-5699(96)80600-1. [DOI] [PubMed] [Google Scholar]
- Caput D, Laurent P, Kaghad M, Lelias JM, Lefort S, Vita N, Ferrara P. Cloning and characterization of a specific interleukin (IL) -13 binding protein structurally related to the IL-5 receptor alpha chain. J Biol Chem. 1996;271:16921–6. doi: 10.1074/jbc.271.28.16921. [DOI] [PubMed] [Google Scholar]
- Donaldson DD, Whitters MJ, Fitz LJ, et al. The murine IL-13 receptor alpha 2: molecular cloning, characterization, and comparison with murine IL-13 receptor alpha 1. J Immunol. 1998;161:2317–24. [PubMed] [Google Scholar]
- Zhang JG, Hilton DJ, Willson TA, et al. Identification, purification, and characterization of a soluble interleukin (IL)-13-binding protein. Evidence that it is distinct from the cloned IL-13 receptor and IL-4 receptor alpha-chains. J Biol Chem. 1997;272:9474–80. doi: 10.1074/jbc.272.14.9474. [DOI] [PubMed] [Google Scholar]
- Luttmann W, Matthiesen T, Matthys H, Virchow JC., Jr Synergistic effects of interleukin-4 or interleukin-13 and tumor necrosis factor-alpha on eosinophil activation in vitro. Am J Respir Cell Mol Biol. 1999;20:474–80. doi: 10.1165/ajrcmb.20.3.3326. [DOI] [PubMed] [Google Scholar]
- Hellman C, Hallden G, Hylander B, Lundahl J. Regulation of the interleukin-5 receptor alpha-subunit on peripheral blood eosinophils from healthy subjects. Clin Exp Immunol. 2003;131:75–81. doi: 10.1046/j.1365-2249.2003.02051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory B, Kirchem A, Phipps S, Gevaert P, Pridgeon C, Rankin SM, Robinson DS. Differential regulation of human eosinophil IL-3, IL-5, and GM-CSF receptor alpha-chain expression by cytokines. IL-3, IL-5, and GM-CSF down-regulate IL-5 receptor alpha expression with loss of IL-5 responsiveness, but up-regulate IL-3 receptor alpha expression. J Immunol. 2003;170:5359–66. doi: 10.4049/jimmunol.170.11.5359. [DOI] [PubMed] [Google Scholar]
- Zheng T, Zhu Z, Liu W, Lee CG, Chen Q, Homer RJ, Elias JA. Cytokine regulation of IL-13Ralpha2 and IL-13Ralpha1 in vivo and in vitro. J Allergy Clin Immunol. 2003;111:720–8. doi: 10.1067/mai.2003.1383. [DOI] [PubMed] [Google Scholar]
- Hartnell A, Robinson DS, Kay AB, Wardlaw AJ. CD69 is expressed by human eosinophils activated in vivo in asthma and in vitro by cytokines. Immunology. 1993;80:281–6. [PMC free article] [PubMed] [Google Scholar]
- Luttmann W, Opfer A, Dauer E, et al. Differential regulation of CD95 (Fas/APO-1) expression in human blood eosinophils. Eur J Immunol. 1998;28:2057–65. doi: 10.1002/(SICI)1521-4141(199807)28:07<2057::AID-IMMU2057>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Wedi B, Raap U, Lewrick H, Kapp A. IL-4-induced apoptosis in peripheral blood eosinophils. J Allergy Clin Immunol. 1998;102:1013–20. doi: 10.1016/s0091-6749(98)70340-9. [DOI] [PubMed] [Google Scholar]
- Tang DC, DeVit M, Johnston SA. Genetic immunization is a simple method for eliciting an immune response. Nature. 1992;356:152–4. doi: 10.1038/356152a0. [DOI] [PubMed] [Google Scholar]
- Thompson J, Lang S. Genetic immunization for antibody production. Tutorial: GENOVAC's technological shortcut from gene to antibody. Genet Eng News. 2002;22:62–3. [Google Scholar]
- Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, Felgner PL. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–8. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- Luttmann W, Dauer E, Schmidt S, Marx O, Hossfeld M, Matthys H, Virchow JC., Jr Effects of interferon-gamma and tumour necrosis factor-alpha on CD95/Fas ligand-mediated apoptosis in human blood eosinophils. Scand J Immunol. 2000;51:54–9. doi: 10.1046/j.1365-3083.2000.00645.x. [DOI] [PubMed] [Google Scholar]
- Kimura I, Moritani Y, Tanizaki Y. Basophils in bronchial asthma with reference to reagin-type allergy. Clin Allergy. 1973;3:195–02. doi: 10.1111/j.1365-2222.1973.tb01321.x. [DOI] [PubMed] [Google Scholar]
- Luttmann W, Franz P, Schmidt S, Barth J, Matthys H, Virchow JC., Jr Inhibition of HLA-DR expression on activated human blood eosinophils by transforming growth factor-beta1. Scand J Immunol. 1998;48:667–71. doi: 10.1046/j.1365-3083.1998.00446.x. [DOI] [PubMed] [Google Scholar]
- Liu LY, Sedgwick JB, Bates ME, et al. Decreased expression of membrane IL-5 receptor alpha on human eosinophils. II. IL-5 down-modulates its receptor via a proteinase-mediated process. J Immunol. 2002;169:6459–66. doi: 10.4049/jimmunol.169.11.6459. [DOI] [PubMed] [Google Scholar]
- Fraser JD, Irving BA, Crabtree GR, Weiss A. Regulation of interleukin-2 gene enhancer activity by the T cell accessory molecule CD28. Science. 1991;251:313–16. doi: 10.1126/science.1846244. [DOI] [PubMed] [Google Scholar]
- Kehrl JH, Wakefield LM, Roberts AB, Jakowlew S, Alvarez-Mon M, Derynck R, Sporn MB, Fauci AS. Production of transforming growth factor beta by human T lymphocytes and its potential role in the regulation of T cell growth. J Exp Med. 1986;163:1037–50. doi: 10.1084/jem.163.5.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl SM, Hunt DA, Wong HL, et al. Transforming growth factor-beta is a potent immunosuppressive agent that inhibits IL-1-dependent lymphocyte proliferation. J Immunol. 1988;140:3026–32. [PubMed] [Google Scholar]
- Alam R, Forsythe P, Stafford S, Fukuda Y. Transforming growth factor beta abrogates the effects of hematopoietins on eosinophils and induces their apoptosis. J Exp Med. 1994;179:1041–5. doi: 10.1084/jem.179.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battegay EJ, Raines EW, Seifert RA, Bowen-Pope DF, Ross R. TGF-beta induces bimodal proliferation of connective tissue cells via complex control of an autocrine PDGF loop. Cell. 1990;63:515–24. doi: 10.1016/0092-8674(90)90448-n. [DOI] [PubMed] [Google Scholar]
- Iwamoto I, Nakajima H, Endo H, Yoshida S. Interferon gamma regulates antigen-induced eosinophil recruitment into the mouse airways by inhibiting the infiltration of CD4+ T cells. J Exp Med. 1993;177:573–6. doi: 10.1084/jem.177.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima H, Iwamoto I, Yoshida S. Aerosolized recombinant interferon-gamma prevents antigen-induced eosinophil recruitment in mouse trachea. Am Rev Respir Dis. 1993;148:1102–4. doi: 10.1164/ajrccm/148.4_Pt_1.1102. [DOI] [PubMed] [Google Scholar]
- Valerius T, Repp R, Kalden JR, Platzer E. Effects of IFN on human eosinophils in comparison with other cytokines. A novel class of eosinophil activators with delayed onset of action. J Immunol. 1990;145:2950–8. [PubMed] [Google Scholar]