Abstract
The transporter associated with antigen processing 1 and 2 (TAP1 and TAP2) genes belong to the ATP-binding cassette family of transporter genes. They provide peptides necessary for the assembly of major histocompatibility complex (MHC) class I molecules by transporting these peptides into the endoplasmic reticulum. As MHC class I protein expression increases with age, we have explored the effect of age on the transcription of MHC class I genes (Kb) and TAP1 and TAP2 genes in C57BL/6 mice. Blood and spleen lymphocytes were isolated from mice aged from 3 months to over 24 months. RNA was extracted and mRNA for Kb, TAP1, TAP2 was quantified using slot-blot hybridization followed by densitometry. There was a parallel age-related increase (1·5-fold) in blood lymphocyte mRNA of these genes from 3 months to 21 months. In mice over 24 months old there was a decrease in Kb and TAP1 mRNA, but an increase in TAP2 mRNA. In spleen lymphocytes an age-related increase in all three mRNA species occurred throughout life. While MHC class I and Tap genes underwent very similar age-related changes, MHC class I mRNA was about 50 times more abundant than either TAP1 or TAP2 mRNA.
Keywords: age, MHC, mouse, Tap genes, transcription
Introduction
The transporter associated with antigen processing 1 and 2 (TAP1 and TAP2) genes have been discovered in several species.1,2 They are located in the class II region of the major histocompatibility complex (MHC). Evidence has been accumulating showing that MHC class I proteins present antigens of viral or intracellular origin.3 Unlike MHC class II, MHC class I protein stability depends on its binding of peptide antigen. This binding in vivo occurs intracellularly, as reviewed by Townsend et al.4 The discovery of Tap genes constitutes a leap forward in the understanding of the processing of MHC class I genes. Monaco et al. have published a review of the structure and function of the Tap genes.5 The mouse T-cell line RMA-S, which has normal MHC genes but does not express class I, was the major tool used in the discovery of Tap genes.6 This T-cell line lacks a TAP1 gene in the MHC class II region, which is associated with a lack of MHC class I expression. MHC class I expression in an RMA-S cell line was restored by the transfection of the TAP1 gene.6,7 The Tap genes have the characteristic structure of the widely distributed ATP-binding cassette (ABC) gene, and include molecules like the protein encoded by the multiple drug-resistant genes. Their function is ATP-driven transport across membranes. Tap proteins have been found to be located in the endoplasmic reticulum (ER).5 They are believed to transport peptides into the ER lumen to make them available to bind MHC class I molecules. The peptides are proposed to result from a proteolysis of self-protein or viral protein by a low-molecular-weight proteasome (LMP).8 LMP protein is a large multimer composed of several subunits. The genes for two LMP subunits (LMP-2 and LMP-7) have been mapped to the MHC class II region, next to the Tap genes.
We have reported that the cell surface expression of MHC class I protein increases with age.9 Since MHC class I surface expression depends on Tap genes, we sought to investigate the relationship between age and the transcription of Tap genes. We have also explored the transcription of MHC class I (Kb) genes as a function of age, with the aim of elucidating the relationship between MHC class I gene expression and Tap gene expression. This work may constitute a basis for the study of the regulation of the expression of MHC and Tap genes and improve understanding of age-related changes in immune function.
Materials and methods
Mice
C57BL/6 mice from our own colony were selected for this study. The colony consists of the offspring of C57BL/6 male and female mice purchased from the Jackson Laboratory (Bar Harbor, ME) twice a year. Two sets of mice were studied. Experiments on peripheral blood mononuclear cells (PBMCs) were carried out on one set of 24 mice, obtained by randomly selecting six mice from each of the following age groups: 2–4, 8–10, 18–21 and > 24 months. Experiments on spleen lymphocytes were carried out on a second set of mice, containing 36 individuals obtained by randomly selecting nine mice from each of the following age groups: 3–6, 9–12, 18–21 and > 24 months. All the groups were used in the study of MHC (Kb) and Tap gene expression in lymphocytes.
Probes
The probes used are summarized in Table 1.
Table 1.
Characteristics of probes used in the study
| Probe | Host bacterium | Vector | Size of insert (bp) |
|---|---|---|---|
| Actin | E. coli | Plasmid (pRSαA3) | 1500 |
| Kb | NA | NA | 260 |
| TAP1 | E. coli | Plasmid (Pgem3Zf+) | 1400 |
| TAP2 | E. coli | Plasmid (Pgem3Zf+) | 900 |
NA, not applicable.
Actin probe
An actin cDNA probe in the pRSαA3 plasmid (Table 1) obtained from Dr Peter A. Rubeinstein (University of Iowa, Iowa City, IA) was prepared following a modification of the procedure described by Sambrook et al.10 Escherichia coli containing the plasmid pRSαA3 was cultured in 500 ml of 2x Luria–Bertani (LB) medium and ampicillin (Sigma Aldrich, St Louis, MO) contained in a 2-l flask at 37° with constant agitation for 18 hr. The medium was then centrifuged at 5300 g at 4° for 10 min. The supernatant was discarded and the pellet was resuspended in 10 ml of an ice-cold 10% sucrose, 50 mm Tris-HCl solution. Plasmids containing inserts were digested by the HindIII restriction enzyme. A 1% agarose gel was then used to separate electrophoretically the insert from the plasmid. The portion of the gel containing the insert was excised and the insert was then retrieved by electroelution as described by Sambrook et al.10 Fifty to 100 ng was then used for each hybridization reaction.
TAP1 and TAP2 probes
The mouse TAP1 and TAP2 probes (Table 1) in plasmids in E. coli were obtained from John Monaco (Virginia Commonwealth University, Richmond, VA). E. coli was grown and the plasmid was extracted as described above for the actin probe. Each plasmid was then digested with the EcoRI restriction enzyme and the probe insert was isolated by agarose gel electrophoresis followed by electroelution.
Kb probe
DNA template preparation and polymerase chain reaction
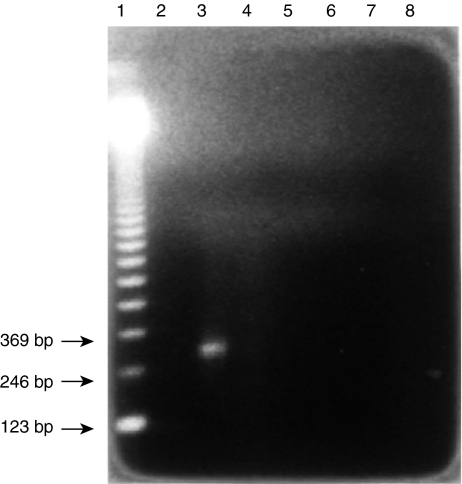
The Kb probe was prepared using polymerase chain reaction (PCR). The DNA template was prepared by the method described by Kawasaki.11 The PCR for the amplification of exon 2 of the Kb gene was carried out as previously described.12–14 Each tube received 10 µl of Taq polymerase buffer from Promega (Madison, WI), 0·1 µg of primer #10 (5′-GCCCACACTCGCTGAGGTAT-3′) in 10 µl, 0·1 µg of primer #11 (5′-CTGGTTGTAGTAGCCGAGCA-3′) in 10 µl, 1 µl of 25 mm dNTP, 0·5 µl of Taq DNA polymerase from Promega, and 10 µl of DNA template solution obtained as described above. The total volume was brought to 100 µl with sterile water. The primers were designed to produce a 260-bp PCR product (from nucleotide +185 to nucleotide +444) (Table 1). Paraffin oil (100 µl) was added to each tube before starting the PCR using a Perkin-Elmer Cetus Thermocycler (Perkin-Elmer, Norwalk, CT). The PCR was carried out for 35 cycles with the following settings: denaturation at 95° for 90 s, annealing at 66° for 1 min, extension at 72° for 2 min, plus 5 s per additional cycle. The 260-bp PCR product was then isolated by electrophoresis on a 1% agarose gel followed by electroelution (Fig. 1).
Figure 1.
Electrophoresis of the product of PCR amplification of exon 2 of the Kb gene. Lane 1, 123-bp ladder. Lane 3, PCR product showing a 260-bp product.
DNA sequencing
The sequencing reaction was carried out using the DNA sequencing kit from Gibco BRL (Life Technologies Inc., Gaitherburg, MD) following the manufacturer's instructions and the procedure of Innis et al.15 A partial sequence was then read and compared to the published sequence of exon 2 of the H-2 Kb gene using the Microgenie program (Beckman, Fullerton, CA). Our sequence for the H-2 Kb PCR product was identical to the published one.12
Isolation of peripheral blood mononuclear cells
Blood was withdrawn from a retro-orbital sinus of the mice using heparinized capillary tubes. Briefly, 0·3 ml of blood was mixed with mixed 7 ml of a phosphate-buffered saline (PBS) solution containing 0·1% ethyleneglycoltetraacetic acid (EGTA) and 25 units of heparin (Sigma). This suspension was then layered on a Ficoll-Hypaque solution (Sigma) and centrifuged. The white cell layer was then removed and resuspended in ammonium chloride to lyse the red blood cells. The white blood cells were washed, and were then ready for use in the RNA extraction procedure.
RNA extraction
A total of 106 cells were resuspended in 300 µl of lysis buffer (NTE-SDS; sodium chloride, Tris, EDTA-sodium dodecyl sulphate) at 37° for 30 min. The lysate was then extracted with 300 µl of phenol:chloroform:isoamyl alcohol (50 : 50 : 1). The samples were then mixed and centrifuged for 3 min at 13 500 g. The upper aqueous phase was collected into 1·5-ml Eppendorf tubes and saved on ice. One hundred µl of NTE-SDS was added to the phenol phase and re-extracted as above. The second aqueous phase was collected and pooled with the first. Forty µl of 3 m sodium acetate (pH 5·2) and 1100 µl of −20° 95% ethanol were added to each tube. The tubes were then mixed and incubated overnight at −70°. Samples were then centrifuged at 13 500 g for 30 min at 4°. The ethanol was then discarded and the pellet was dried using a Speed Vac (GMI Inc, Abbotsville, MN). Each pellet was then resuspended in 15 µl of 0·1 NaCl, 10 mm Tris (pH 7·5), 5 mm MgCl2 and 20 µg/ml DNAse I. The DNA digestion was carried out at 37° for 15 min. The resulting samples were used as the source of RNA for the slot-blot hybridization protocols.
Slot blot of RNA and hybridization
Slot blot was carried out following a procedure previously described.16 RNA samples were first denatured with 20 mm Na2HPO4, 1 mm EDTA, 12·5 µl of 12·3 m formaldehyde and 35 µl of formamide, and heated at 60° for 5 min. Each sample was diluted with 30x sodium chloride, sodium citrate and blotted onto a nylon filter using a slot blotter from Schleicher & Schuell (Keene, NH). Filters were air-dried and kept dried until hybridization. Probes were labelled with 32P by the random labelling method, using the oligolabelling kit from Pharmacia (Piscataway, NJ). Hybridization was performed according to published methods.10,17 Samples were placed in sealed plastic bags containing prehybridization solution (5 ml formamide, 1 ml 10% SDS solution, 2 ml 50% dextran sulfate solution and 2 ml H2O). The radiolabelled probe (107 dpm) was then added to the prehybridization solution in the bag. The hybridization was allowed to proceed with constant agitation in a water bath at 42° for 12–15 hr. The filters were then washed and exposed to X-ray film. The quantification of mRNA was performed by reading the X-ray film with a GS 300 scanning densitometer (Hoefer Scientific Instruments, San Francisco, CA) linked to an IBM PC computer (IBM, Whit). The results, expressed as areas, were subsequently stastistically analysed.
Statistical analysis
Results were analysed statistically using analysis of variance in the statgraphics computer program (STSC, IBM) for the IBM-PC.
Results
Standard DNA probes
Slot-blot hybridization of each probe (actin, exon 2 of Kb, TAP1 and TAP2) showed a linear relationship between the amount of probe used and the density of the signal on the autoradiogram. These results were used to ensure that the correct exposure time was used for each autoradiogram as well as to estimate the amount of mRNA in the samples.
TAP1 mRNA expression in blood PBMCs according to age
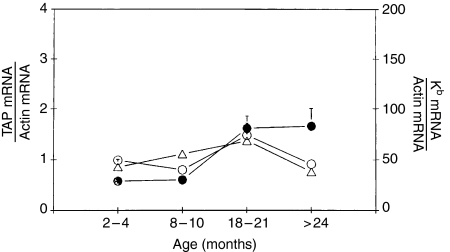
Because actin mRNA levels have been shown to remain constant with age,16 we have normalized the results for TAP1, TAP2 and Kb gene transcripts in each sample to actin mRNA content. Transcripts of TAP1 genes increased with age in C57BL/6 mice from 3 to 24 months. The amount of TAP1 mRNA divided by the amount of actin showed a significant increase with age (1·5–2-fold). However, this amount decreased in very old mice. (Table 2; Fig. 2).
Table 2.
Effect of age on mRNA levels of MHC class I (Kb), TAP1, and TAP2 in peripheral blood mononuclear cells
| mRNA level | ||||
|---|---|---|---|---|
| Age | ||||
| (months) | n | MHC class I | TAP1 | TAP2 |
| 2–4 | 9 | 43 ± 7·5 | 0·99 ± 0·13 | 0·58 ± 0·13 |
| 8–10 | 9 | 55·5 ± 3·5 | 0·80 ± 0·05 | 0·60 ± 0·06 |
| 18–21 | 9 | 68·5 ± 8 | 1·48 ± 0·24 | 1·62 ± 0·23 |
| >24 | 9 | 38 ± 5 | 0·91 ± 0·10 | 1·66 ± 0·34 |
| P-value | <0·02 | ns | <0·01 | |
ns, not significant.
Figure 2.
Summary of changes in MHC class I Kb mRNA (○), TAP1 mRNA (○), and TAP2 mRNA (•) in resting C57BL/6 mouse PBMCs according to age.
TAP2 mRNA expression in blood PBMCs according to age
TAP2 mRNA also significantly increased with age. The ratio of TAP2 mRNA to actin mRNA increased steadily with age to reach a level three times higher in > 24 month old mice than in 4 month old mice (Table 2; Fig. 2).
Kb mRNA expression in PBMCs according to age
Kb mRNA in PBMCs was 1·5-fold higher in mice 18–21 months old than in mice 2–4 months old. However, this increase was reversed in old mice, in which mRNA levels were similar to those found in very young mice (Table 2; Fig. 2).
TAP1 mRNA expression in spleen cells according to age
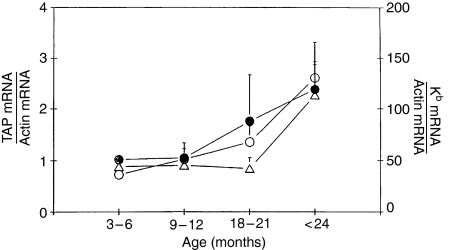
Transcripts of TAP1 genes increased with age in spleen cells of C57BL/6 mice from 3 to 24 months. The amount of TAP1 mRNA divided by the amount of actin showed a significant (3-fold) increase with age (Table 3; Fig. 3).
Table 3.
Effect of age on mRNA levels of MHC class I (Kb), TAP1, and TAP2 in spleen lymphocytes
| mRNA level | ||||
|---|---|---|---|---|
| Age | ||||
| (months) | n | MHC class I | TAP1 | TAP2 |
| 3–6 | 6 | 44 ± 6·5 | 0·73 ± 0·08 | 1·02 ± 0·13 |
| 9–12 | 6 | 45·5 ± 8 | 1·02 ± 0·21 | 1·05 ± 0·29 |
| 18–21 | 6 | 42·5 ± 10·5 | 1·36 ± 0·31 | 1·76 ± 0·91 |
| >24 | 6 | 113·5 ± 30 | 2·61 ± 0·69 | 2·38 ± 0·55 |
| P-value | <0·02 | <0·03 | ns | |
ns, not significant.
Figure 3.
Summary of changes in MHC class I Kb mRNA (○), TAP1 mRNA (○), and TAP2 mRNA (•) in resting C57BL/6 mouse spleen lymphocytes according to age.
TAP2 mRNA expression in spleen cells according to age
TAP2 mRNA also significantly increased with age in mouse spleen cells. The ratio of TAP2 mRNA to actin mRNA increased steadily with age to reach levels 2·5 times higher in > 24 month old mice than in 3–6 month old mice (Table 3; Fig. 3).
Kb mRNA expression in spleen cells according to age
Kb mRNA in spleen cells was 1·5-fold higher in mice 18–21 months old than in mice 3–6 months old. The ratio of Kb mRNA to actin mRNA increased steadily with age to reach levels 2·5 times higher in > 24 month old mice than in 3–6 month old mice (Table 3; Fig. 3).
Comparison of the levels of MHC class I Kb mRNA and Tap mRNA
The time needed to visualize mRNA from filters probed with either the TAP1 probe or the TAP2 was about 3 times longer than the time needed to visualize mRNA for Kb. The standard curve obtained by hybridizing a known amount of probe blotted onto filters gives an estimation of the approximate amount of message in the samples. While the levels of TAP1 mRNA and TAP2 mRNA were similar, they were about 50 times lower than the levels of MHC class I mRNA (Figs 2 and 3).
Discussion
Numerous studies have analysed MHC class I transcription.18–23 However, only a few studies have addressed changes in MHC transcription with age.10,16 MHC class I protein expression increases with age in mice.16 This increase is also observed at the transcription level. Janick-Buckner et al. showed a 6–9-fold increase in MHC mRNA in old mice.16 Our data reveal a 3-fold increase with age in MHC mRNA. We may have obtained a lower increase because we used a different probe (exon 2 of Kb), which is more specific for Kb mRNA than the all-class-I probe used by Janick-Buckner et al.16 However, the increase in cell surface MHC protein was lower (1·5–2-fold). This suggests that there is a post-transcriptional age-dependent regulation of MHC class I expression. Our mRNA study and our protein study using an antibody specific for Kb offer a focused way of analysing the expression of the Kb gene. The stability of MHC class I protein depends on β2 microglobulin and peptide.7,24–26 MHC class I proteins are known to assemble in the lumen of the endoplasmic reticulum.27,28 Peptide transporters play an important role in this system by bringing peptide to the rough endoplasmic reticulum where the association takes place.
The TAP1 and TAP2 genes belonging to the ATP-binding cassette family of transporter genes, and encode proteins which provide peptides necessary for the assembly of MHC class I molecules by transporting these peptides into the endoplasmic reticulum. As MHC class I expression changes with age, we explored the effect of age on the transcription of MHC class I (Kb), TAP1 and TAP2 genes in C57BL/6 mice. We found that peptide transporter gene expression also increased with age. There was a parallel age-related increase (1·5-fold) in blood lymphocyte mRNA for these genes from 3 to 21 months. However, in mice over 24 months old there was a decrease in Kb and TAP1 mRNA, but an increase in TAP2 mRNA. In spleen lymphocytes there was an increase in all three mRNA species throughout life. The difference between blood lymphocyte and spleen may be attributed to the difference in the T-cell/B-cell ratio. Also, a preferential homing effect of lymphocytes expressing high levels of Kb mRNA, TAP1 and TAP2 mRNA in the spleen may explain the difference. While MHC class I and Tap genes underwent similar age-related changes, MHC class I mRNA was about 50 times more abundant than either TAP1 or TAP2 mRNA. This increase was in parallel with the increase in MHC class I expression. The transporter genes may be under the same type of regulation as the MHC class I genes. In fact, like MHC genes, their transcription is dramatically increased by gamma interferon.5 The body distribution of their expression mirrors the distribution of MHC class I expression. Their role in the expression of MHC protein is crucial as it has been found that mutant cells having a deletion in TAP1 or TAP2 lack cell surface MHC class I proteins.6,25 Polymorphisms in both TAP1 and TAP2 genes have been found.29–31 As it has been shown that certain MHC haplotypes are associated with long life and others are associated with short life,32–38 it would be interesting to explore whether there is any relationship between Tap alleles and lifespan.
It is possible that coordinated expression of MHC class I and TAP1 or TAP2 may predispose an animal to better antigen presentation and a longer life. In addition to transporter genes, genes of the proteasome, the enzyme complex responsible for generating peptide by cutting proteins, should be investigated.39 Two of these genes, LMP-7 and LMP-2, have been located in the MHC complex.40,41 Much more work is needed on proteasomes; for example, the existence of polymorphism in proteasomes needs to be explored. The peptide transporters and the proteasomes may work in close association to make available to the MHC class I protein the right type of peptide which is crucial both for MHC protein stability and for antigen presentation. Defect or polymorphism in Tap genes has been associated various diseases, such as cystic fibrosis and Sjogren's disease.42,43 Regulation of the expression of Tap genes is influenced by various cytokines, such as interferon (IFN)-γ, IFN-α and tumour necrosis factor (TNF)-α.43,44 However, in a recent report, a defect in Tap gene expression was found to remain asymptomatic, raising the possibility of compensatory mechanisms.45 Our study shows a parallel up-regulation of MHC and Tap genes. It is possible that the age-related up-regulation of Tap transcription followed by translation would allow an increase in the assembly of MHC molecules and therefore an increase in the density of these molecules on the cell surface.
Acknowledgments
This work was supported by NIH grant AG02440. Dr Alain G. Assounga was a recipient of a Fellowship award from John L. Archibald.
References
- 1.Spies T, Bresnahan M, Bahram S, Arnold D, Blanck G, Mellins E, Pious D, DeMars R. A gene in the human major histocompatibility complex class II region controlling the class I antigen presentation pathway. Nature. 1990;348:744–7. doi: 10.1038/348744a0. [DOI] [PubMed] [Google Scholar]
- 2.Deverson EV, Gow IR, Coadwell WJ, Monaco JJ, Butcher GW, Howard JC. MHC class II region encoding proteins related to the multidrug resistance family of transmembrane transporters. Nature. 1990;348:738–41. doi: 10.1038/348738a0. [DOI] [PubMed] [Google Scholar]
- 3.Roitt I, Brostoff J, Male D, editors. Immunology. 6th edn. London: Gower Medical Publishing; 2001. [Google Scholar]
- 4.Townsend A, Bodmer H. Antigen recognition by class I-restricted T lymphocytes. Annu Rev Immunol. 1989;7:601–24. doi: 10.1146/annurev.iy.07.040189.003125. [DOI] [PubMed] [Google Scholar]
- 5.Monaco JJ. A molecular model of MHC class-I- restricted antigen processing. Immunol Today. 1992;13:125–32. doi: 10.1016/0167-5699(92)90122-N. [DOI] [PubMed] [Google Scholar]
- 6.Spies T, DeMars R. Restored expression of major histocompatibility class I molecules by gene transfer of a putative peptide transporter. Nature. 1991;351:323–4. doi: 10.1038/351323a0. [DOI] [PubMed] [Google Scholar]
- 7.Attaya M, Jameson S, Martinez CK, et al. Ham-2 corrects the class I antigen-presenting defect in RMA-S cells. Nature. 1992;355:647–9. doi: 10.1038/355647a0. [DOI] [PubMed] [Google Scholar]
- 8.Goldberg AL, Rock KL. Proteolysis, proteasome and antigen presentation. Nature. 1992;357:375–9. doi: 10.1038/357375a0. [DOI] [PubMed] [Google Scholar]
- 9.Janick-Buckner D, Briggs CJ, Meyer TE, Harvey N, Warner CM. Major histocompatibility complex antigen expression on lymphocytes from strain A mice. Growth Devel Aging. 1991;55:53–62. [PubMed] [Google Scholar]
- 10.Sambrook J, Fritsch EF, Maniatis T, editors. Molecular Cloning: A Laboratory Manual. 2nd edn. Vol. 1. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. pp. 6.1–6.62. [Google Scholar]
- 11.Kawasaki ES. Sample preparation from blood, cells, and other fluids. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR ProtocolsA Guide to Methods and Applications. San Diego, CA: Academic Press; 1990. pp. 146–52. [Google Scholar]
- 12.Weiss E, Golden L, Zakut R, Mellor A, Fahrner K, Kvist S, Flavell RA. The DNA sequence of the H-2 Kb gene: evidence for gene conversion as a mechanism for the generation of polymorphism in histocompatibility antigens. EMBO J. 1983;2:453–62. doi: 10.1002/j.1460-2075.1983.tb01444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gyllenten UB. PCR and DNA sequencing. Biotechniques. 1989;7:700–8. [PubMed] [Google Scholar]
- 14.Innis MA, Gelfand DH. Optimization of PCRs. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. San Diego, CA: Academic Press; 1990. pp. 3–20. [Google Scholar]
- 15.Innis MA, Myambo KB, Gelfand DH, Brow MAD. DNA sequencing with thermus acquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc Natl Acad Sci USA. 1988;85:9436–40. doi: 10.1073/pnas.85.24.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Janick-Buckner D, Warner CM. An analysis of class I and class II major histocompatibility antigen expression on C57BL/6 lymphocytes during aging. In: Harrison D, editor. Genetic Effects on Aging II. Caldwell, NJ: Telford Press; 1990. pp. 413–27. [Google Scholar]
- 17.Beck Keeney J, Hansen TH. Cis-acting elements determine the locus-specific shutoff of class I major histocompatibility genes in murine S49 lymphoma sublines. Proc Natl Acad Sci USA. 1989;86:6288–92. doi: 10.1073/pnas.86.16.6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korber B, Mermod N, Hood L, Stroynowski I. Regulation of gene expression by interferons. Control of H-2 promoter responses. Science. 1988;239:1302–6. doi: 10.1126/science.3125612. [DOI] [PubMed] [Google Scholar]
- 19.Singer DS, Maguire JE. Regulation of the expression of class I MHC genes. Crit Rev Immunol. 1990;10:235–7. [PubMed] [Google Scholar]
- 20.David-Watine B, Israel A, Kourilsky P. The regulation and expression of MHC class I genes. Immunol Today. 1990;11:286–92. doi: 10.1016/0167-5699(90)90114-o. [DOI] [PubMed] [Google Scholar]
- 21.Burke PA, Hirschfeld Shirayoshi Y, Kasik JW, Hamada K, Appella E, Ozato K. Developmental and tissue-specific expression of nuclear proteins that bind the regulatory element of the major histocompatibility complex class I gene. J Exp Med. 1989;169:1309–21. doi: 10.1084/jem.169.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drezen JM, Babinet C, Morello D. Transcriptional control of MHC class I and beta 2 microglobulin genes in vivo. J Immunol. 1992;150:2805–13. [PubMed] [Google Scholar]
- 23.Powis SJ, Townsend ARM, Deverson EV, Bastin J, Butcher GW, Howard JC. Restoration of antigen presentation to the mutant cell line RMA-S by an MHC-linked transporter. Nature. 1991;354:528–31. doi: 10.1038/354528a0. [DOI] [PubMed] [Google Scholar]
- 24.Degen E, Cohen-Doyle MF, Williams DB. Efficient dissociation of the p88 chaperone from major histocompatibility complex class I molecules requires both beta 2 microglobulin and peptide. J Exp Med. 1992;175:1653–61. doi: 10.1084/jem.175.6.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dobberstein B. Who needs peptide transporters? Nature. 1992;355:109–10. doi: 10.1038/355109a0. [DOI] [PubMed] [Google Scholar]
- 26.Cox JH, Yewdell JW, Eisenlohr LC, Johnson PR, Bennink JR. Antigen presentation requires transport of MHC class I molecules from the endoplasmic reticulum. Science. 1990;247:715–21. doi: 10.1126/science.2137259. [DOI] [PubMed] [Google Scholar]
- 27.Monaco JJ, Cho S, Attaya M. Transport protein genes in the murine MHC. Possible implications for antigen processing. Science. 1990;250:1723–6. doi: 10.1126/science.2270487. [DOI] [PubMed] [Google Scholar]
- 28.Powis SJ, Deverson EV, Coadwell WJ, Ciruela A, Huskisson NS, Smith H, Butcher GW, Howard JC. Effect of polymorphism of an MHC-linked transporter on the peptides assembled in a class I molecule. Nature. 1992;357:211–5. doi: 10.1038/357211a0. [DOI] [PubMed] [Google Scholar]
- 29.Powis SH, Mockridge I, Kelly A, Kerr LA, Glynne R, Gileadi U, Beck S, Trowsdale J. Polymorphism in a second ABC transporter gene located within the class II region of the human major histocompatibility complex. Proc Natl Acad Sci USA. 1992;89:1463–7. doi: 10.1073/pnas.89.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carrington M, Colonna M, Spiers T, Stephens JC, Mann D. 1. Haplotype variation of the transporter associated with antigen processing (TAP) genes and their extension of HLA class II region haplotypes. Immunogenetics. 1993;36:1–8. doi: 10.1007/BF00187452. [DOI] [PubMed] [Google Scholar]
- 31.Walford RL. Antibody diversity, histocompatibility systems, disease states, and ageing. Lancet. 1970;ii:1226–9. doi: 10.1016/s0140-6736(70)92184-7. [DOI] [PubMed] [Google Scholar]
- 32.Walford RL. Immunologic theory of aging: current status. Federation Proc. 1974;33:2020–7. [PubMed] [Google Scholar]
- 33.Yunis EJ, Greenberg LJ. Immunopathology of aging. Federation Proc. 1974;33:2017–9. [PubMed] [Google Scholar]
- 34.Smith GS, Walford R. Influence of the main histocompatibility complex on ageing in mice. Nature. 1977;270:727–9. doi: 10.1038/270727a0. [DOI] [PubMed] [Google Scholar]
- 35.Popp DM, Popp RA. Genetics. In: Kay MB, Makinodan T, editors. CRC Handbook of Immunology and Aging. Boca Raton, FL: CRC Press; 1981. pp. 15–60. [Google Scholar]
- 36.Gelman R, Watson A, Bronston R, Yunis E. Murine chromosomal regions correlated with longevity. Genetics. 1988;118:693–704. doi: 10.1093/genetics/118.4.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer TE, Armstrong MJ, Warner CM. Effects of H-2 haplotype and gender on the lifespan of A and C57BL/6 mice and their F1, F2, and backcross offspring. Growth Dev Aging. 1989;53:175–83. [PubMed] [Google Scholar]
- 38.Brown MG, Driscoll J, Monaco JJ. Structural and serological similarity of MHC-linked LMP and proteasome (multicatalytic proteinase) complexes. Nature. 1991;353:355–7. doi: 10.1038/353355a0. [DOI] [PubMed] [Google Scholar]
- 39.Martinez CK, Monaco JJ. Homology of proteasome subunits to a major histocompatibility complex-linked LMP gene. Nature. 1991;353:664–7. doi: 10.1038/353664a0. [DOI] [PubMed] [Google Scholar]
- 40.Monaco JJ, McDevitt HO. H-2-linked low-molecular weight polypeptide antigens assemble into an unusual macromolecular complex. Nature. 1984;309:797–9. doi: 10.1038/309797a0. [DOI] [PubMed] [Google Scholar]
- 41.Bauer D, Tampe R. Herpes viral proteins blocking the transporter associated with antigen processing TAP-from genes to function and structure. Curr Top Microbiol Immunol. 2002;269:87–99. [PubMed] [Google Scholar]
- 42.Anaya JM, Correa PA, Mantilla RD, Arcos-Burgos M. TAP, HLA-DQB1, HLA-DRB1 polymorphism in Colombian patients with primary Sjogren's syndrome. Semin Arthritis Rheum. 2002;31:396–405. doi: 10.1053/sarh.2002.32557. [DOI] [PubMed] [Google Scholar]
- 43.Abarca-heidenmann K, Friederichs S, Klamp T, Boehm U, Guethlein LA, Ortmann B. Regulation of the expression of mouse TAP-associated glycoprotein tapasin by cytokines. Immunol Lett. 2002;83:197–207. doi: 10.1016/s0165-2478(02)00104-9. [DOI] [PubMed] [Google Scholar]
- 44.Schiffer R, Baron J, Dagtekin G, Jahnen-dechent W, Zwaldo-Klarwasse G. Differential regulation of the expression of transporters associated with antigen processing, TAP1 and TAP2. Inflamm Rest. 2002;51:403–8. doi: 10.1007/pl00000321. [DOI] [PubMed] [Google Scholar]
- 45.De la Salle H, Saulquin X, Mansour I, et al. Asymptomatic deficiency in the peptide transporters associated to antigen processing (TAP) Clin Exp Immunol. 2002;128:525–31. doi: 10.1046/j.1365-2249.2002.01862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]