Abstract
Mycoplasmas are known to enhance human immunodeficiency virus (HIV) replication, and mycoplasma-derived lipid extracts have been reported to activate nuclear factor-κB (NF-κB) through Toll-like receptors (TLRs). In this study, we examined the involvement of TLRs in the activation of HIV long-terminal repeats (LTR) by mycoplasma and their active components responsible for the TLR activation. Lipid-associated membrane proteins (LAMPs) from two species of mycoplasma (Mycoplasma fermentans and M. penetrans) that are associated with acquired immune-deficiency syndrome (AIDS), were found to activate HIV LTRs in a human monocytic cell line, THP-1. NF-κB deletion from the LTR resulted in inhibition of the activation. The LTR activation by M. fermentans LAMPs was inhibited by a dominant negative (DN) construct of TLR1 and TLR6, whereas HIV LTR activation by M. penetrans LAMPs was inhibited by DN TLR1, but not by DN TLR6. These results indicate that the activation of HIV LTRs by M. fermentans and M. penetrans LAMPs is dependent on NF-κB, and that the activation of HIV LTR by M. fermentans LAMPs is mediated through TLR1, TLR2 and TLR6. In contrast, the LTR activation by M. penetrans LAMPs is carried out through TLR1 and TLR2, but not TLR6. Subsequently, the active component of M. penetrans and M. fermentans LAMPs was purified by reverse-phase high-performance liquid chromatography (HPLC). Interestingly, the purified lipoprotein of M. penetrans LAMPs (LPMp) was able to activate NF-κB through TLR1 and TLR2. On the other hand, the activation of NF-κB by purified lipoprotein of M. fermentans LAMPs (LPMf) was mediated through TLR2 and TLR6, but not TLR1.
Keywords: HIV, lipoprotein, mycoplasma, Toll-like receptor
Introduction
Human immunodeficiency virus (HIV) is recognized as the aetiological agent of acquired immune-deficiency syndrome (AIDS). However, the progression of AIDS is highly variable in different individuals, and several factors, such as viral strains or host factors, have been attributed as the possible cause of such variations. Infectious agents, including various viruses, parasites and bacteria, are considered to be cofactors in the progression of AIDS.1 Mycoplasmas are wall-less parasitic Gram-positive bacteria, and the smallest organisms capable of self-replication.2Mycoplasma fermentans and M. penetrans have been isolated from the tissues and urine of patients with AIDS3–5 and were shown to enhance the cytopathic effect of HIV-1 infection.6,7 In addition, mycoplasmas and acholeplasmas have been reported to enhance HIV-1 replication in vitro.8,9 Thus, mycoplasmas might be reasonable cofactor candidates in the progression of AIDS.
A nuclear transcription factor – nuclear factor-κB (NF-κB) – is thought to play a major role in the regulation of HIV-1 gene expression.10 Although the HIV long-terminal repeat (LTR) alone can serve as its own promoter, early mRNA transcription appears to rely primarily on the binding of cellular transcription factors, including NF-κB, to the LTR.11 The activation of cytoplasmic NF-κB by various cytokines, including interleukin (IL)-2, IL-6 and tumour necrosis factor-α (TNF-α), or after infection with other viruses, induces HIV replication.12–17 These findings indicate increased rates of HIV replication, probably through NF-κB-mediated regulation of the HIV LTR. TNF-α induced by mycoplasma had been thought to induce NF-κB and HIV replication; however, anti-TNF-α immunoglobulin failed to inhibit the enhancement of HIV replication by mycoplasma.18 Although activation of NF-κB has been implicated in the mycoplasma-induced enhancement of HIV replication, the receptor(s) and the pathways of signal transduction via NF-κB have not been clearly defined.
Recently, it has been reported that Toll-like receptors (TLRs) are pattern-recognition receptors in the innate immune system and play important roles in early innate recognition and inflammatory responses by the host to microbial challenges.19 Among nine TLR family members reported, TLR2, 4, 5 and 9 have been implicated in the recognition of different bacterial components. Pepidoglycan, lipoarabinomannan, zymosan and lipoproteins from various micro-organisms are recognized by TLR2.20–28 On the other hand, lipopolysaccharide (LPS), bacterial flagellin and bacterial DNA are recognized by TLR4, TLR5 and TLR9, respectively.29–33 These TLR family members have been shown to activate NF-κB via IL-1R-associated signal molecules, including myeloid differentiation protein (MyD88), IL-1R-activated kinase (IRAK), TNFR-associated factor 6 (TRAF6), and NF-κB-inducing kinase (NIK).34 However, the precise mechanisms by which mycoplasma activate HIV LTR have not been fully clarified.
In this study, we examined the involvement of TLRs in the activation of HIV LTRs by mycoplasmas and their active components responsible for the TLR activation. We observed that lipid-associated membrane proteins (LAMPs) from the AIDS-associated mycoplasmas, M. penetrans and M. fermentans, activated the HIV LTR in a human monocytic cell line (THP-1) through NF-κB. Activation of the HIV LTR by LAMPs from M. fermentans (referred to as M. fermentans LAMPs hereafter) was apparently dependent on TLR1, TLR2 and TLR6. In contrast, activation of the HIV LTR by LAMPs from M. penetrans (referred to as M. penetrans LAMPs hereafter) was dependent on TLR1 and TLR2, but not on TLR6. Furthermore, the active components of M. penetrans and M. fermentans LAMPs were purified by reverse-phase HPLC. The activity of purified lipoprotein from M. penetrans LAMPs (LPMp) to induce NF-κB was dependent on TLR1 and TLR2. On the other hand, the activity of purified lipoprotein from M. fermentans LAMPs (LPMf) was dependent on TLR2 and TLR6, but not on TLR1.
Materials and methods
Cells
Cells of a human monocytic cell line, THP-1, were cultured in RPMI-1640 containing 10% fetal calf serum (FCS; Mitsubishi Chemical, Tokyo, Japan), 2 mm l-glutamine, 100 U/ml penicillin G and 100 µg/ml streptomycin. Cells of a human kidney cell line, 293T, were cultured in Dulbecco's modified Eagle's minimal essential medium (DMEM) containing 10% FCS, 2 mm l-glutamine, 100 U/ml penicillin G and 100 µg/ml streptomycin.
Antibodies
The mouse anti-human TLR2 monoclonal antibodies (mAbs) ABM-8320 and IMG-416 were obtained from Cascade Bioscience (Winchester, MA) and Imgenex (San Diego, CA).35 Normal mouse immunoglobulin G (IgG)2a was purchased from PharMingen (San Diego, CA).
Pathogen-associated molecular patterns (PAMPs)
(S)-[2,3-Bis(palmitoyloxy)-(2-RS)-propyl]-N-palmitoyl-(R)-Cys-(S)-Ser-(S)-Lys4-OH.3HCl (Pam3CSK4) was purchased from Calbiochem (Darmstadt, Germany). M. fermentans macrophage-activating lipopeptide 2 (MALP-2) was kindly provided by Dr M. Matsumoto (Osaka Medical Center for Cancer and Cardiovascular Diseases, Osaka, Japan).36,37
Preparation of LAMPs from M. fermentans and M. penetrans
M. fermentans and M. penetrans were cultured in PPLO medium and SP-4 medium, respectively, to the start of stationary phase, and then pelleted by centrifugation for 10 min at 12 000 g. Preparation of LAMPs was performed as described previously by Feng et al.38,39 Briefly, a mycoplasma pellet was suspended in Tris-buffered saline (TBS) (50 mm Tris, 0·15 m NaCl, pH 8·0) containing 1 mm EDTA (TBSE), solubilized by adding TX-114 to a final concentration of 2% and incubated at 4° for 1 hr. The lysate was incubated at 37° for 10 min prior to phase separation. After centrifugation at 10 000 g for 20 min, the upper aqueous phase was removed and replaced with the same volume of TBSE. The procedure of phase separation was repeated twice. The final TX-114 phase was resuspended in TBSE to the original volume, 2·5 volumes of ethanol were added to precipitate membrane components and the phase was incubated at −20° overnight. After centrifugation, the pellet was suspended in phosphate-buffered saline (PBS) followed by sonication for 30 seconds at output 5 (Sonifier cell disruptor 200; Branson, Danbury, CT). The protein concentration of the suspension was measured by using the Coomassie Protein Assay Regent (Pierce, Rockford, IL).
Expression vectors
To prepare TLR1, TLR2 and TLR6 expression vectors (pFLAG-TLR1, pFLAG-TLR2, and pFLAG-TLR6, respectively), the coding regions of TLR1, TLR2 and TLR6, minus the respective N-terminal signal sequences, were amplified by polymerase chain reaction (PCR) from a cDNA of THP-1 and cloned into the expression vector pFLAG-CMV1 (Sigma, St Louis, MO), in which a preprotrypsin leader precedes an N-terminal FLAG epitope. Dominant negative (DN) TLR1 and TLR6 expression vectors were constructed by subcloning TIR (Toll and interleukin 1 receptor) homology domain-deleted TLR1 and TLR6 fragments into pFLAG-CMV1 (pFLAG-dTLR1 and pFLAG-dTLR6). pHIV-LTR-luc, a mutant lacking the NF-κB-binding site (pHIV-LTRΔκB-luc), a mutant lacking the SP-1-binding site (pHIV-LTRΔSP1-luc), and a mutant lacking both NF-κB- and SP-1-binding sites (pHIV-LTRΔκBSP1-luc) were gifts from Dr Y. Koyanagi (Tohoku University Graduate School of Medicine, Sendai, Japan).40 The NF-κB Cis-Reporting System, containing pNF-kB-luc, a plasmid in which the luciferase reporter gene is fused to the NF-κB enhancer, was purchased from Stratagene (La Jolla, CA).
Transfection and luciferase assay
Transient transfection was performed by using FuGENE6 (Roche, Basel, Switzerland), according to the manufacturer's instructions. A total of 4 × 105 THP-1 cells, or 1 × 105 293 T cells, were transfected with 0·1 µg of pFLAG-TLR2, 0·01 µg of pHIV-LTR-luc, 0·01 µg of the pRL-TK internal control plasmid (Promega, Madison, WI), and DN TLRs expressing plasmid, in 24-well plates. After 20 hr, transfected cells were stimulated with 1·0 µg/ml M. fermentans LAMPs or 0·5 µg/ml M. penetrans LAMPs. After a further 24 hr of incubation, cells were lysed and assayed for luciferase activity using a Dual-Luciferase Reporter Assay System (Promega). Both firefly and Renilla luciferase activity were monitored using a Lumat LB9507 luminometer (Berthold, Wildbad, Germany). Normalized reporter activity is expressed as the firefly luciferase value divided by the Renilla luciferase value. Relative fold induction is calculated as the normalized reporter activity of the test samples divided by the unstimulated samples.
Reverse-phase high-performance liquid chromatography (HPLC)
LAMPs were dissolved in 6 m guanidine hydrochloride, and 100 µg of LAMPs were applied on µBondasphere C18 300A (Waters, Milford, MA). Elution was carried out using a 0–100% linear water/2-propanol gradient. The flow rate was 1·0 ml/min. Each fraction was dried in vacuo at room temperature and dissolved in 25 mm n-octyl-β-gulucopyranoside. Protein concentration was measured by using the Coomassie Protein Assay Regent (Pierce).
Lipoprotein lipase treatment
Approxmately 100 ng/ml LPMf and LPMp, separated from M. fermentans and M. penetrans LAMPs, respectively, were treated with 100 µg/ml lipoprotein lipase (Sigma) at 37° for 2 hr. 293T cells transfected with 0·02 µg/ml pNF-kB-luc and 0·2 µg/ml pFLAG-TLR2 were stimulated with 10 ng/ml of the lipoprotein lipase-treated LPMf and LPMp. Luciferase activity was measured as described above.
Results
Activation of HIV LTR by LAMPs
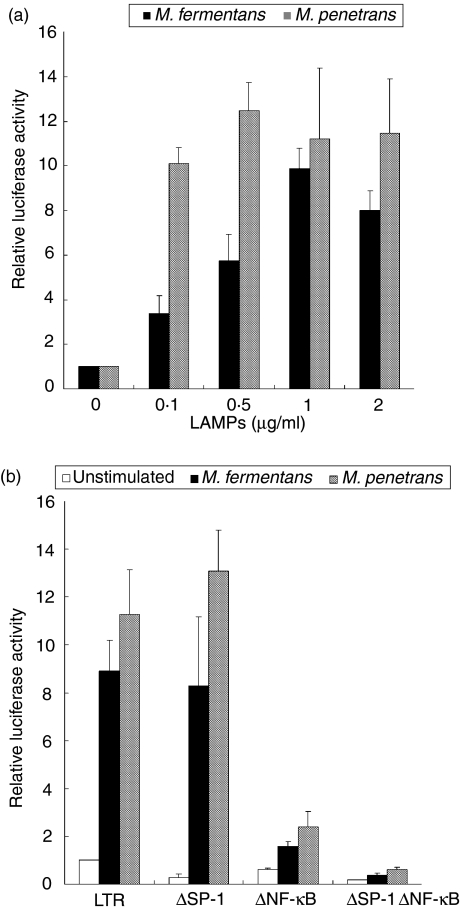
LAMPs of M. penetrans have been reported to stimulate macrophages.38,39 We therefore initially examined whether LAMPs from M. fermentans and M. penetrans can enhance HIV replication in macrophages. To determine the enhancement of HIV replication, THP-1 cells were first transfected with a plasmid in which the luciferase reporter gene was fused to HIV LTR (pHIV-LTR-luc) and then were stimulated with M. fermentans and M. penetrans LAMPs. The level of luciferase expression was enhanced by M. fermentans or M. penetrans LAMPs in a dose-dependent manner (Fig. 1a). When 0·5 µg/ml M. penetrans LAMPs was added, the luciferase expression was maximal and ≈ 12-fold higher than that of the unstimulated control. In contrast, the expression of THP-1 cells was maximal when stimulated with 1·0 µg/ml M. fermentans LAMPs, being ≈ 10-fold higher compared with the control. These results indicate that LAMPs from M. fermentans and M. penetrans can enhance HIV replication.
Figure 1.
Enhancement of long-terminal repeat (LTR) activation by lipid-associated membrane proteins (LAMPs) through nuclear factor-kappa B (NF-κB). (a) THP-1 cells were transfected with 0·1 µg/ml pHIV-LTR-luc and 0·01 µg/ml pRL-TK. The cells were stimulated with the indicated concentrations of LAMPs. All values represent the mean and standard deviation (SD) of three assays. (b) THP-1 cells were transfected with 0·1 µg/ml pHIV-LTR-luc (LTR), pHIV-LTRΔSP1-luc (ΔSP-1), pHIV-LTRΔκB-luc (ΔNFκB) or pHIV-LTRΔκBSP1-luc (ΔSP-1ΔNF-κB) in combination with 0·01 µg/ml pRL-TK. The cells were stimulated with 0·5 µg/ml Mycoplasma penetrans LAMPs and 1·0 µg/ml M. fermentans LAMPs. All values represent the mean and SD of three assays.
Activation of HIV LTR through NF-κB
HIV LTR contains various binding sites of cellular transcription factors, including NF-κB, SP-1, AP2, TCF-1, USF-1 and Ets; in particular, NF-κB and SP-1 are thought to be major transcription factors.41 To examine the roles of NF-κB and SP-1 in the activation of HIV LTR, pHIV-LTRΔκB-luc and pHIV-LTRΔSP1-luc (in which the NF-κB- and SP-1-binding sites, respectively, have been deleted from pHIV-LTR-luc) were prepared (Fig. 1b). When pHIV-LTRΔκB-luc-transfected THP-1 cells were stimulated with M. fermentans and M. penetrans LAMPs, the level of luciferase expression was lower than that of the control. In contrast, the level of luciferase expression was almost constant when pHIV-LTRΔSP1-luc was transfected. Moreover, the deletion of both NF-κB- and SP-1-binding sites (pHIV-LTRΔκBSP1-luc) resulted in a decrease of the expression level down to the level of the unstimulated control. These results indicate that NF-κB may be a major transcription factor induced by LAMPs.
Inhibition of LTR activation by anti-TLR2 mAb
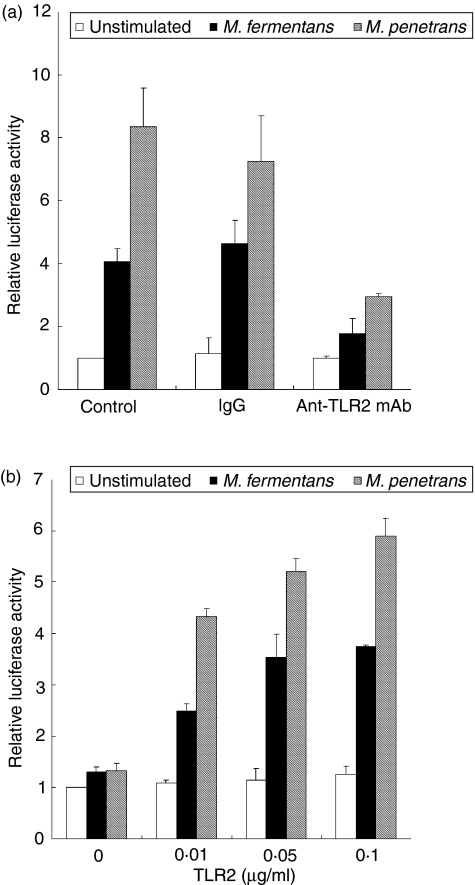
It was reported that a lipopeptide of M. fermentans– MALP-2 – can activate NF-κB through TLR2.28 We therefore examined whether the enhancement of HIV LTR activation with LAMPs is mediated through TLR2. Anti-human TLR2 mAb (ABM-8320)-pretreated THP-1 cells were transfected with pHIV-LTR-luc, followed by stimulation with M. fermentans and M. penetrans LAMPs. Pretreatment with anti-TLR2 mAb decreased the expression level of luciferase, and control antibody (mouse IgG2a) had no effect on luciferase (Fig. 2a). These results indicate that the activation of HIV LTR by LAMPs is TLR2-mediated.
Figure 2.
Enhancement of long-terminal repeat (LTR) activation through Toll-like receptor 2 (TLR2). (a) THP-1 cells were transfected with 0·1 µg/ml pHIV-LTR-luc and 0·01 µg/ml pRL-TK. The cells were treated with anti-TLR2 monoclonal antibody (mAb) followed by stimulation with 0·5 µg/ml Mycoplasma penetrans lipid-associated membrane proteins (LAMPs) and 1·0 µg/ml M. fermentans LAMPs. All values represent the mean and standard deviation (SD) of three assays. (b) 293T cells were transfected with the indicated concentrations of pFLAG-TLR2, 0·01 µg/ml pHIV-LTRΔSP1-luc, and 0·01 µg/ml pRL-TK. The cells were stimulated with 0·5 µg/ml M. penetrans LAMPs and 1·0 µg/ml M. fermentans LAMPs. All values represent the mean and SD of three assays.
Activation of HIV LTR through TLR2
To confirm whether the activation of NF-κB by LAMPs is mediated through TLR2, we constructed a TLR2 expression vector (pFLAG-TLR2). 293T cells were transfected with both pFLAG-TLR2 and pHIV-LTRΔSP1-luc. In this experiment, we used pHIV-LTRΔSP1-luc instead of pHIV-LTR-luc, because the binding site for SP-1 on the LTR resulted in a high level of luciferase activity (data not shown). When 293T cells were transfected with a high dose of pFLAG-TLR2 and then stimulated with M. fermentans and M. penetrans LAMPs, the levels of luciferase expression were augmented in a dose-dependent manner (Fig. 2b). In contrast, the level of luciferase expression was the same as that of the unstimulated control when 293T cells were transfected with the empty vector pFLAG-CMV1. This suggests that M. fermentans and M. penetrans LAMPs activate HIV LTR through TLR2.
Co-operation of TLR6 and TLR2 for LTR activation
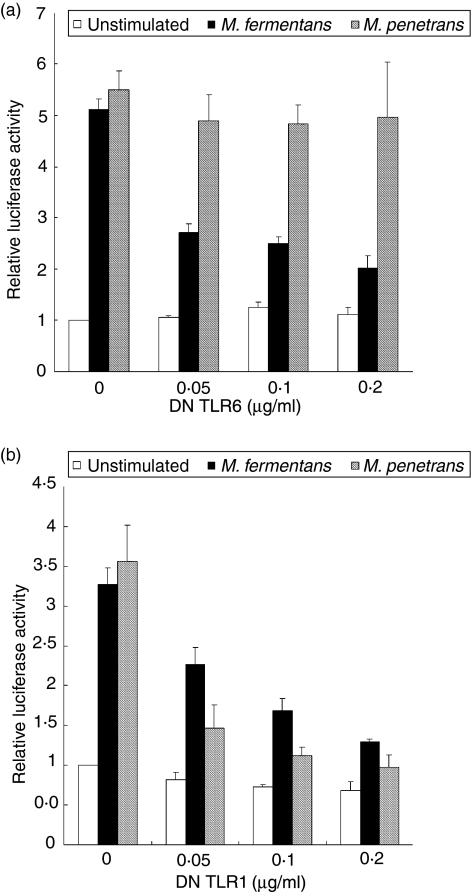
Mouse TLR6 has been reported to recognize diacylated lipopeptides, such as MALP-2, co-operatively with TLR2.42 To investigate whether M. fermentans and M. penetrans LAMPs are also recognized by both TLR2 and TLR6 for the activation of HIV LTR, we constructed a plasmid encoding DN TLR6 (pFLAG-dTLR6). 293T cells were transfected with pFLAG-TLR2, pHIV-LTRΔSP1-luc and various concentrations of pFLAG-dTLR6. Initially, the effect of DN TLR6 on the expression of TLR2 was analysed by flow cytometry. The level of TLR2 expression was almost constant, irrespective of the expression of DN TLR6 or of DN TLR1 (data not shown). When the transfected cells were stimulated with M. fermentans LAMPs, the level of luciferase expression decreased in a dose-dependent manner (Fig. 3a). Upon transfection with 0·2 µg/ml pFLAG-dTLR6, the expression level decreased to a level similar to that of the unstimulated control. In contrast, the level of luciferase expression was almost constant when the transfected 293T cells were stimulated with M. penetrans LAMPs. These results suggest that the LTR activation by M. fermentans LAMPs is dependent on TLR2 and TLR6, but the activation by M. penetrans LAMPs is not dependent on TLR6. To further examine whether TLR6 alone can mediate the activation of LTR by LAMPs, 293T cells were transfected with a TLR6 expression vector (pFLAG-TLR6). Although the transfected cells were stimulated with M. fermentans and M. penetrans LAMPs, the level of luciferase expression was not augmented (data not shown). These results indicate that both TLR2 and TLR6 co-operatively mediate the LTR activation by M. fermentans LAMPs, but not by M. penetrans LAMPs.
Figure 3.
Cooperation of Toll-like receptor (TLR)1, TLR6 and TLR2 for long-terminal repeat (LTR) activation by lipid-associated membrane proteins (LAMPs). 293T cells were transfected with the indicated concentrations of pFLAG-dTLR6 (a) or pFLAG-dTLR1 (b), 0·1 µg/ml pFLAG-TLR2, 0·01 µg/ml pHIV-LTRΔSP1-luc and 0·01 µg/ml pRL-TK. The cells were stimulated with 0·5 µg/ml Mycoplasma penetrans LAMPs or 1·0 µg/ml M. fermentans LAMPs. All values represent the mean and standard deviation (SD) of three assays. DN, dominant negative.
Co-operation of TLR1 and TLR2 for LTR activation
Triacylated bacterial lipopeptides, such as Pam3CSK4, were reported to be recognized by murine TLR1, in association with TLR2.43 The above results (Fig. 3a) suggest that M. penetrans LAMPs might contain different active components from M. fermentans LAMPs and, like the triacylated lipopeptides, M. penetrans LAMPs might be recognized by TLR1 and TLR2. We therefore next determined whether M. fermentans and M. penetrans LAMPs are recognized by both TLR1 and TLR2 for the activation of HIV LTR. To achieve this, we transfected a plasmid encoding DN TLR1 (pFLAG-dTLR1) into 293T cells containing both pFLAG-TLR2 and pHIV-LTRΔSP1-luc. When the transfected cells were stimulated with M. penetrans LAMPs, the level of luciferase expression was decreased in a dose-dependent manner (Fig. 3b). Unexpectedly, the level of luciferase expression of the cells stimulated with M. fermentans LAMPs was also decreased. Upon transfection with 0·2 µg/ml pFLAG-dTLR1, the level of expression in both cells decreased down to almost control levels. These results suggest that the LTR activation by M. fermentans and M. penetrans LAMPs is dependent on both TLR1 and TLR2. To further examine whether TLR1 alone can mediate the activation of LTR by LAMPs, 293T cells were transfected with a TLR1 expression vector (pFLAG-TLR1). Like the TLR6 expression in 293T cells, as mentioned previously, the level of luciferase expression of the transfected cells was not augmented by stimulation with M. fermentans and M. penetrans LAMPs (data not shown). These results indicate that the co-operation of TLR1 and TLR2 is required for the LTR activation with M. fermentans and M. penetrans LAMPs.
Purification of active components of LAMPs
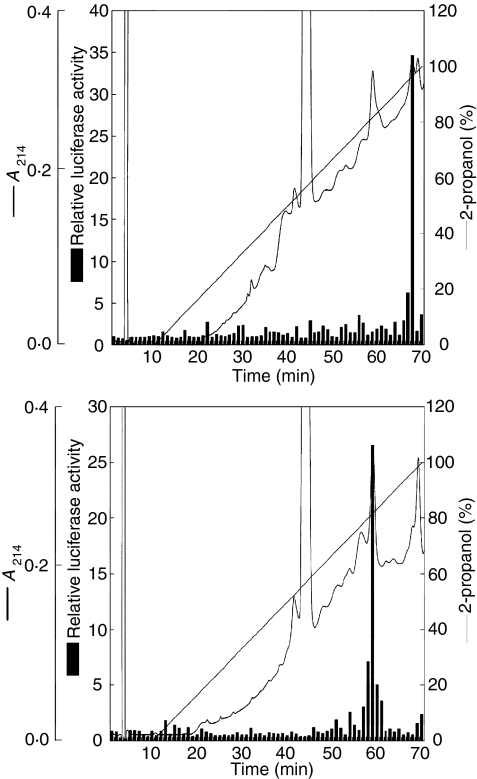
To purify the active components of LAMPs, M. penetrans and M. fermentans LAMPs were fractionated using reverse-phase HPLC with a linear gradient of isopropanol. To measure the activity of fractions to induce NF-κB, each fraction was added to 293T cells transfected with pFLAG-TLR2 and pNF-κB-luc. As shown in Fig. 4(a), the active component of M. penetrans LAMPs (LPMp) was eluted by ≈ 97% isopropanol, whereas the active component of M. fermentans LAMPs (LPMf) was eluted by ≈ 80% isopropanol (Fig. 4b).
Figure 4.
Isolation of active components of lipid-associated membrane proteins (LAMPs). Mycoplasma penetrans (a) and M. fermentans (b) LAMPS were dissolved in 6 m guanidine hydrochloride, and 100 µg of LAMPs was separated by reverse-phase high-performance liquid chromatography (HPLC). Elution was performed using a 0–100% linear gradient of water/2-propanol. Each fraction was added to THP-1 cells transfected with 0·1 µg/ml pFLAG-TLR2 and 0·1 µg/ml pNF-κB-luc. These results are representative examples of the three independent experiments.
Analysis of active components of LAMPs
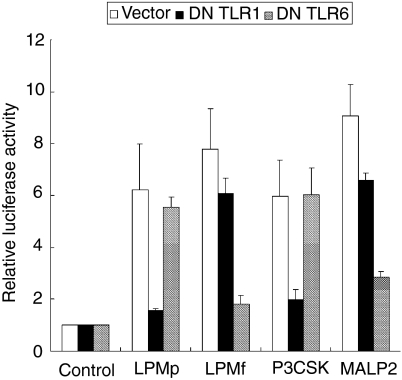
We next examined whether LPMp and LPMf separated from LAMPs are recognized by TLR2 and TLR6, or TLR1 and TLR2. 293T cells transfected with pFLAG-TLR2, pNF-κB-luc and pFLAG-dTLR1 or pFLAG-dTLR6 were stimulated with LPMp, LPMf, Pam3CSK4 or MALP-2. As reported previously by Takeuchi et al.,42,43 both DN TLR1 and DN TLR6 suppressed the activity of Pam3CSK4 and MALP-2, respectively, to induce NF-κB (Fig. 5). When the cells were stimulated with LPMp, the relative luciferase activity was reduced by the expression of DN TLR1 (Fig. 6), which is consistent with the results obtained using M. penetrans LAMPs (Fig. 3b). In contrast, the 293T cells stimulated with LPMf showed a relatively suppressed level of the luciferase activity when DN TLR6, but not DN TLR1, was expressed, inconsistent with the results that M. fermentans LAMPs was recognized by TLR1, TLR2 and TLR6 (Figs 3a and 5).
Figure 5.
Toll-like receptor (TLR) usage of Mycoplasma penetrans lipid-associated membrane proteins (LPMp) and M. fermentans lipid-associated membrane proteins (LPMf). 293T cells transfected with 0·01 µg/ml pFLAG-TLR2, 0·01 µg/ml pNF-κB-luc, and 0·2 µg/ml pFLAG-dTLR1 or pFLAG-dTLR6, were stimulated with 10 ng/ml LPMp, 10 ng/ml LPMf, 1 nm MALP-2, and 1 µg of Pam3CSK4 (P3CSK). All values represent the mean and standard deviation (SD) of three assays. DN, dominant negative.
Figure 6.
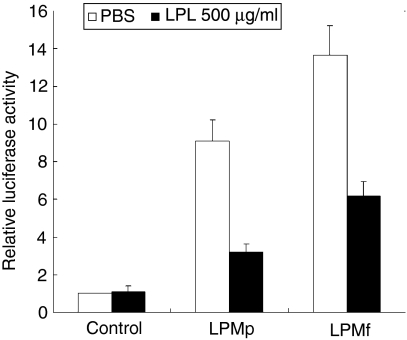
Lipoprotein lipase (LPL) treatment of Mycoplasma penetrans lipid-associated membrane proteins (LPMp) and M. fermentans lipid-associated membrane proteins (LPMf). One microgram of LPMp and LPMf was treated with 100 µg/ml LPL at 37° for 2 hr. 293T cells transfected with 0·01 µg/ml pNF-κB-luc and 0·01 µg/ml pFLAG-TLR2 were stimulated with 10 ng/ml LPL-treated LPMp and LPMf. All values represent the mean and standard deviation (SD) of three assays. PBS, phosphate-buffered saline.
To analyse the chemical components of LPMf and LPMp, they were treated with lipoprotein lipase and proteinase K. Treatment with proteinase K failed to decrease the activity of LPMf and LPMp (data not shown), while lipoprotein lipase treatment decreased the ability to induce NF-κB (Fig. 6). These results suggest that lipid moiety, but not protein moiety, is required for activity.
Discussion
In this study, we demonstrated that LAMPs from M. fermentans and M. penetrans activated the HIV LTR through NF-κB. Activation of the LTR by M. fermentans LAMPs was TLR1-, TLR2- and TLR6 dependent, while the activation by M. penetrans LAMPs was TLR1- and TLR2 dependent. The active components of M. fermentans and M. penetrans LAMPs were purified by reverse-phase HPLC. The purified lipoprotein from M. penetrans LAMPs (LPMp) was recognized by TLR1 and TLR2. Although the recognition of M. fermentans LAMPs involved TLR1, TLR2 and TLR6 (Fig. 3), the purified lipoprotein from M. fermentans LAMPs (LPMf) was recognized only by TLR2 and TLR6. These results indicate that M. fermentans LAMPs may contain several active components, in which one component is recognized by TLR2 and TLR6, and other components might be recognized by TLR1 and TLR2. Both LPMf and LPMp showed resistance to proteinase K treatment, in agreement with the previous report by Feng et al.39 In addition, the ability of LPMf and LPMp to induce NF-κB was reduced by treatment with lipoprotein lipase. These results suggest that the active components are attributable to lipid moieties. MALP-2 constitutes a lipopeptide, isolated from M. fermentans, which has been well documented as an activator of NF-κB.44,45 Subsequently, a 44-kDa membrane-bound lipoprotein of M. salivarium has been reported to induce TNF-α production in THP-1.46 To our knowledge, there still seems to be little information on the activity in innate immune responses of lipoproteins derived from mycoplasmas such as M. fermentans, M. penetrans and M. salivarium, although a variety of mycoplasma strains have been shown to exhibit diverse bioactivities in interaction with eukaryotic cells.47,48
In TLR-deficient mice, bacterial lipopeptides containing three acyl chains were reported to be recognized by TLR1 and TLR2,43 whereas MALP-2, containing two acyl chains, were recognized by TLR2 and TLR6.42 These findings, and our results, suggest that LPMp might be similar to lipoprotein(s) containing three acyl chains. In contrast, LPMf separated from M. fermentans in this study may possess components comparable to MALP-2, as MALP-2 is a lipopeptide derived from M. fermentans.
In mycoplasmas, acylated proteins are abundant cell-surface antigens, and many putative lipoprotein-encoding genes have been identified in the sequenced mycoplasma genomes.49,50 It is, at present, controversial as to whether or not mycoplasmas have triacylated lipoprotein. Chemically identified lipoproteins from M. fermentans,44M. hyorhinis,51M. salivarium46 and M. gallisepticum52 are not N-acylated, nor has an N-acyltransferase gene been found in M. pneumoniae,53M. genitalium54 or M. penetrans55 genomes. To date, the presence of proteins with N-acyltransferase activity has not been clearly established. However, the study on the ratio of N-amide and O-ester bonds in M. gallisepticum and M. mycoides may indicate the presence of diacylated and triacylated lipoproteins.56 The resistance to Edoman degradation of proteins from M. mycoides also indicates the presence of N-acylation.50 In this study, we found that the lipoprotein separated from M. penetrans induced NF-κB through TLR1 and TLR2. Triacylated lipoproteins, such as Pam3-CSK4, have been reported to be recognized by TLR1 and TLR2,43 whereas diacylated lipoproteins, such as MALP-2, have been shown to be recognized by TLR2 and TLR6.42 Interestingly, synthetically triacylated MALP-2, N-palamitoyl-MALP-2, was not recognized by TLR6.57 These findings may indicate the existence of triacylated lipoproteins in mycoplasma species.
Our results indicate that the lipoproteins from M. fermentans and M. penetrans can activate NF-κB in HIV LTR, leading to the enhancement of HIV replication. The activation of NF-κB was also observed following stimulation with bacterial components, including LPS30 and peptidoglycan.24 We have previously reported that glycolipids from Acholeplasma laidlawii, binding to both HIV and macrophages, enhance HIV replication.58,59 In addition to the ability of lipoproteins to induce NF-κB, glycolipids from mycoplasma might, in concert, enhance the replication of HIV. We assume that lipoproteins and glycolipids residing in the mycoplasma membrane can efficiently attach to the surface of HIV-infected cells, as mycoplasmas are completely wall-less bacteria.2 Moreover, mycoplasmas contain various surface proteins that tend to show high-frequency variation, suggesting that mycoplasmas can escape from immune surveillance and establish a persistent infection.60 These findings suggest that mycoplasma, rather than bacteria with cell walls, might play an important role in the progression of HIV infection.
We previously reported that a variety of mycoplasma strains can induce TNF-α production in mouse macrophages61 and THP-1 cells.62,63M. penetrans LAMPs have been reported to induce TNF-α production in mouse thioglycolate exudate peritoneal macrophage cells.39 Moreover, TNF-α has been shown to activate HIV LTR through NF-κB.64 These findings suggest that TNF-α produced by macrophages may activate NF-κB. However, we observed that anti-TNF-α mAb failed to inhibit the activation of LTR by LAMPs (data not shown). It is therefore unlikely that TNF-α produced by LAMPs-stimulated THP-1 cells may directly contribute to the activation of NF-κB.
In summary, we demonstrated that lipoproteins from the AIDS-associated mycoplasmas, M. fermentans and M. penetrans, can enhance HIV LTR activity in THP-1 cells through NF-κB, and that this enhancement is dependent on TLRs. The enhancement of the HIV LTR activation induced by M. fermentans LAMPs was dependent on TLR1, TLR2 and TLR6. Interestingly, LPMf separated from M. fermentans LAMPs activated NF-κB through TLR2 and TLR6, but not TLR1. In contrast, the enhancement of the NF-κB activation induced by M. penetrans LAMPs, as well as LPMp separated from M. penetrans LAMPs, was dependent on TLR1 and TLR2, but not TLR6. Clarifying the mechanisms by which various bacteria, including mycoplasma, enhance HIV replication may have therapeutic values in preventing the progression of AIDS during opportunistic infection.
Acknowledgments
We thank Dr Koyanagi for gifts of plasmids pHIV-LTR-luc, pHIV-LTRΔκB-luc, pHIV-LTRΔSP1-luc and pHIV-LTRΔκBSP1-luc. We also thank Dr Matsumoto for the gift of MALP-2. This work was supported, in part, by a grant from the Ishibashi Foundation.
References
- 1.Blanchard A, Montagnier L, Gougeon ML. Influence of microbial infections on the progression of HIV disease. Trends Microbiol. 1997;5:326–31. doi: 10.1016/S0966-842X(97)01089-5. [DOI] [PubMed] [Google Scholar]
- 2.Weisburg WG, Tully JG, Rose DL, et al. A phylogenetic analysis of the mycoplasmas: basis for their classification. J Bacteriol. 1989;171:6455–67. doi: 10.1128/jb.171.12.6455-6467.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lo SC, Dawson MS, Wong DM, et al. Identification of Mycoplasma incognitus infection in patients with AIDS. an immunohistochemical, in situ hybridization and ultrastructural study. Am J Trop Med Hyg. 1989;41:601–16. doi: 10.4269/ajtmh.1989.41.601. [DOI] [PubMed] [Google Scholar]
- 4.Lo SC, Hayes MM, Wang RY, Pierce PF, Kotani H, Shih JW. Newly discovered mycoplasma isolated from patients infected with HIV. Lancet. 1991;338:1415–8. doi: 10.1016/0140-6736(91)92721-d. [DOI] [PubMed] [Google Scholar]
- 5.Lo SC, Hayes MM, Tully JG, Wang RY, Kotani H, Pierce PF, Rose DL, Shih JW. Mycoplasma penetrans sp. nov., from the urogenital tract of patients with AIDS. Int J Syst Bacteriol. 1992;42:357–64. doi: 10.1099/00207713-42-3-357. [DOI] [PubMed] [Google Scholar]
- 6.Lo SC, Tsai S, Benish JR, Shih JW, Wear DJ, Wong DM. Enhancement of HIV-1 cytocidal effects in CD4+ lymphocytes by the AIDS-associated mycoplasma. Science. 1991;251:1074–6. doi: 10.1126/science.1705362. [DOI] [PubMed] [Google Scholar]
- 7.Lemaitre M, Henin Y, Destouesse F, Ferrieux C, Montagnier L, Blanchard A. Role of mycoplasma infection in the cytopathic effect induced by human immunodeficiency virus type 1 in infected cell lines. Infect Immun. 1992;60:742–8. doi: 10.1128/iai.60.3.742-748.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowdhury IH, Munakata T, Koyanagi Y, Kobayashi S, Arai S, Yamamoto N. Mycoplasma can enhance HIV replication in vitro: a possible cofactor responsible for the progression of AIDS. Biochem Biophys Res Commun. 1990;170:1365–70. doi: 10.1016/0006-291x(90)90545-x. [DOI] [PubMed] [Google Scholar]
- 9.Chowdhury MI, Munakata T, Koyanagi Y, Arai S, Yamamoto N. Mycoplasma stimulates HIV-1 expression from acutely- and dormantly- infected promonocyte/monoblastoid cell lines. Arch Virol. 1994;139:431–8. doi: 10.1007/BF01310804. [DOI] [PubMed] [Google Scholar]
- 10.Ou SHI, Gaynor RB. Intracellular factors involved in gene expression of human retroviruses. In: Levy JA, editor. The Retroviridae. Vol. 4. New York: The Plenum Press; 1995. p. 97. [Google Scholar]
- 11.Nabel G, Baltimore D. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature. 1987;326:711–3. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 12.Skolnik PR, Kosloff BR, Hirsch MS. Bidirectional interactions between human immunodeficiency virus type 1 and cytomegalovirus. J Infect Dis. 1988;157:508–14. doi: 10.1093/infdis/157.3.508. [DOI] [PubMed] [Google Scholar]
- 13.Golden MP, Kim S, Hammer SM, Ladd EA, Schaffer PA, DeLuca N, Albrecht MA. Activation of human immunodeficiency virus by herpes simplex virus. J Infect Dis. 1992;166:494–9. doi: 10.1093/infdis/166.3.494. [DOI] [PubMed] [Google Scholar]
- 14.Poli G, Fauci AS. Cytokine modulation of HIV expression. Semin Immunol. 1993;5:165–73. doi: 10.1006/smim.1993.1020. [DOI] [PubMed] [Google Scholar]
- 15.Fauci AS. Host factors and the pathogenesis of HIV-induced disease. Nature. 1996;384:529–34. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 16.Wahl SM, Orenstein JM. Immune stimulation and HIV-1 viral replication. J Leukoc Biol. 1997;62:67–71. doi: 10.1002/jlb.62.1.67. [DOI] [PubMed] [Google Scholar]
- 17.Sulkowski MS, Chaisson RE, Karp CL, Moore RD, Margolick JB, Quinn TC. The effect of acute infectious illnesses on plasma human immunodeficiency virus (HIV) type 1 load and the expression of serologic markers of immune activation among HIV-infected adults. J Infect Dis. 1998;178:1642–8. doi: 10.1086/314491. [DOI] [PubMed] [Google Scholar]
- 18.Iyama K, Ono S, Kuwano K, Ohishi M, Shigematsu H, Arai S. Induction of tumor necrosis factor alpha (TNF alpha) and enhancement of HIV-1 replication in the J22HL60 cell line by Mycoplasma penetrans. Microbiol Immunol. 1996;40:907–14. doi: 10.1111/j.1348-0421.1996.tb01159.x. [DOI] [PubMed] [Google Scholar]
- 19.Kopp EB, Medzhitov R. The Toll-receptor family and control of innate immunity. Curr Opin Immunol. 1999;11:13–8. doi: 10.1016/s0952-7915(99)80003-x. [DOI] [PubMed] [Google Scholar]
- 20.Aliprantis AO, Yang RB, Mark MR, et al. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science. 1999;285:736–9. doi: 10.1126/science.285.5428.736. [DOI] [PubMed] [Google Scholar]
- 21.Brightbill HD, Libraty DH, Krutzik SR, et al. Host defense mechanisms triggered by microbial lipoproteins through toll-like receptors. Science. 1999;285:732–6. doi: 10.1126/science.285.5428.732. [DOI] [PubMed] [Google Scholar]
- 22.Lien E, Sellati TJ, Yoshimura A, et al. Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. J Biol Chem. 1999;274:33419–25. doi: 10.1074/jbc.274.47.33419. [DOI] [PubMed] [Google Scholar]
- 23.Means TK, Lien E, Yoshimura A, Wang S, Golenbock DT, Fenton MJ. The CD14 ligands lipoarabinomannan and lipopolysaccharide differ in their requirement for Toll-like receptors. J Immunol. 1999;163:6748–55. [PubMed] [Google Scholar]
- 24.Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning CJ. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J Biol Chem. 1999;274:17406–9. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 25.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of Gram-negative and Gram-positive bacterial cell wall components. Immunity. 1999;11:443–51. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 26.Underhill DM, Ozinsky A, Hajjar AM, Stevens A, Wilson CB, Bassetti M, Aderem A. The Toll-like receptor 2 is recruited to macrophage phagosomes and discriminates between pathogens. Nature. 1999;401:811–5. doi: 10.1038/44605. [DOI] [PubMed] [Google Scholar]
- 27.Yoshimura A, Lien E, Ingalls RR, Tuomanen E, Dziarski R, Golenbock D. Cutting edge. recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J Immunol. 1999;163:1–5. [PubMed] [Google Scholar]
- 28.Takeuchi O, Kaufmann A, Grote K, Kawai T, Hoshino K, Morr M, Muhlradt PF, Akira S. Cutting edge: preferentially the R-stereoisomer of the mycoplasmal lipopeptide macrophage-activating lipopeptide-2 activates immune cells through a toll-like receptor 2- and MyD88-dependent signaling pathway. J Immunol. 2000;164:554–7. doi: 10.4049/jimmunol.164.2.554. [DOI] [PubMed] [Google Scholar]
- 29.Poltorak A, He X, Smirnova I, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–8. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 30.Chow JC, Young DW, Golenbock DT, Christ WJ, Gusovsky F. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J Biol Chem. 1999;274:10689–92. doi: 10.1074/jbc.274.16.10689. [DOI] [PubMed] [Google Scholar]
- 31.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol. 1999;162:3749–52. [PubMed] [Google Scholar]
- 32.Hemmi H, Takeuchi O, Kawai T, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–5. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 33.Hayashi F, Smith KD, Ozinsky A, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 34.Medzhitov R, Preston-Hurlburt P, Kopp E, Stadlen A, Chen C, Ghosh S, Janeway CA., Jr MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol Cell. 1998;2:253–8. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- 35.Flo TH, Halaas O, Lien E, Ryan L, Teti G, Golenbock DT, Sundan A, Espevik T. Human toll-like receptor 2 mediates monocyte activation by Listeria monocytogenes, but not by group B streptococci or lipopolysaccharide. J Immunol. 2000;164:2064–9. doi: 10.4049/jimmunol.164.4.2064. [DOI] [PubMed] [Google Scholar]
- 36.Nishiguchi M, Matsumoto M, Takao T, et al. Mycoplasma fermentans lipoprotein M161Ag-induced cell activation is mediated by Toll-like receptor 2: role of N-terminal hydrophobic portion in its multiple functions. J Immunol. 2001;166:2610–6. doi: 10.4049/jimmunol.166.4.2610. [DOI] [PubMed] [Google Scholar]
- 37.Seya T, Matsumoto M. A lipoprotein family from Mycoplasma fermentans confers host immune activation through Toll-like receptor 2. Int J Biochem Cell Biol. 2002;34:901–6. doi: 10.1016/s1357-2725(01)00164-9. [DOI] [PubMed] [Google Scholar]
- 38.Feng SH, Lo SC. Induced mouse spleen B-cell proliferation and secretion of immunoglobulin by lipid-associated membrane proteins of Mycoplasma fermentans incognitus and Mycoplasma penetrans. Infect Immun. 1994;62:3916–21. doi: 10.1128/iai.62.9.3916-3921.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng SH, Lo SC. Lipid extract of Mycoplasma penetrans proteinase K-digested lipid-associated membrane proteins rapidly activates NF-kappaB and activator protein 1. Infect Immun. 1999;67:2951–6. doi: 10.1128/iai.67.6.2951-2956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cann AJ, Koyanagi Y, Chen IS. High efficiency transfection of primary human lymphocytes and studies of gene expression. Oncogene. 1988;3:123–8. [Google Scholar]
- 41.Coffin JM, Rabson AB, Graves BJ. Synthesis and processing of viral RNA. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. New York: Cold Spring Harbor Laboratory Press; 1997. pp. 205–61. [PubMed] [Google Scholar]
- 42.Takeuchi O, Kawai T, Muhlradt PF, Morr M, Radolf JD, Zychlinsky A, Takeda K, Akira S. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int Immunol. 2001;13:933–40. doi: 10.1093/intimm/13.7.933. [DOI] [PubMed] [Google Scholar]
- 43.Takeuchi O, Sato S, Horiuchi T, Hoshino K, Takeda K, Dong Z, Modlin RL, Akira S. Cutting edge: role of toll-like receptor 1 in mediating immune response to microbial lipoproteins. J Immunol. 2002;169:10–4. doi: 10.4049/jimmunol.169.1.10. [DOI] [PubMed] [Google Scholar]
- 44.Muhlradt PF, Kiess M, Meyer H, Sussmuth R, Jung G. Isolation, structure elucidation, and synthesis of a macrophage stimulatory lipopeptide from Mycoplasma fermentans acting at picomolar concentration. J Exp Med. 1997;185:1951–8. doi: 10.1084/jem.185.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaufmann A, Muhlradt PF, Gemsa D, Sprenger H. Induction of cytokines and chemokines in human monocytes by Mycoplasma fermentans-derived lipoprotein MALP-2. Infect Immun. 1999;67:6303–8. doi: 10.1128/iai.67.12.6303-6308.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shibata K, Hasebe A, Into T, Yamada M, Watanabe T. The N-terminal lipopeptide of a 44-kDa membrane-bound lipoprotein of Mycoplasma salivarium is responsible for the expression of intercellular adhesion molecule-1 on the cell surface of normal human gingival fibroblasts. J Immunol. 2000;165:6538–44. doi: 10.4049/jimmunol.165.11.6538. [DOI] [PubMed] [Google Scholar]
- 47.Cole BC, Washburn LR. Mycoplasma arthritidis pathogenicity. membranes, MAM, and MAV1. In: Razin S, Herrmann R, editors. Molecular Biology and Pathogenicity of Mycoplasmas. New York: Kluwer Academic/Plenum Publishers; 2002. pp. 473–89. [Google Scholar]
- 48.Muhlradt PF. Immunomodulation by mycoplasmas: artifacts, facts and active molecules. In: Razin S, Herrmann R, editors. Molecular Biology and Pathogenicity of Mycoplasmas. New York: Kluwer Academic/Plenum Publishers; 2002. pp. 445–72. [Google Scholar]
- 49.Wieslamder A, Boyer MJ, Wroblewski H. Membrane protein structure. In: Maniloff J, McElhaney RN, Finch LR, Baseman JB, editors. Mycoplasmas – Molecular Biology and Pathogenesis. Washington, DC: American Society for Microbiology; 1992. pp. 93–112. [Google Scholar]
- 50.Chambaud I, Wroblewski H, Blanchard A. Interactions between mycoplasma lipoproteins and the host immune system. Trends Microbiol. 1999;7:493–9. doi: 10.1016/s0966-842x(99)01641-8. [DOI] [PubMed] [Google Scholar]
- 51.Muhlradt PF, Kiess M, Meyer H, Sussmuth R, Jung G. Structure and specific activity of macrophage-stimulating lipopeptides from Mycoplasma hyorhinis. Infect Immun. 1998;66:4804–10. doi: 10.1128/iai.66.10.4804-4810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jan G, Brenner C, Wroblewski H. Purification of Mycoplasma gallisepticum membrane proteins p52, p67 (pMGA), and p77 by high-performance liquid chromatography. Protein Expr Purif. 1996;7:160–6. doi: 10.1006/prep.1996.0023. [DOI] [PubMed] [Google Scholar]
- 53.Fraser CM, Gocayne JD, White O, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 54.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li BC, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–49. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sasaki Y, Ishikawa J, Yamashita A, et al. The complete genomic sequence of Mycoplasma penetrans, an intracellular bacterial pathogen in humans. Nucleic Acids Res. 2002;30:5293–300. doi: 10.1093/nar/gkf667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jan G, Fontenelle C, Verrier F, Le Henaff M, Wroblewski H. Selective acylation of plasma membrane proteins of Mycoplasma mycoides subsp. mycoides SC, the contagious bovine pleuropneumonia agent. Curr Microbiol. 1996;32:38–42. doi: 10.1007/s002849900007. [DOI] [PubMed] [Google Scholar]
- 57.Morr M, Takeuchi O, Akira S, Simon MM, Muhlradt PF. Differential recognition of structural details of bacterial lipopeptides by toll-like receptors. Eur J Immunol. 2002;32:3337–47. doi: 10.1002/1521-4141(200212)32:12<3337::AID-IMMU3337>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 58.Toujima S, Kuwano K, Zhang Y, et al. Binding of glycoglycerolipid derived from membranes of Acholeplasma laidlawii PG8 and synthetic analogues to lymphoid cells. Microbiology. 2000;146:2317–23. doi: 10.1099/00221287-146-9-2317. [DOI] [PubMed] [Google Scholar]
- 59.Shimizu T, Arai S, Imai H, Oishi T, Hirama M, Koito A, Kida Y, Kuwano K. Glycoglycerolipid from the membranes of Acholeplasma laidlawii binds to human immunodeficiency virus-1 (HIV-1) and accelerates its entry into cells. Curr Microbiol. 2004;48:182–8. doi: 10.1007/s00284-003-4101-x. [DOI] [PubMed] [Google Scholar]
- 60.Yogev D, Browning GF, Wise KS. Genetic mechanisms of surface variation. In: Razin S, Herrmann R, editors. Molecular Biology and Pathogenicity of Mycoplasma. New York: Plenum Publishers; 2002. pp. 417–44. [Google Scholar]
- 61.Arai S, Furukawa M, Munakata T, Kuwano K, Inoue H, Miyazaki T. Enhancement of cytotoxicity of active macrophages by mycoplasma: role of mycoplasma-associated induction of tumor necrosis factor-alpha (TNF-alpha) in macrophages. Microbiol Immunol. 1990;34:231–43. doi: 10.1111/j.1348-0421.1990.tb01006.x. [DOI] [PubMed] [Google Scholar]
- 62.Sugama K, Kuwano K, Furukawa M, Himeno Y, Satoh T, Arai S. Mycoplasmas induce transcription and production of tumor necrosis factor in a monocytic cell line, THP-1, by a protein kinase C-independent pathway. Infect Immun. 1990;58:3564–7. doi: 10.1128/iai.58.11.3564-3567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nishimoto M, Akashi A, Kuwano K, Tseng CC, Ohizumi K, Arai S. Gene expression of tumor necrosis factor alpha and interferon gamma in the lungs of Mycoplasma pulmonis-infected mice. Microbiol Immunol. 1994;38:345–52. doi: 10.1111/j.1348-0421.1994.tb01789.x. [DOI] [PubMed] [Google Scholar]
- 64.Duh EJ, Maury WJ, Folks TM, Fauci AS, Rabson AB. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proc Natl Acad Sci USA. 1989;86:5974–8. doi: 10.1073/pnas.86.15.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]