Abstract
The memory T-cell population is a heterogeneous population, including both effector cells, which exert a direct secondary immune response, and resting or intermediate cells, which serve as a reservoir and exert a possible regulatory role. To further dissect the T-cell memory population residing in the CD4+ CD45RO+ T-cell pool, we studied the functional properties of memory populations identified by the CD27 marker. This marker clearly divides the memory population into two groups. One group consists of effector cells lacking CD27 and displaying a high antigen recall response. The other group consists of an intermediate memory population, displaying CD27. This latter group lacks an antigen recall response and requires costimulation for T-cell receptor triggering. To evaluate the function of the CD27+ memory pool, we analysed the transcriptional profile, using high-density microarray technology. These gene data strongly support the different functional profiles of CD27+ and CD27− memory populations, in terms of protein expression and the capacity to respond to antigen.
Keywords: cellular activation, human, T lymphocytes, Th1/Th2 cells
Introduction
T-cell memory is a crucial property of the immune system, characterized by an increased frequency of antigen-specific T cells with direct effector activity.1 Memory T cells differ from their naïve counterparts in phenotypic profiles as well as in functionality. Human peripheral blood CD4+ T lymphocytes can be divided into naïve CD45RA+ cells and memory CD45RO+ cells.2,3 These two populations differ in their recall response to antigen, with a higher proliferative response in the memory population than in the naïve population.4,5 Furthermore, memory T cells produce high levels of interleukin (IL)-2, IL-4 and interferon-γ (IFN-γ), whereas naïve T cells produce high levels of IL-2 but only trace amounts of IL-4 and IFN-γ.1 Surface molecules, apart from CD45, which are differentially regulated during T-cell activation and thus memory formation, are CD62L and CD44.1 When T lymphocytes home to secondary lymphoid organs, they roll on high endothelial venules using CD62L (also known as L-selectin) before migrating across the endothelium.6 This is facilitated by secretion of the chemokines CCL19 and CCL21 by endothelial cells at high endothelial venules, which attracts chemokine receptor CCR7+ T cells.7–9 CCR7 is expressed by all naïve CD4+ T cells, but only on a subpopulation of the human CD4+ memory T-cell pool.10 The CCR7− memory T cells are also suggested to act as effector cells migrating through peripheral tissues as compared to the central memory cells (CCR7+), which home to lymph nodes to mount a secondary signal.11 Another activation marker that can divide the memory pool is CD27, belonging to the tumour necrosis factor/nerve growth factor receptor (TNF/NGF-R) family.12 CD27 is expressed by all naïve CD4+ T cells and by ≈ 80% of the CD4+ memory population.13 The loss of CD27 has been reported to be a characteristic of the induced effector phenotype of CD4+ T cells13 and CD8+ T cells.14
To further dissect the T-cell memory phenotype, we sorted these two memory populations according to the marker CD27 and demonstrate here that CD27 provides a clear division of memory into responding versus resting cells, based on functional properties as well as transcriptional profiles. The results clearly demonstrate that the CD27− memory population is more differentiated, with high-level secretion of a broad panel of effector cytokines and a strong antigen-recall response compared with the CD27+ population. The CD27+ population, on the other hand, lacks both responsiveness towards different antigens and T-cell receptor triggering, without the help of costimulation. Furthermore, the transcriptional profiles of CD27+ and CD27− populations demonstrate that CD27− cells are more differentiated, based on their differential expression of, e.g. transcription factors, chemokines and activation markers, than CD27+ cells.
Materials and methods
Cell purification protocols
Blood mononuclear cells were isolated from buffy coats obtained from the University Hospital Lund (Lund, Sweden) and Novozyme A/S (Copenhagen, Denmark), by Ficoll-Isopaque (Amersham, Biosciences, Uppsala, Sweden) density centrifugation. CD4+ T cells were purified by negative selection, using CD4+ T-cell isolation MACS kits (Miltenyi Biotec, Bergisch Gladbach, Germany). The enrichment of CD4+ T cells was > 95%. Naïve CD4+ T cells were also isolated by negative selection. The cells were first incubated with anti-CD45RO monoclonal antibody (mAb) and anti-glycophorin A mAb (DakoCytomation, Glostrup, Denmark) for 30 min at 4°. Thereafter, pan mouse IgG immunomagnetic beads (Dynal, A.S., Oslo, Norway) were added for 30 min at 4° to deplete memory cells. The isolated naïve CD4+ T cells contained < 1% contaminating CD45RO+ T cells. Memory CD4+ T cells were also isolated by negative selection, using CD45RA mAb, anti-glycophorin A mAb (DakoCytomation) and CXCR5 mAb (R & D Systems Inc., Minneapolis, MN), to deplete activated cells, followed by pan mouse IgG immunomagnetic beads (Dynal, A.S.). The isolated memory CD4+ T cells contained < 1% contaminating CD45RA+ T cells. The memory CD4+ T cells were separated into CD27+ and CD27− cells, using phycoerythrin (PE)-conjugated CD27 mAb (DakoCytomation) or CD27-antibody-coated MACS beads (Miltenyi Biotec). The cells were washed, and CD27+ and CD27− T cells were obtained by cell sorting using a FACS Vantage SE (Becton Dickinson, Franklin Lakes, NJ) or with a MACS isolation kit/column, rendering > 98% CD27+ T cells and > 89% CD27− T cells. Irradiated (2·000 rads) peripheral blood mononuclear cells (PBMC) were used as antigen-presenting cells. Phosphate-buffered saline (PBS) (Ca/Mg-free) (Sigma-Aldrich, St Louis, MO), containing 1% bovine serum albumin (BSA) (Sigma-Aldrich) was used in all cell labelling and washing steps.
Reagents
Tetanus toxoid (TT) (25 µg/ml) (SBL, Uppsala, Sweden), streptokinase (200 U/ml) (Sigma-Aldrich), Candida albicans enolase (10 µg/ml) (TaKaRa Shuzo Co., Outso, Japan) or lipase (lipolase) (1 µg/ml) (Novozyme A/S) were used for antigen-specific stimulation. For the T-cell polarization assay, phytohaemagglutinin (PHA) (1 µg/ml) (Sigma-Aldrich) and IL-2 (10 ng/ml) were used as costimulators. IL-4 (20 ng/ml) and anti-IFN-γ (0·5 µg/ml) were used in T helper 2 (Th2) polarization, and IL-12 (5 ng/ml), anti-IL-4 (5 µg/ml) and anti-IL-4-receptor (0·5 µg/ml) (all from R & D Systems Inc.) were used in T helper 1 (Th1) polarization. The cytokines and antibodies were added to the culture once weekly. Th1 polarization was allowed to continue for 20 days and the Th2 polarization for 28 days, to achieve maximum polarization.15
Proliferation assay
Proliferation assays were performed with purified CD27+ CD4+ or CD27− CD4+ memory T cells, or with CD4+ naïve T cells (5 × 104 cells), co-cultured with autologous irradiated PBMC (1 × 105). The cells were pulsed with TT, streptokinase, Candida enolase or lipase, in U-bottomed 96-well plates, using RPMI-1640 (Sigma-Aldrich) supplemented with 10% (v/v) fetal calf serum (FCS), 2 mm l-glutamine, non-essential amino acids (Life Technologies, Grand Island, NY) and 2 × 10−5 m 2-mercaptoethanol (Merck, Whitehouse, NJ, USA). After 8 days of incubation, each well was pulsed with 2·5 µCi/ml [3H]thymidine (Amersham Biosciences). Samples were harvested and radioactivity determined in a scintillation counter (Microbeta 1450; Wallac, Turku, Finland). Samples were run in four replicates. Purified CD27+ CD4+ or CD27− CD4+ memory T cells, or CD4+ naïve T cells (2 × 104 cells), were stimulated with plastic-bound, anti-CD3 mAb (Okt-3) (Ortho Biotech, Raritan, NJ) (concentrations from 0·001 to 10 µg/ml), in the absence of antigen-presenting cells, with or without the addition of anti-CD28 immunoglobulin (0·5 µg/ml) (Pharmingen, San Diego, CA). After 6 days, each well was pulsed for 16 hr with 2·5 µCi/ml [3H]thymidine and analysed.
Flow cytometry analyses
For intracellular cytokine staining, the cells were incubated with phorbol 12-myristate 13-acetate (PMA) (50 ng/ml), ionomycin (500 ng/ml) and brefeldin A (10 µg/ml) (all from Sigma-Aldrich) for 5 hr, washed twice in PBS and fixed with 2% paraformaldehyde (Sigma-Aldrich) for 15 min. The cells were first washed in PBS and then in 0·5% saponin (Sigma-Aldrich) in PBS containing 0·5% BSA. The cells were incubated with PE-conjugated anti-IL-4 (Becton Dickinson), fluorescein isothiocyanate (FITC)-conjugated anti-IFN-γ, FITC-conjugated anti-IL-2, PE-conjugated anti-IL-13 and PE-conjugated anti-IL-10 (Pharmingen) for 30 min at 4°. Thereafter, the cells were washed twice in PBS containing 0·5% saponin and 0·5% BSA, and once in PBS containing 0·5% BSA, but no saponin, and then analysed on a FACScan (Becton Dickinson). Chemokine receptor Th2 (CR-Th2)-conjugated biotin mAb was a generous gift from Professor Kinya Nagatas (R & D Center, BML, Kawagoe Saitama, Japan). The cells were incubated with CR-Th2-conjugated biotin for 30 min at 4°, washed twice and incubated with Alexa 488 conjugated-streptavidin (Molecular Probes, Eugene, OR) for 30 min at 4°, and then analysed on a FACScan (Becton Dickinson) after a final wash.
Preparation of cRNA and gene chip hybridization
Total RNA was isolated from CD27+ and CD27− CD4+ memory T cells from two different donors. Cell samples were lysed in TRIzol Reagent (Invitrogen, Paisley, UK) and stored at −20° until further RNA isolation. Fragmentation, hybridization and scanning of the human U133A arrays were performed according to the manufacturer's protocol (Affymetrix Inc., Santa Clara, CA). Preparation of labelled cRNA was performed according to the small-sample labelling protocol vII (Affymetrix Inc.). Briefly, cDNA was generated from total RNA (0·25–2·4 µg), using SuperScript II (Invitrogen) and a T7-Oligo(dT) promoter primer (Affymetrix Inc.). After a second-strand cDNA synthesis and clean-up with ethanol precipitation, cDNA was converted to cRNA by an in vitro transcription reaction (MEGAscript T7 kit; Ambion, Austin, TX). Thereafter, the cRNA was purified using an RNeasy Mini Kit (Qiagen, Valencia, CA) and the yield controlled with spectrophotometry. A second cycle of cDNA sysnthesis was performed, followed by clean-up (as described above) and a second in vitro transcription reaction cycle with biotin-labelled ribonucleotides and T7 RNA polymerase (ENZO, Farmingdale, NY). Labelled cRNA was purified using an RNeasy Mini Kit (Qiagen) and denatured at 94° before hybridization. The samples were hybridized to the Human Genome U133A arrays at 45° for 16 hr by rotation (60 r.p.m.) in an oven. The arrays were then washed, stained with Steptavidin-PE (Molecular Probes), washed again and scanned with a GeneArray™ Scanner (Affymetrix Inc.).
Microarray data analysis
The fluorescence intensity was analysed using the Microarray Suite Software 5·0 (Affymetrix, Inc.). The arrays were scaled based on average intensity on the chips, and a comparative analysis was performed on the CD27+ and CD27− cell populations. Differentially expressed genes were selected based on the increase/decrease call, absent/present call and a fold change in the expression level ±2. Thus, a SLR of 1·0 indicates a twofold increase of the transcript level.
Results
Antigen recall response could only be detected within the CD27− CD4+ memory population
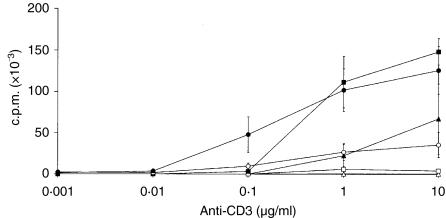
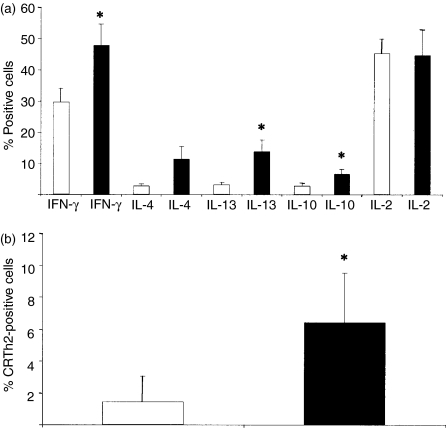
The CD4+ T cells were divided into naïve, CD27+ and CD27− memory cells, which were co-cultured with autologous irradiated PBMC pulsed with TT, streptokinase, Candida enolase or recombinant lipase. The CD27− memory population was the only cell population that displayed a clear-cut recall response to all four antigens (Fig. 1a). The CD27+ memory population showed a recall response as low as the naïve population. Requirements for triggering the T-cell response were further studied by seeding cells on plastic-bound anti-CD3 immunoglobulin (in the absence of antigen-presenting cells) with or without the additional stimulus of anti-CD28 immunoglobulin (Fig. 2). T-cell-receptor triggering, with anti-CD3 immunoglobulin but without CD28 costimulation, demonstrated responsiveness only of the CD27− cells. The CD27+ and the naïve cells did not display any proliferation in response to anti-CD3 stimulation. In contrast, the further addition of CD28 mediated costimulation and full responsiveness of the CD27+ cells. The naïve cells also responded to costimulation with both CD3 and CD28. However, a dose-dependent difference was observed, as the naïve cells required 10 µg/ml of anti-CD3 immunoglobulin, CD27+ cells required 1 µg/ml and CD27− cells only 0·1 µg/ml of anti-CD3 immunoglobulin to respond by proliferation. Figure 2 shows data after 6 days of stimulation, but the same result was also evident after 4 days of stimulation (data not shown). To further investigate the functional properties of the different T-cell memory populations, the CD27+ and CD27− cells were analysed regarding their production of IFN-γ, IL-4, IL-13 and IL-10, using intracellular cytokine staining and flow cytometry analysis (Fig. 3a). A significantly higher percentage (1·4–3·2-fold) of the CD27− cell population produced the cytokines IFN-γ, IL-13 and IL-10, as compared to the CD27+ cell population. However, no significant difference in IL-2 production was detected between the two populations (Fig. 3a). The two different populations were also analysed for surface expression of the Th2 marker CR-Th2 (Fig. 3b). The CD27− population showed a significantly higher expression of CR-Th2 compared with the CD27+ population. Taken together, these data support the interpretation that the CD27− memory T cells are of an effector phenotype, while the CD27+ population is a non-responding phenotype.
Figure 1.
CD4+ naïve and memory CD27+ and CD27− cells were co-cultured with irradiated autologous peripheral blood mononuclear cells (PBMC) and stimulated with medium (white bar) or one of four different antigens (black bar): (a) tetanus toxoid; (b) streptokinase C; (c) Candida enolase; or (d) lipase. The total incubation time was 8 days. The average of four replicate cultures ± standard deviation is shown, and the data are representative of two to four experiments. Statistical analysis was performed by analysis of variance (anova) (**P < 0·01; ***P < 0·001). c.p.m., counts per minute.
Figure 2.
Proliferative response of CD4+ naïve (triangles), CD27+ (squares) and CD27− (circles) memory T cells to different concentrations of plastic-bound anti-CD3 immunoglobulin (in the absence of antigen-presenting cells) in the absence (white symbols) or presence (black symbols) of anti-CD28 immunoglobulin. The average of three individual experiments ± standard deviation is shown. CD27− cells proliferate at a significantly higher rate (P < 0·01) than CD27+ cells without the addition of co-stimulation, at both 1 and 10 µg/ml concentrations of anti-CD3, in all individual experiments. c.p.m., counts per minute.
Figure 3.
(a) CD27+ (white bars) and CD27− (black bars) memory T cells were stimulated directly with phorbol 12-myristate 13-acetate (PMA) and ionomycin for 5 hr in the presence of Brefeldin A and assayed for their intracellular cytokine production. The cells were stained for interferon-γ (IFN-γ), interleukin (IL)-4, IL-13, IL-10 and IL-2. The average of four to six independent experiments ± standard error is shown. Statistical analysis was performed by Tukey's test (*P < 0·05). (b) Surface expression of the T helper 2 (Th2) marker, chemokine receptor Th2 (CR-Th2) in CD27+ (white bars) and CD27− (black bars) memory T cells that were analysed by flow cytometry. The average of five independent experiments ± standard deviation is shown. Statistical analysis was performed by analysis of variance (anova) (*P < 0·05).
Differential gene expression of CD27+ versus CD27− cells
In an attempt to explain the functional differences in CD27+ and CD27− memory populations, we performed gene expression profiling. A comparative analysis revealed 79 genes to be differentially expressed (Table 1), where homing markers such as CCR7 and CD62L (L-selectin) were up-regulated in the CD27+ cells.
Table 1.
Differential gene expression in CD27+ compared with CD27− memory CD4+ T cells
| GenBank | ||||
|---|---|---|---|---|
| Accession No. | FC* | Description | Definition | |
| AV711904 | −59·7 | LYS | Lysozyme precursor | |
| NM_002964 | −45·3 | S100A8 | S100 calcium-binding protein A8 | |
| AL554008 | −42·2 | GPR56 | G protein-coupled receptor 56 | |
| NM_002432 | −24·3 | MNDA | Myeloid cell nuclear differentiation ag | |
| NM_018434 | −21·1 | G1RZFP | Goliath protein | |
| U20350 | −14·9 | CCRL1 | CX3CR1 | |
| M21121 | −13·9 | CCL5 | RANTES | |
| NM_002003 | −12·1 | FCNM | Ficolin 1 | |
| M27487 | −9·2 | HLA-DPA1 | MHC class II, DP alpha 1 | |
| D14705 | −8·6 | CTNNA1 | Catenin alpha 1 | |
| AJ297586 | −8·6 | HLA-DRB5 | MHC, class II, DR beta 5 | |
| M60334 | −8·0 | HLA-DRA1 | MHC class II, DR alpha | |
| U65585 | −8·0 | HLA-DR1B | MHC class II, DR beta 1 | |
| NM_000632 | −7·0 | CD11b | Integrin alpha M | |
| NM_002121 | −7·0 | HLA-DP1B | MHC class II, DP beta 1 | |
| AB059408 | −6·1 | HOP | Homeodomain-only protein | |
| NM_002664 | −6·1 | PLEK | Pleckstrin | |
| AL563460 | −5·7 | GATA2 | GATA2 | |
| NM_002125 | −5·3 | HLA-DRB5 | MHC class II, DR beta 5 | |
| AF031824 | −5·3 | CST7 | Cystatin F | |
| AB018580 | −4·3 | AKR1C3 | Aldo-keto reductase family 1, C3 | |
| BC003143 | −4·0 | DUSP6 | Dual specificity phosphatase 6 | |
| NM_021983 | −3·7 | HLA-DRB4 | MHC class II, DR beta 4 | |
| BC005912 | −3·7 | FCE1A | Fc fragment of IgE, high affinity I, a | |
| NM_002350 | −3·7 | LYN | Lyn | |
| NM_003243 | −3·7 | TGF-βR3 | TGF-β receptor III | |
| NM_003853 | −3·5 | ACPL | IL-18 receptor accessory protein | |
| NM_014795 | −3·5 | SIP1 | Zinc finger homeobox 1B | |
| NM_003930 | −3·5 | RA70 | Src family associated phosphoprot 2 | |
| NM_013351 | −3·2 | T-BET | T-box 21 | |
| NM_004776 | −3·2 | B4GALT5 | b GlcNAc b 1,4 galactosyltransferase 5 | |
| NM_000024 | −3·0 | ADRB2R | Adrenergic, beta-2-, receptor | |
| NM_007182 | −2·8 | RDA32 | Ras association domain family 1 | |
| BC000182 | −2·6 | ANX4 | Annexin A4 | |
| NM_004642 | −2·6 | DOC1 | CDK2-associated protein 1 | |
| NM_005655 | −2·6 | TIEG | TGF-β inducible early growth response | |
| NM_003595 | −2·6 | TPST2 | Tyrosylprotein sulfotransferase 2 | |
| NM_002984 | −2·6 | MIP-1β | Macrophage inflammatory protein-1β | |
| NM_003189 | −2·6 | TCL5 | T-cell acute lymphocytic leukaemia 1 | |
| NM_018641 | −2·6 | C4S-2 | Chondroitin 4-O-sulfotransferase 2 | |
| NM_002166 | −2·5 | ID2 | Inhibitor of DNA-binding 2 | |
| NM_021822 | −2·5 | APOBEC3G | Apolipoprotein B editing enzyme | |
| NM_002961 | −2·5 | S100A4 | S100 calcium-binding protein A4 | |
| AF020314 | −2·5 | CMRF-35-H9 | Leucocyte membrane antigen | |
| NM_003670 | −2·3 | BHLHB2 | Basic helix-loop-helix domain | |
| AF034607 | −2·3 | NCC27 | Chloride intracellular channel 1 | |
| NM_001814 | −2·1 | CTSC | Cathepsin C | |
| BC004188 | −2·1 | TUBB2 | Tubulin, beta, 2 | |
| AF087942 | −2·1 | GYG | Glycogenin | |
| AL161952 | −2·1 | GLUL | Glutamate-ammonia ligase | |
| NM_002118 | −2·1 | RING7 | MHC, class II, DM beta | |
| NM_005904 | −2·1 | SMAD7 | MAD homologue 7 | |
| NM_001780 | −2·0 | CD63 | CD63 | |
| AF313911 | −2·0 | TXN | Thioredoxin | |
| NM_004052 | 2·0 | NIP3 | BCL2/adenovirus interacting prot 3 | |
| NM_020987 | 2·0 | ANK3 | Ankyrin 3, node of Ranvier | |
| NM_001974 | 2·1 | EMR1 | Hormone receptor-like sequence 1 | |
| NM_003983 | 2·1 | LAT-2 | Solute carrier family 7 member 6 | |
| AF261135 | 2·1 | GPR18 | G protein-coupled receptor 18 | |
| AJ223321 | 2·1 | ZNF238 | Zinc finger protein 238 | |
| NM_000130 | 2·3 | PCCF | Coagulation factor V | |
| AJ250014 | 2·3 | CYLD1 | Cylindromatosis | |
| D79994 | 2·3 | KANK | Kidney ankyrin repeat-containing protein | |
| NM_000210 | 2·5 | CD49f | Integrin alpha 6 | |
| AF113682 | 2·5 | TMPO | Thymopoietin | |
| AW166711 | 2·5 | PIP3-E | Phosphoinositide-binding protein PIP3-E | |
| NM_020651 | 2·5 | PELI1 | Pellino (Drosophila) homologue 1 | |
| NM_006720 | 2·5 | ABLIM1 | Actin binding LIM protein 1 | |
| NM_000655 | 2·6 | CD62L | L-selectin | |
| NM_016340 | 2·6 | RA-GEF-2 | Rap guanine nucleotide exchange factor | |
| AW514267 | 2·6 | NY-REN-7 | NY-REN-7 | |
| AF288571 | 2·6 | TCF1ALPHA | Lymphoid enhancer-binding factor 1 | |
| NM_006159 | 2·8 | NRP2 | Nel-like 2 | |
| NM_002371 | 3·0 | MAL | Mal, T-cell differentiation protein | |
| NM_003328 | 3·2 | BTKL | TXK tyrosine kinase | |
| AI650848 | 3·5 | TBC1D4 | TBC1 domain family, member 4 | |
| D50925 | 4·0 | STK37 | PAS-serine/threonine kinase | |
| NM_001242 | 4·9 | CD27 | CD27 | |
| NM_001838 | 4·9 | CCR7 | EBI1 |
Negative and positive fold change (FC) represent higher levels of transcripts in CD27− and CD27+ cells, respectively.
Genes expressed in the CD27− population were related more to cellular effector functions and the polarization status of the T cells (Table 1). These genes include the two transcription factors T-bet and GATA-2, the chemokine receptor CX3CR1 (expressed by Th1 cells),16 the chemokines RANTES (regulated on activation, normal, T-cell expressed, and secreted) and macrophage inflammatory protein-1β (MIP-1β), and the IL-18 accessory protein (AcPL) (highly expressed in IL-12-secreting T cells).17 Activation markers such as human leucocyte antigen (HLA)-DR and HLA-DP were also up-regulated in the CD27− cells, as were the adhesion molecules CD11b and catenin alpha 1. Taken together, these data demonstrate a much more polarized profile of the CD27− population as compared to the CD27+ population.
Polarization of CD27+ cells as compared to naïve T cells
The naïve population and the memory CD27+ population were stimulated with PHA and IL-2, or by conditions that drive Th1 (IL-12, anti-IL-4 and anti-IL-4R antibodies) or Th2 (IL-4 and anti-IFN-γ antibodies) polarization. The cells were restimulated with PMA, ionomycin and brefeldin A, and the intracellular production of different cytokines was analysed. Both naïve and CD27+ T cells, stimulated under Th1 conditions, demonstrated a strong polarization towards a Th1 cytokine profile, with an increased percentage of cells producing IFN-γ(Fig. 4a). Furthermore, CD27+ cells stimulated under the Th2 conditions resulted in an increased percentage of cells producing the cytokines IL-4 and IL-13 (Fig. 4b). Interestingly, a higher level of IFN-γ, IL-4 and IL-13 production was observed in the memory CD27+ population compared with the naïve population, after stimulation with only IL-2 and PHA. These data demonstrate that the CD27+ memory population could indeed be polarized and display a high degree of plasticity.
Figure 4.
Naïve and CD27+ memory cells were stimulated with phytohaemagglutinin (PHA), interleukin (IL)-2 and either (a) a T helper 1 (Th1) cocktail (IL-12, anti-IL-4 and anti-IL-4R immunoglobulin) for 20 days, or (b) a T helper 2 (Th2) cocktail (IL-4 and anti-IFN-γ immunoglobulin) for 28 days. The cytokines and the antibodies in the different cocktails were distributed once weekly. The data are representative of three similar experiments.
Discussion
During an immune response, a memory T-cell pool is formed. This memory pool can be divided into either effector memory T cells or resting, non-polarized T cells.18 We have compared the functional properties of two different memory populations, defined by expression of the marker CD27 as well as their transcriptional profiles. To define the functional differences in the two memory subpopulations, we examined the recall response against four different antigens (TT, streptokinase, Candida enolase and lipase). In responding donors, the only antigenic recall response was observed in the CD27− memory population, while the CD27+ population had a proliferative response to all four antigens that was as low as the naïve population. Amyes et al. recently demonstrated that the CD27+ CD4+ memory population did not respond to the viral antigen cytomegalovirus (CMV), but responded to Epstein–Barr virus (EBV).19 However, others have demonstrated that the CD27+ cell population showed a much lower percentage of responding cells towards a CMV peptide than the CD27− population.20 In the present study, we also studied the T-cell receptor-mediated response, with or without additional costimulation, to determine differences in activation thresholds in the two memory subpopulations. We found that only the CD27− population responded to anti-CD3 stimulation without additional costimulation, while CD27+ and naïve cells needed additional CD28-mediated stimulation. This result demonstrates that the CD27+ population responded as intermediate cells and not as a true memory phenotype, as the latter does not require costimulation.1 In contrast, in an antigen-specific response, Bitsmansour et al. demonstrated that the CD27+ population did not require costimulation to a higher extent than the CD27− population.20 In the comparative analysis, we found twice as many up-regulated genes in the CD27− population than in the CD27+ population, supporting the interpretation that CD27− cells are in a more activated state. In the CD27+ population, higher expression was found of the homing marker, CCR7, confirming data showing overlapping expression of CD27 and CCR7.21 The homing marker CD62L (L-selectin) is also more highly expressed in the CD27+ population. Kaech et al. have shown, in mice, that CD62L is down-regulated in the more differentiated effector CD8+ T-cell pool.22 CD4+ effector cells down-regulate the expression of both CD62L and CCR7.23 The CD62L has also been demonstrated to become down-regulated in cells down-regulating CD27 upon CD27–CD70 interaction.24 Genes up-regulated in the CD27− population are more involved in cell differentiation. Interesting genes are, for example, T-bet (known as a Th1 transcription factor binding to the ifng promotor)25 and GATA-2 (known to induce GATA-3, a Th2 transcription factor, and to enhance IL-4 and IL-5 production).26–28 These findings support our functional data, demonstrating that CD27− cells secrete the effector cytokines IFN-γ, IL-4, IL-10 and IL-13 to a higher degree and also have higher expression of the Th2 marker CR-Th2,29 as compared to cells in the CD27+ population. Another gene of interest, found in CD27− cells, is the chemokine receptor CX3CR1. This chemokine receptor is preferentially expressed in Th1 cells compared with Th2 cells.16 It has also been demonstrated that the CX3CR1 is expressed almost exclusively by activated HLA-DR+ CD4+ T cells in the CD45RO+ population.30 In addition, we showed that six different major histocompatibility complex (MHC) class II transcripts are detected in the CD27− (CX3CR1+ population) compared with the CD27+ population. The chemokines, regulated on activation, normal, T-cell expressed, and secreted (RANTES) and macrophage inflammatory protein-1β (MIP-1β) are also expressed in the CD27− population. CD4+ Th1 cells have recently been demonstrated to secrete RANTES.31 Finally, the transcripts of AcPL, an accessory protein to the IL-18 receptor,32 are expressed in CD27− cells. A recent study has also shown that AcPL transcripts are expressed at high levels in IL-12-polarized T cells.17 Taken together, these data demonstrate that the CD27− population defines a more polarized cellular state than the CD27+ population within the memory T-cell pool.
In this work, we also demonstrate that the CD27+ memory population has the same ability to polarize towards both Th1 and Th2 as the naïve population, indicating that even though they do not respond to antigen, the CD27+ memory population has a high degree of plasticity. Thus, the CD27+ memory population might act as a backup to CD27− cells during an active immune response, as stimulation of CD27+ memory cells results in loss of the CD27 marker (data not shown). The naïve population and the CD27+ memory population also differ in their cytokine production when stimulated under non-polarizing conditions. A larger pool of cells in the non-polarized CD27+ memory population secretes effector cytokines, such as IL-4, IL-13 and IFN-γ, as compared to the naïve population.
In summary, the CD27− cell population are effector memory cells, as demonstrated at both transcriptional and protein levels. CD27+ CD45RO+ cells, on the other hand, are of a more resting phenotype, and act as a non-memory cell type, according to transcriptional and functional analysis, displaying an inability to respond to antigen and T-cell receptor triggering without costimulation.
Acknowledgments
This work was supported by grants from the European Commission (project number QLK3-CT-2000-00270) and by Knowledge Foundation in Sweden. We would like to thank Kristin Holmgren, Ann-Charlott Olsson and Claus R. Johnsen for excellent technical assistance.
Abbreviations
- BSA
bovine serum albumin
- CCR
chemokine receptor
- CR-Th2
chemokine receptor Th2
- FITC
fluorescein isothiocyanate
- IL
interleukin
- IFN-γ
interferon-γ
- mAb
monoclonal antibody
- PBMC
peripheral blood mononuclear cells
- PBS
phosphate-buffered saline
- PE
phycoerythrin
- PHA
phytohaemagglutinin
- PMA
phorbol 12-myristate 13-acetate
- Th1
T helper 1
- Th2
T helper 2
- TT
tetanus toxoid
References
- 1.Dutton RW, Bradley LM, Swain SL. T cell memory. Annu Rev Immunol. 1998;16:201–23. doi: 10.1146/annurev.immunol.16.1.201. [DOI] [PubMed] [Google Scholar]
- 2.Sanders ME, Makgoba MW, Sharrow SO, Stephany D, Springer TA, Young HA, Shaw S. Human memory T lymphocytes express increased levels of three cell adhesion molecules (LFA-3, CD2, and LFA-1) and three other molecules (UCHL1, CDw29, and Pgp-1) and have enhanced IFN-gamma production. J Immunol. 1988;140:1401–7. [PubMed] [Google Scholar]
- 3.Terry LA, Brown MH, Beverley PC. The monoclonal antibody, UCHL1, recognizes a 180,000 MW component of the human leucocyte-common antigen, CD45. Immunology. 1988;64:331–6. [PMC free article] [PubMed] [Google Scholar]
- 4.Akbar AN, Terry L, Timms A, Beverley PC, Janossy G. Loss of CD45R and gain of UCHL1 reactivity is a feature of primed T cells. J Immunol. 1988;140:2171–8. [PubMed] [Google Scholar]
- 5.Merkenschlager M, Terry L, Edwards R, Beverley PC. Limiting dilution analysis of proliferative responses in human lymphocyte populations defined by the monoclonal antibody UCHL1: implications for differential CD45 expression in T cell memory formation. Eur J Immunol. 1988;18:1653–61. doi: 10.1002/eji.1830181102. [DOI] [PubMed] [Google Scholar]
- 6.Spertini O, Kansas GS, Reimann KA, Mackay CR, Tedder TF. Function and evolutionary conservation of distinct epitopes on the leukocyte adhesion molecule-1 (TQ-1, Leu-8) that regulate leukocyte migration. J Immunol. 1991;147:942–9. [PubMed] [Google Scholar]
- 7.Yoshida R, Imai T, Hieshima K, et al. Molecular cloning of a novel human CC chemokine EBI1-ligand chemokine that is a specific functional ligand for EBI1 Ccr7. J Biol Chem. 1997;272:13803–9. doi: 10.1074/jbc.272.21.13803. [DOI] [PubMed] [Google Scholar]
- 8.Campbell JJ, Bowman EP, Murphy K, et al. 6-C-kine (SLC), a lymphocyte adhesion-triggering chemokine expressed by high endothelium, is an agonist for the MIP-3beta receptor CCR7. J Cell Biol. 1998;141:1053–9. doi: 10.1083/jcb.141.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gunn MD, Tangemann K, Tam C, Cyster JG, Rosen SD, Williams LT. A chemokine expressed in lymphoid high endothelial venules promotes the adhesion and chemotaxis of naive T lymphocytes. Proc Natl Acad Sci USA. 1998;95:258–63. doi: 10.1073/pnas.95.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–12. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 11.Sallusto F, Lanzavecchia A. Exploring pathways for memory T cell generation. J Clin Invest. 2001;108:805–6. doi: 10.1172/JCI14005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camerini D, Walz G, Loenen WA, Borst J, Seed B. The T cell activation antigen CD27 is a member of the nerve growth factor/tumor necrosis factor receptor gene family. J Immunol. 1991;147:3165–9. [PubMed] [Google Scholar]
- 13.De Jong R, Brouwer M, Hooibrink B, Van der Pouw-Kraan T, Miedema F, Van Lier RA. The CD27− subset of peripheral blood memory CD4+ lymphocytes contains functionally differentiated T lymphocytes that develop by persistent antigenic stimulation in vivo. Eur J Immunol. 1992;22:993–9. doi: 10.1002/eji.1830220418. [DOI] [PubMed] [Google Scholar]
- 14.Hamann D, Baars PA, Rep MH, Hooibrink B, Kerkhof-Garde SR, Klein MR, van Lier RA. Phenotypic and functional separation of memory and effector human CD8+ T cells. J Exp Med. 1997;186:1407–18. doi: 10.1084/jem.186.9.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cousins DJ, Lee TH, Staynov DZ. Cytokine coexpression during human Th1/Th2 cell differentiation. Direct evidence for coordinated expression of Th2 cytokines. J Immunol. 2002;169:2498–506. doi: 10.4049/jimmunol.169.5.2498. [DOI] [PubMed] [Google Scholar]
- 16.Fraticelli P, Sironi M, Bianchi G, et al. Fractalkine (CX3CL1) as an amplification circuit of polarized Th1 responses. J Clin Invest. 2001;107:1173–81. doi: 10.1172/JCI11517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sareneva T, Julkunen I, Matikainen S. IFN-alpha and IL-12 induce IL-18 receptor gene expression in human NK T cells. J Immunol. 2000;165:1933–8. doi: 10.4049/jimmunol.165.4.1933. [DOI] [PubMed] [Google Scholar]
- 18.Lanzavecchia A, Sallusto F. Dynamics of T lymphocyte responses: intermediates, effectors, and memory cells. Science. 2000;290:92–7. doi: 10.1126/science.290.5489.92. [DOI] [PubMed] [Google Scholar]
- 19.Amyes E, Hatton C, Montamat-Sicotte D, Gudgeon N, Rickinson AB, McMichael AJ, Callan MF. Characterization of the CD4+ T cell response to Epstein-Barr virus during primary and persistent infection. J Exp Med. 2003;198:903–11. doi: 10.1084/jem.20022058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bitmansour AD, Douek DC, Maino VC, Picker LJ. Direct ex vivo analysis of human CD4(+) memory T cell activation requirements at the single clonotype level. J Immunol. 2002;169:1207–18. doi: 10.4049/jimmunol.169.3.1207. [DOI] [PubMed] [Google Scholar]
- 21.Campbell JJ, Murphy KE, Kunkel EJ, et al. CCR7 Expression and memory T cell diversity in humans. J Immunol. 2001;166:877–84. doi: 10.4049/jimmunol.166.2.877. [DOI] [PubMed] [Google Scholar]
- 22.Kaech SM, Hemby S, Kersh E, Ahmed R. Molecular and functional profiling of memory Cd8 T cell differentiation. Cell. 2002;111:837–51. doi: 10.1016/s0092-8674(02)01139-x. [DOI] [PubMed] [Google Scholar]
- 23.Roman E, Miller E, Harmsen A, Wiley J, Von Andrian UH, Huston G, Swain SL. CD4 effector T cell subsets in the response to influenza: heterogeneity, migration, and function. J Exp Med. 2002;196:957–68. doi: 10.1084/jem.20021052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arens R, Tesselaar K, Baars PA, et al. Constitutive CD27/CD70 interaction induces expansion of effector-type T cells and results in IFNgamma-mediated B cell depletion. Immunity. 2001;15:801–12. doi: 10.1016/s1074-7613(01)00236-9. [DOI] [PubMed] [Google Scholar]
- 25.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–69. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 26.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–96. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 27.Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J Biol Chem. 1997;272:21597–603. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- 28.Ranganath S, Murphy KM. Structure and specificity of GATA proteins in Th2 development. Mol Cell Biol. 2001;21:2716–25. doi: 10.1128/MCB.21.8.2716-2725.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cosmi L, Annunziato F, Galli MIG, Maggi RME, Nagata K, Romagnani S. CR-Th2 is the most reliable marker for the detection of circulating human type 2 Th and type 2 T cytotoxic cells in health and disease. Eur J Immunol. 2000;30:2972–9. doi: 10.1002/1521-4141(200010)30:10<2972::AID-IMMU2972>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 30.Foussat A, Coulomb-L'Hermine A, Gosling J, et al. Fractalkine receptor expression by T lymphocyte subpopulations and in vivo production of fractalkine in human. Eur J Immunol. 2000;30:87–97. doi: 10.1002/1521-4141(200001)30:1<87::AID-IMMU87>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 31.Dorner BG, Scheffold A, Rolph MS, Huser MB, Kaufmann SH, Radbruch A, Flesch IE, Kroczek RA. MIP-1alpha, MIP-1beta, RANTES, and ATAC/lymphotactin function together with IFN-gamma as type 1 cytokines. Proc Natl Acad Sci USA. 2002;99:6181–6. doi: 10.1073/pnas.092141999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Born TL, Thomassen E, Bird TA, Sims JE. Cloning of a novel receptor subunit, AcPL, required for interleukin-18 signaling. J Biol Chem. 1998;273:29445–50. doi: 10.1074/jbc.273.45.29445. [DOI] [PubMed] [Google Scholar]