Abstract
The world-wide effort to identify susceptibility genes for allergic diseases is motivated by the conviction that the identification of disease genes may permit the design of new classes of anti-inflammatory compounds. Molecules concerned with the allergic reaction, such as cytokines, chemokines, their receptors, major histocompatibility complex molecules, and transcription factors, could provide the candidate genes of the allergic diseases. On the basis of genetic studies, multiple research groups have attempted to identify a susceptibility gene for allergy using the candidate gene approach and/or genome-wide screening. Both of these approaches suggest genetic heterogeneity of allergic diseases. Many variants of candidate genes are or are not associated with particular diseases in different ethnic groups and the function of variants is now being investigated. Based on the information accumulated thus far and the information on the human genome sequence, future advances in research on genetic factors for allergic diseases will be likely lead to the establishment of more effective prophylaxis and therapy for these diseases.
Introduction
Allergic diseases are characterized by elevated serum immunoglobulin E (IgE) levels and hypersensitivity to normally innocuous antigen. Approximately one-third of the population of the world is affected with allergic diseases and morbidity from allergic diseases in the inner city is rising steadily. There is now overwhelming support for a genetic component for allergic diseases. Information on the genetics of allergic diseases is valuable not only for analysing the molecular basis of allergic diseases but also for new drug development. However, the identification of susceptibility genes is not straightforward. The following are reasons that introduce complexity into the analyses: (1) participation of various molecules in distinct phases of allergic reaction, (2) participation of multiple environmental factors, and (3) multiple clinical phenotypes (i.e. asthma, allergic rhinitis, atopic dermatitis, or allergic conjunctivitis) present in allergic patients, and the definition of these phenotypes is complex.
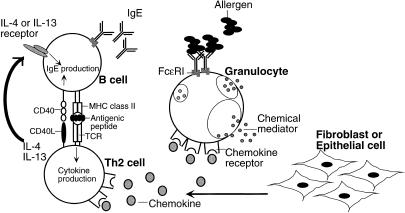
A particular allergen first encounters antigen-processing cells (APC) such as dendritic cells or macrophage directly, or as part of an immune complex with immunogloublin. The allergen captured by the APC is processed and presented to CD4+ T cells as a peptide fragment in the context of major histocompatibility complex (MHC) class II molecules. CD4+ T cells are polarized into T helper type 1 (Th1) cells, producing interferon-γ (IFN-γ) and interleukin-2 (IL-2), and Th2 cells, producing IL-4, IL-5, IL-6, IL-10 and IL-13. IL-4 and IL-13 stimulate immunoglobulin class switching, leading to the production of IgE, which binds its high-affinity receptor (FcεRI) on the surface of mast cells or basophiles. The association of allergen with IgE bound to FcεRI results in the activation of signal transduction in these cells and rapidly leads to the release of inflammatory chemical mediators, such as cytokines, histamine and leukotrienes. Th2-type cytokines also trigger the production of chemokines such as monocyte chemoattractant protein-1 (MCP-1), eotaxin-1, or RANTES in tissue fibroblast or epithelial cells, which results in infiltration of inflammatory cells into the site of allergen exposure (see Fig. 1 for schematic of events in the sensitization and acute-phase reactions during an allergic response).
Figure 1.
Schematic representation of the cell and molecular events participating in the sensitization and acute-phase reactions during an IgE-mediated allergic response. These include cytokine/cytokine receptor signalling events involved in the generation of antigen-specific IgE, and the downstream events such as mast cell/basophil degranulation associated with acute diseases. As amplified signals at each stage may predispose individuals for allergic disease, it is perhaps not surprising that gene products in each of these pathways have been implicated as potential allergy susceptibility genes.
As described above, various molecules participate during the phases of the allergic reaction. This means that many genes encoding molecules relating to the allergic reaction, such as cytokines, chemokines, their receptors, MHC molecules and transcription factors, are potential candidate genes for allergy-predisposing genes. In addition, the aetiology of allergic diseases involves interactions between multiple genes and a variety of environmental factors, such as protein allergens, air pollutants, and viral or bacterial proteins. Significant effort has been invested in the search for allergic susceptibility genes and much useful information has been accumulated. In this manuscript, we review several of the strong candidate genes for susceptibility to allergic diseases, especially gene-encoding molecules regulating IgE production and inflammatory responses.
Background
In the early 1920s, it was already suspected that there was a strong genetic component to the susceptibility to allergic diseases.1,2 At present, this has been well-established on the basis of many investigations of allergic subjects and/or in monozygonic versus dizygotic twins: i.e. the risk of children with no family history is 20%, the risk of an individual with a history of a single parent is from 25% to 30%, and the risk of an individual with a dual parental history is from 50% to 75%.
The development of molecular biological techniques in the 1970s greatly contributed to the identification of candidate susceptibility genes for allergic diseases. In 1989, the first evidence of linkage was reported by the group of Cookson et al. of Oxford University.3 They showed that there is a striking linkage of chromosome 11q with the IgE response underlying asthma and rhinitis and named the allergic susceptibility gene an ‘atopy gene’. Atopy is defined as a general predisposition to develop an allergic reaction to an innocuous antigen. The group of Cookson/Hopkin then reported, in 1996, linkages between (1) IgE level of serum and chromosomes 7, 11 and 16, (2) cutaneous reactions and chromosome 11, and (3) atopic predisposition and chromosomes 6 and 13 (Table 1).4 The first study of Cookson's group stimulated researchers in the field of allergic research. So far, genome-wide screens for susceptibility genes to asthma or related phenotypes have been conducted in ethnically different populations (Table 1). Based on the information about these reports, many susceptibility genes on these chromosomes are currently being investigated.
Table 1. Region showing evidence for linkage to asthma and allergic phenotypes in genome-wide screening.
| Locus | Australian4 | CSGA5,6 | Dutch7 | French8 | German9 | Japanese10 | Hutterites11 |
|---|---|---|---|---|---|---|---|
| 1p | – | Asthma (Hispanic)6 | – | Asthma | Eosinophils | – | Skin test |
| Total IgE | Asthma | ||||||
| Specific IgE | |||||||
| 2p | – | – | – | – | Asthma | – | – |
| BHR | |||||||
| Total IgE | |||||||
| RAST | |||||||
| Specific IgE | |||||||
| 2q | – | Asthma (Hispanic)5 | – | – | Total IgE | – | Skin test |
| 3p | – | – | BHR | – | – | – | Asthma |
| Skin test | |||||||
| 4q | BHR | – | – | – | RAST | Mite-sensitive atopic asthma | – |
| 5p | – | Asthma (African American)5 | – | – | Specific IgE | – | BHR |
| 5q | – | – | BHR | – | – | Mite-sensitive atopic asthma | Asthma |
| Asthma symptoms | |||||||
| 6p | Eosinophils | Asthma (Caucasian)6 | – | – | Asthma | Mite-sensitive atopic asthma | – |
| Atopy | RAST | ||||||
| Total IgE | |||||||
| Eosinophils | |||||||
| 7p | BHR | – | – | – | RAST | – | – |
| Total IgE | |||||||
| 8p | – | – | – | – | – | – | Asthma |
| 9p | – | – | – | – | BHR | – | Asthma symptoms |
| 9q | – | – | – | – | Asthma | – | – |
| 10q | – | – | Total IgE | – | Specific IgE | – | – |
| 11p | – | Asthma (Caucasian)5 | – | Total IgE | – | – | Skin test |
| 11q | Total IgE | Asthma (African ″American)6 | – | Total IgE | Eosinophils | – | – |
| Skin test | |||||||
| 12q | – | Asthma (Caucasian)5 | Total IgE | Eosinophils | – | Mite-sensitive atopic asthma | Asthma |
| Skin test | |||||||
| 13q | Atopy | Asthma (Caucasian)5 | – | Eosinophils | – | Mite-sensitive atopic asthma | Asthma symptoms |
| 14q | – | Asthma (Caucasian)5 | – | – | – | – | Asthma |
| 15q | – | – | – | – | Total IgE | – | – |
| Specific IgE | |||||||
| 16q | Total IgE | – | – | – | – | – | Skin test |
| 17p | – | Asthma (African ″American)5 | – | – | – | – | – |
| 17q | – | – | Total IgE | Asthma | – | – | – |
| Skin test | |||||||
| 18p | – | – | – | – | – | – | Asthma symptoms |
| 19q | – | Asthma (Caucasian)5 | – | – | – | – | BHR |
| 20p | – | – | – | – | – | – | Skin test |
| 21q | – | Asthma (Caucasian)5 | – | – | – | – | – |
| Xq | – | – | – | – | Total IgE | – | – |
| Eosinophils |
Only values P<0·01 are shown.
Genome-wide screening and candidate gene approach
Two representative methods are used to identify susceptibility genes for multifactorial diseases: genome-wide screening and the candidate gene approach. In genome-wide screening, the presumptive location of the target gene is defined, and then the target gene(s) is sequenced to determine the polymorphisms contributing to disease susceptibility. To define the presumptive location of the target gene, highly polymorphic microsatellite repeats (whose chromosomal locations are already known) are used. A study of a family with a specific disease or trait and subsequent linkage analysis of the inheritance of these marker genes and disease phenotypes together make it possible to deduce the chromosomal region of the target gene. All studies indicated in Table 1 involve genome-wide screening and linkage analysis for allergy susceptibility genes. The technical feature of genome-wide screening is that one can identify the disease gene with little information about the properties of the disease gene product. When genome-wide screening is applied to identify susceptibility genes, subjects need to be carefully selected considering phenotypes, because the susceptibility genes are different between ethnic groups and disease phenotypes as shown in Table 1. In our study, evidence for genetic linkage of allergic conjunctivitis in USA populations was obtained for chromosome 5, 16 and 17.12 According to reports by the Collaborative Study on the Genetics of Asthma (CSGA), USA, the human chromosomes 5, 6, 11, 12 and 14 have been implicated in asthma and bronchial hyperresponsiveness.5 These data clearly demonstrate that both shared and distinct genetic loci control susceptibility to the various allergic diseases, and underscore the importance of performing genome-wide screening for disease susceptibility genes in well-defined patient populations, enriched for each allergic disease.
In the candidate gene approach, the pathogenesis of a disease is analysed first, and then the protein potentially involved in the development of the disease is identified. Subsequently, the gene for the protein is sequenced and controls are found to investigate the relationship between DNA mutations and the disease or trait. If an unknown protein or gene controls disease phenotypes, this strategy fails to discover the susceptibility gene. Moreover, when many genes are involved in a disease, it is necessary to search for candidate genes one after another. In common practice, the approximate chromosomal location of a disease gene is determined by genome-wide screening, then potential candidate genes in that genetic region are analysed.
FcεRI β subunit (Chromosome 11)
In 1989, the first genetic linkage for allergy was reported by the group of Cookson/Hopkin at Oxford University.3 They found a striking linkage of chromosome 11q with the IgE response underlying asthma and rhinitis via genome-wide screening and defined the gene on chromosome 11q as an ‘atopy gene’. Cookson et al. also found that the atopy gene was transmitted through maternal inheritance and was located in 11q13, in close proximity to the gene encoding the β-subunit of the high-affinity IgE receptor, FcεRI.13,14 This receptor, FcεRI, is expressed on the mast cells and eosinophils, and is composed of an α-subunit, a β-subunit, and a dimer of the γ-subunit. The β-subunit is an intrinsic signal amplifier and could be considered to be one of the control factors in the effector phase of type-I allergic reaction.
In subsequent studies, a point mutation was found in exon 6 of the gene encoding the β-subunit in families with asthma and rhinitis.15 This mutation results in the substitution of isoleucine to leucine at position 181 located in the 4th transmembrane region of the β-subunit. Moreover, this variant was maternally inherited, consistent with the initial mapping into chromosome 11q, the data strongly suggesting that an atopy gene might be FcεRI-β subunit itself. Following this work, many research groups have confirmed linkage of chromosome 11q13 with high serum IgE level, asthma, and/or atopic dermatitis in African American,16 Australian,17 Japanese,18 Germany,19 Italian,20 or Swedish population.21 Other variants of the FcεRI β-subunit, Val183Leu and Glu237Gly, were also found. Variant Val183Leu (the substitution of valine to leucine at position 183) was associated with asthma in the black South African population.22 Variant Glu237Gly (the substitution of glutamic acid to glycine at position 237) was highly associated with skin test positivity in an Australian study,23 with asthma in a Caucasian South African population,22 or with high serum IgE in French-Canadians.24 The exact function of these variants remains unclear.
Although the data from other groups supported the results of Cookson/Hopkin, many research groups were unable to obtain evidence for an atopy gene on chromosome 11q. The linkage of 11q13 with asthma and/or high IgE has not been observed in studies in Australia,25 USA,26,27 Britain,28 Japan,29 Germany,9 or Spain.30 These findings emphasize the genetic heterogeneity of allergy and point to multiple mechanisms prediposing individuals to develop allergic diseases. It is clear that the gene of the FcεRI β-subunit is one susceptibility gene for allergy. However, further studies are needed to clarify the role of particular variants in susceptibility to allergic diseases among different ethnic groups.
IL-4 and IL-13 (chromosome 5)
The genomic location attracting the next greatest interest is human chromosome 5q31–5q33. Many studies have indicated linkage between 5q31–5q33 and asthma, atopic dermatitis and/or these associated traits,21,31–35 while some research groups have reported no linkage (as was the case with 11q).30,36–39 The human chromosome 5q31–5q33 encodes the IL-4, IL-5, IL-9, IL-13, granulocyte–macrophage colony-stimulating factor cluster of cytokine genes, and the β chain gene of IL-12. In addition, T-cell and airway phenotype regulator (Tapr) has been identified as an allergy/asthma-susceptibility locus on 5q33, which encodes T-cell membrane proteins (TIMs) and controls the development of airway hyperreactivity and T-cell production of IL-4 and IL-13.40 IL-4 and IL-13 are of course essential for IgE production and promote Th2 differentiation. Considering their roles in the allergic reaction, IL-4 and IL-13 genes are regarded as strong candidates of allergic susceptibility genes.
IL-4
In 1994, Marsh and co-workers at Johns Hopkins first reported linkage between markers within the IL-4 gene locus and elevated IgE concentrations in Amish families.41 Following the initial report of an association of the IL-4 gene with serum IgE level, many researchers have asked whether polymorphisms within IL-4 regulatory elements might contribute to disease susceptibility by mediating overexpression of the gene, and subsequently amplifying Th2 differentiation and class switching to IgE. In fact, our group demonstrated that several polymorphic nucleotides in the IL-4 promoter region (observed in laboratory B-cell lines) could affect the affinity of transcription factors for their cognate cis-elements, thus mediating the overexpression of the gene.42 However, the polymorphic nucleotides we observed in the B-cell lines do not appear to be common variants in the general population. Thus far, multiple nucleotide variants in human IL-4 promoter region such as −34C/T, −590C/T and + 33C/T have been reported in allergic individuals.43–45 From studies on these individuals, −590C/T seems to be an important variant. The linkage between the promoter polymorphism −C590T of the IL-4 gene and asthma/atopic eczema in the US46 or Japanese populations47 have been reported, while data from studies on UK,48 French,49 Hutterite,11 Kuwaiti Arab,50 or Australian populations51 have shown no linkage. −C590T associates in vitro with IL-4 activity, and shows higher binding affinity to nuclear transcription factors.46 However, it remains unclear whether −C590T plays a direct role in the allergic condition.
IL-13
IL-13 shares a receptor component and signalling pathway with IL-4 and exerts biological effects similar to IL-4 in IgE production and IgE-based mucosal inflammation. In addition, IL-13 induces the pathophysiological features of asthma through the stimulation of bronchial epithelial mucus secretion and smooth muscle hyper-reactivity in an IL-4-independent manner.52,53 Multiple promoter variants and coding region variants in allergic patients were reported.54–60 From them, Gln110Arg (the substitution of arginine to glutamine at position 110 of IL-13) seems to be important. The group of Martinez reported the association of coding region variant Arg130Gln with increased serum IgE level.56 Deichmann and co-workers reported that variant Arg130Gln is associated with asthma but not with IgE levels in Britain and Japan populations.55 These data clearly need to be investigated further.
IL-4 receptor α-chain (chromosome 16)
A component of IL-4 receptor, the α-chain has also been regarded as a strong candidate for an allergic susceptibility gene. The IL-4 receptor is composed of an α-chain and β-chain. The gene encoding the IL-4 receptor α-chain is localized on chromosome 16p11–12, which is a site associated with asthma or high IgE level in ethnically different populations.4,61
The IL-4 receptor α-chain is also used as a component of the IL-13 receptor. Upon stimulation of IL-4 or IL-13, Janus tyrosine kinase-1 and 3 are activated, followed by activation of the signal transducer and activator of transcription 6 (STAT6), which has a central role in IgE production. These findings have led investigators to direct their attention to the IL-4 receptor α-chain gene as the susceptibility gene to allergy. The group of Chatila conducted pioneering research in this field.62 They identified a variant Arg576Glu (the substitution of glutamine to arginine at position 551 of mature IL-4R α-chain located in the intracellular domain of α-chain) in an American population with allergic inflammatory disorder (hyper-IgE syndrome or atopic dermatitis). The association of Arg576Glu with atopic dermatitis was also found in a Japanese population.63 Functional analysis using monocytes of human allergy patients with Arg576Glu indicated that the variant induced higher expression of CD23 and impaired the binding of the negative regulator protein tyrosine phosphatase SHIP1 by the human IL-4 stimulation.62 However, Arg576Glu did not induce a significant effect on IL-4 signal transduction such as STAT6 activation.64 A large cohort of patients with the hyper-IgE syndrome in USA showed no increase in frequency of Arg576Glu as compared with that in normal subjects.65 This was in accord with negative findings in a Japanese or an Italian population with asthma or atopy.20,66 From these observations, the linkage of Arg576Glu with allergic diseases has been controversial.
Following Chatila's report, Izuhara's group found that the substitution of Val to Ile at position 50 of mature protein (Ile50Val) in the extracellular domain of the α-chain is associated with atopic asthma in a Japanese population.67 They demonstrated that Ile50Val augments STAT6 activation, expression of CD23, germline ε transcription, and IgE production upon stimulation with IL-4 in a transfectant of human B cells harbouring the Ile50Val variant.68 Surprisingly, a negative effect of Arg576Glu on serum IgE level was also reported. It has been indicated that Arg576Glu is in tight linkage disequilibrium with a second variant Pro503Ser (the substitution Ser to Pro at position 478 of mature protein) of IL-4 α-chain in a German population with lower IgE levels.69 In contrast, linkage disequilibrium between Ser786Pro (the substitution Pro to Ser at position 761 of mature protein) and Gln576Arg is associated with high IgE levels in an American population with asthma.70 The Ser786Pro variant in isolation did not affect IL-4R function when tested by cDNA transfection into a mouse B lymphoma cell line. These observations suggest that each mutant in the gene of IL-4 receptor α-chain affects IL-4 signalling and IgE production through different mechanisms and that multiple mutations in one gene may change the structure of the receptor, leading to altered signal transduction.
MHC molecules (Chromosome 6)
The MHC comprises a family of highly polymorphic genes encoding a set of transmembrane proteins that present peptide epitope to a specific antigen receptor [ T-cell receptor; (TCR)] on T cells. The extensive polymorphism of these MHC-encoded proteins defines the repertoire of an antigenic determinant to which each of us is capable of responding. From this consideration, human leucocyte antigen (HLA) genes are major contributors to the genetic susceptibility underlying human diseases, not only immune diseases such as allergic diseases, but also cancer and infectious diseases.
The MHC is localized on human chromosome 6p21.3–23. Genome-wide searches for an asthma-susceptibility gene in ethnically diverse populations have provided evidence for linkage between 6p21.3–23 and asthma or its associated traits.4,5,9–11,71 Also, a number of studies have reported the association of HLA class II (HLA-DR, DQ, or DP) alleles with IgE responsiveness to a number of different allergen (for review see ref. 72). Some studies have indicated that HLA class II alleles have a protective effect on the development of a sensitization to allergen, asthma, or other allergic diseases.73–76 However, these studies have often yielded conflicting results for multiple reasons: (1) ethnic background, phenotype definitions, or HLA typing methods are different in each study; (2) a large allergen contains multiple T-cell epitopes; and (3) T-cell responsiveness and IgE production to specific allergen are influenced by the presence and concentration of allergen. It seems clear that HLA class II molecules do control immune responses to particular allergens in many cases. However, significant and consistent linkage of HLA class II alleles with allergy might be observed in the carefully controlled studies.
CC Chemokines (chromosome 17) and their receptor (chromosome 3)
CC chemokines participate in the activation and recruitment of granulocytes, monocytes, and T-cell subsets, and play a crucial role in allergic inflammation. The CC chemokine cluster is localized on chromosome 17p12–17p11.2. The USA CSGA reported evidence for linkage of asthma with chromosome 17p12–17q11.2 in African Americans.5 Our group also observed the linkage of allergic conjunctivitis with the site.12 These observations suggest that a CC chemokine may be a strong candidate for an allergic susceptibility gene. RANTES, one of the most extensively studied CC chemokines in allergic diseases, attracts Th2 cells and eosinophils via the chemokine receptors CCR1, CCR3 and CCR5. Following the paradigm set with IL-4 promoter polymorphism, the Johns Hopkins group identified a functional mutation in the proximal promoter of the RANTES gene, −A401G, which creates a near consensus-binding site for the GATA transcription factor family. This promoter polymorphism of RANTES gene was associated with atopic dermatitis or skin test positivity, but was not associated with asthma and IgE levels in the children of a German Multicentre allergy study.77 Another promoter polymorphism of the RANTES gene, −A403G was also found in Caucasians, which is associated with an increased susceptibility to asthma and atopy – characterized by high serum IgE and skin test positivity.78 Functional analysis using transfectant showed significantly higher transcriptional activity of the mutant promoter with −A401G,77 although biological function of the variants remains unknown. More recently, a promoter polymorphism of MCP-1 (monocyte chemoattractant protein-1) was found in Hungarian children with asthma.79
A gene cluster of CC chemokine receptors is located on chromosome 3p21–24. Evidence for linkage of atopic dermatitis to chromosome 3q21 in Germany has been reported.80 So far, multiple variants of CCR receptor, especially CCR5, have been found in human immunodeficiency virus-infected patients or normal subjects. One of the CCR5 variants, the CCR5-delta 32-deletion polymorphism (CCR5-delta32) in allergic patients was investigated for association with asthma and atopy. No statistically significant linkage of CCR5-delta32 to atopy was observed, although functional evidence might suggest that CCR5 is a good candidate gene for allergy.81 In allergic inflammation involving eosinophils and Th2 cells, not only RANTES and MCP-1 but also other CC chemokines, such as thymus- and activation-regulated chemokine (TARC), macrophage inflammatory protein 1α (MIP-1α), and particularly eotaxin-1 play an important role. Multiple CCRs, such as CCR1, CCR3 and CCR5, are involved in the activation and recruitment of inflammatory cells in the late-phase reaction. A detailed screen for mutation in these chemokines and their receptors is underway in several laboratories around the world.
Potential involvement of lineage commitment and signalling pathways
The control of Th1/Th2 differentiation is clearly germane to our understanding of the molecular basis of allergic diseases. Since allergic diseases are characterized by an exaggerated Th2 response, alleles of lineage commitment factors or transcription factors that bias T cells to the Th2 phenotype are potential allergy susceptibility genes. Our own studies point toward new potential allergy susceptibility genes. First, our genome-wide analysis for susceptibility genes for allergic conjunctivitis has provided evidence for association between the c-maf locus and this disease.12 Since the c-maf transcription factor is essential for Th2 cytokine production in rodents, we are investigating whether particular alleles of the c-maf gene product underlie the association we have observed. Second, we have found that CP2, a factor homologous to Drosophila Efl-1, is also important for IL-4 promoter-driven transcription.82 Overexpressed CP2 markedly enhances IL-4 promoter activity and expression of the endogenous IL-4 gene when tested by transfection of the murine Th2 cell line D10 with the CP2 expression vector. It is interesting that the human CP2 gene maps to the susceptibility locus for allergic conjunctivitis (unpublished data).
Other transcription factors attracting some attention as a candidate gene are BCL6, STAT6, GATA-3 or T-bet. STAT6 is an essential molecule for IL-4 and IL-13 signal transduction. STAT6-deficient mice exhibit impaired IgE production and Th2 inflammatory reactions.83,84 The gene of STAT6 is located on chromosome 12q13–14, another site of genetic linkage to asthma.5,9 Gao et al. have found that the G2964A variant in a 3′ untranslated region of STAT6 is associated with mild atopic asthma characterized by positive antigen-specific IgE or high total IgE in a Japanese population, but is not linked with atopy or asthma in the British population.85
BCL6 is known to encode a zinc-finger transcriptional repressor expressed in both B cells and CD4+ T cells, and contributes to a hyperimmune Th2 cell response. BCL6-deficient mice developed multiple organ inflammation characterized by eosinophilia and an elevation of IgE levels.86 A variant of the BCL6 gene was associated with marked atopy.87 The function in the variant of STAT6 and BCL6 remains unclear.
T-bet, a transcription factor participating in Th1 differentiation, transactivates the IFN-γ gene in Th1 cells and has the unique ability to redirect fully polarized Th2 cells into Th1 cells.88 GATA-3, a transcription factor in Th2 differentiation, regulates the expression of IL-5 and IL-13 genes in Th2 cells.89,90 While T-bet expression is down-regulated in human asthmatic airways,91 GATA-3 expression is increased.92 Although variants of T-bet or GATA-3 have not yet been found in allergic patients, these are strong candidates for allergy susceptibility genes. Further studies are underway to identify and clarify the role of the transcription factors in susceptibility to allergic diseases.
Conclusions – Diagnostic and Therapeutic Considerations
Although many investigators have reported potential susceptibility genes for allergic diseases (see Table 2 for a partial list of implicated genes), many of the results are controversial. These are presumably related to genetic heterogeneity, differences in family recruitment methods and statistical analysis techniques. However, recent technical advances in genetic analysis are remarkable. In particular, development of cDNA array technology and publication of the entire human genome sequence in 2001 are expected to mark a turning point in the study of genetic factors for allergic diseases.93,94 Based on the information accumulated so far and on information of the genome sequence, the DNA array technique allows the simultaneous analysis of the expression of multiple genes or of single nucleotides polymorphisms (SNIPs). Brutsche et al. have investigated significantly altered gene expression in peripheral blood mononuclear cells derived from patients with allergy and asthma compared with that derived from the healthy subjects using cDNA array.95,96 The differently expressed genes in their study include surface molecules involved in T- and B-cell interaction and activation, apoptosis-related molecules, cytokines, intracellular signalling products, and transcription factors. Using microarray analysis of pulmonary gene expression and SNIP-based genotyping, Karp et al. newly identified the gene encoding complement factor 5 (C5) as a susceptibility locus for allergen-induced airway hyper-responsiveness (AHR) in a murine model of asthma.97 A possible mechanism for the involvement of C5 in AHR is given by the finding that C5a cleaved from C5 may block IL-12 production, and hence might have an exacerbating effect on AHR.97 The researchers studying allergic susceptibility genes have devoted much attention to acquired immunity relating to IgE production. However, Karp's study emphasized that the molecules participating in innate immunity including the complement network are also candidates for the allergy susceptibility gene.
Table 2. Candidate genes for allergic diseases and their function.
| Chromosome | Candidate gene | Function |
|---|---|---|
| 1p31.2 | IL-12 receptors β2 chain | Signal transducer of IL-12 |
| 2q33 | CD28 | Co-stimulator in T-cell activation |
| CTLA-4 | Co-stimulator in T-cell activation | |
| 3p24.2–p22 | CC chemokine receptor | Signal transducer of chemokine |
| 3q27 | BCL6 | Repression of Stat6-activated transcription |
| 5q31 | IL-4 | Differentiation of Th2 cells/induction of IgE production |
| IL-5 | Eosinophils growth and activation/Promotion of IgE production | |
| IL-9 | Mast cell growth factor | |
| IL-13 | Induction of IgE production | |
| 5q31–33 | IL-12 p40 | Inhibition of Th2 activity |
| 5q33 | T-cell membrane proteins (TIMs) | Induction of IL-4 and IL-13 production |
| 5q35 | Leukotriene C4 synthase | Synthesis of leukotriene |
| 6p21 | Major histocompatibility complex (HLA) | Presentation of antigenic peptide |
| Tumour necrosis factors | Induction of inflammation | |
| Transports involved in antigen processingand and presentation (TAP-1 and TAP-2) | Transportation of antigenic peptide | |
| 7p15 | IL-6 | Promotion of IgE production |
| T-cell receptor γ chain | Recognition of antigen | |
| 7q35 | T-cell receptor β chain | Recognition of antigen |
| 9q34 | Complement factor 5 | Induction of IL-12 production |
| 10p14 | GATA3 | Transcription factor in Th2 differentiation |
| 11q13 | FcεRI β subunit | Amplifier of IgE signalling |
| 12q13–14 | STAT6 | Transcription factor in IL-4 signalling |
| 12q14 | Stem cell factor | Mast cell growth factor |
| 12q21 | IFN-γ | Inhibition of Th2 activity/Inhibition of IgE isotype classwitch |
| 12q22 | Leukotriene A4 hydrolase | Synthesis of leukotriene |
| 14q11.2–q13 | T-cell receptor α/δ chain | Recognition of antigen |
| 16p12 | IL-4 receptor α chain | Signal transducer of IL-4 |
| 17p11 | CC chemokine | Recruitment and activation of inflammatory cells |
| 19q13.3 | Complement factor 5a receptor | Induction of IL-12 production |
| Xq13 | IL-13 receptor α1/α2 chain | Signal transducer of IL-13 |
IL-12, interleukin-12; IFN-γ, interferon-γ; Th2, T helper type 2; IgE, immunoglobulin E.
In addition to experiments using cDNA array techniques, studies in proteomics, to analyse the expression pattern of protein in biological systems, have begun.98 Proteomic techniques would enable us to investigate how SNIPs influence the expression and function of proteins and to obtain increased knowledge of mechanisms underlying allergic diseases. A variety of genetic factors are related to allergic diseases, and environmental factors are also involved in disease expression. Therefore, the nature of causative genes and the degree to which they contribute to disease will vary from patient to patient. Elucidation of disease gene expression and protein function based on information about SNIPs at the individual level may some day lead to the development of prophylactic and therapeutic regimens tailored to the individual patient. It is hoped that future advances in research on genetic factors for allergic diseases will lead to the establishment of more effective prophylaxis and therapy for these diseases.
References
- 1.Coca AF, Cooke RA. On the classification of the phenomenon of hypersensitiveness. J Immunol. 1923;8:163–82. [Google Scholar]
- 2.Adkinson J. The behavior of bronchial asthma as an inherited character. Genetics. 1920;5:363–418. doi: 10.1093/genetics/5.4.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cookson WO, Sharp PA, Faux JA, Hopkin JM. Linkage between immunoglobulin E responses underlying asthma and rhinitis and chromosome 11q. Lancet. 1989;1:1292–5. doi: 10.1016/s0140-6736(89)92687-1. [DOI] [PubMed] [Google Scholar]
- 4.Daniels SE, Bhattacharrya S, James A, et al. A genome-wide search for quantitative trait loci underlying asthma. Nature. 1996;383:247–50. doi: 10.1038/383247a0. [DOI] [PubMed] [Google Scholar]
- 5.The Collaborative Study on the Genetics of Asthma. A genome-wide search for asthma susceptibility loci in ethnically diverse populations. The Collaborative Study on the Genetics of Asthma (CSGA) Nat Genet. 1997;15:389–92. doi: 10.1038/ng0497-389. [DOI] [PubMed] [Google Scholar]
- 6.Xu J, Meyers DA, Ober C, et al. Genomewide screen and identification of gene–gene interactions for asthma-susceptibility loci in three U.S. populations: collaborative study on the genetics of asthma. Am J Hum Genet. 2001;68:1437–46. doi: 10.1086/320589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu J, Postma DS, Howard TD, Koppelman GH, Zheng SL, Stine OC, Bleecker ER, Meyers DA. Major genes regulating total serum immunoglobulin E levels in families with asthma. Am J Hum Genet. 2000;67:1163–73. doi: 10.1086/321190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dizier MH, Besse-Schmittler C, Guilloud-Bataille M, et al. Genome screen for asthma and related phenotypes in the French EGEA study. Am J Respir Crit Care Med. 2000;162:1812–18. doi: 10.1164/ajrccm.162.5.2002113. [DOI] [PubMed] [Google Scholar]
- 9.Wjst M, Fischer G, Immervoll T, Jung M, et al. A genome-wide search for linkage to asthma. German Asthma Genetics Group. Genomics. 1999;58:1–8. doi: 10.1006/geno.1999.5806. [DOI] [PubMed] [Google Scholar]
- 10.Yokouchi Y, Nukaga Y, Shibasaki M, et al. Significant evidence for linkage of mite-sensitive childhood asthma to chromosome 5q31-q33 near the interleukin 12 B locus by a genome-wide search in Japanese families. Genomics. 2000;66:152–60. doi: 10.1006/geno.2000.6201. 10.1006/geno.2000.6201. [DOI] [PubMed] [Google Scholar]
- 11.Ober C, Tsalenko A, Parry R, Cox NJ. A second-generation genomewide screen for asthma-susceptibility alleles in a founder population. Am J Hum Genet. 2000;67:1154–62. doi: 10.1016/s0002-9297(07)62946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishimura A, Campbell-Meltzer RS, Chute K, Orrell J, Ono SJ. Genetics of allergic disease: evidence for organ-specific susceptibility genes. Int Arch Allergy Immunol. 2001;124:197–200. doi: 10.1159/000053709. [DOI] [PubMed] [Google Scholar]
- 13.Cookson WO, Young RP, Sandford AJ, et al. Maternal inheritance of atopic IgE responsiveness on chromosome 11q. Lancet. 1992;340:381–4. doi: 10.1016/0140-6736(92)91468-n. [DOI] [PubMed] [Google Scholar]
- 14.Sandford AJ, Shirakawa T, Moffatt MF, et al. Localisation of atopy and beta subunit of high-affinity IgE receptor (Fc epsilon RI) on chromosome 11q. Lancet. 1993;341:332–4. doi: 10.1016/0140-6736(93)90136-5. [DOI] [PubMed] [Google Scholar]
- 15.Shirakawa T, Li A, Dubowitz M, et al. Association between atopy and variants of the beta subunit of the high-affinity immunoglobulin E receptor. Nat Genet. 1994;7:125–9. doi: 10.1038/ng0694-125. [DOI] [PubMed] [Google Scholar]
- 16.Hizawa N, Freidhoff LR, Ehrlich E, et al. Genetic influences of chromosomes 5q31-q33 and 11q13 on specific IgE responsiveness to common inhaled allergens among African American families. Collaborative Study on the Genetics of Asthma (CSGA) J Allergy Clin Immunol. 1998;102:449–53. doi: 10.1016/s0091-6749(98)70134-4. [DOI] [PubMed] [Google Scholar]
- 17.Palmer LJ, Daniels SE, Rye PJ, et al. Linkage of chromosome 5q and 11q gene markers to asthma-associated quantitative traits in Australian children. Am J Respir Crit Care Med. 1998;158:1825–30. doi: 10.1164/ajrccm.158.6.9804037. [DOI] [PubMed] [Google Scholar]
- 18.Shirakawa T, Hashimoto T, Furuyama J, Takeshita T, Morimoto K. Linkage between severe atopy and chromosome 11q13 in Japanese families. Clin Genet. 1994;46:228–32. doi: 10.1111/j.1399-0004.1994.tb04231.x. [DOI] [PubMed] [Google Scholar]
- 19.Folster-Holst R, Moises HW, Yang L, Fritsch W, Weissenbach J, Christophers E. Linkage between atopy and the IgE high-affinity receptor gene at 11q13 in atopic dermatitis families. Hum Genet. 1998;102:236–9. doi: 10.1007/s004390050685. 10.1007/s004390050685. [DOI] [PubMed] [Google Scholar]
- 20.Malerba G, Trabetti E, Patuzzo C, Lauciello MC, Galavotti R, Pescollderungg L, Boner AL, Pignatti PF. Candidate genes and a genome-wide search in Italian families with atopic asthmatic children. Clin Exp Allergy. 1999;29:27–30. [PubMed] [Google Scholar]
- 21.Soderhall C, Bradley M, Kockum I, Wahlgren CF, Luthman H, Nordenskjold M. Linkage and association to candidate regions in Swedish atopic dermatitis families. Hum Genet. 2001;109:129–35. doi: 10.1007/s004390100556. [DOI] [PubMed] [Google Scholar]
- 22.Green SL, Gaillard MC, Song E, Dewar JB, Halkas A. Polymorphisms of the beta chain of the high-affinity immunoglobulin E receptor (Fcepsilon RI-beta) in South African black and white asthmatic and nonasthmatic individuals. Am J Respir Crit Care Med. 1998;158:1487–92. doi: 10.1164/ajrccm.158.5.9707099. [DOI] [PubMed] [Google Scholar]
- 23.Hill MR, Cookson WO. A new variant of the beta subunit of the high-affinity receptor for immunoglobulin E (Fc epsilon RI-beta E237G): associations with measures of atopy and bronchial hyper-responsiveness. Hum Mol Genet. 1996;5:959–62. doi: 10.1093/hmg/5.7.959. [DOI] [PubMed] [Google Scholar]
- 24.Laprise C, Boulet LP, Morissette J, Winstall E, Raymond V. Evidence for association and linkage between atopy, airway hyper-responsiveness, and the beta subunit Glu237Gly variant of the high-affinity receptor for immunoglobulin E in the French-Canadian population. Immunogenetics. 2000;51:695–702. doi: 10.1007/s002510000185. 10.1007/s002510000185. [DOI] [PubMed] [Google Scholar]
- 25.Brereton HM, Ruffin RE, Thompson PJ, Turner DR. Familial atopy in Australian pedigrees: adventitious linkage to chromosome 8 is not confirmed nor is there evidence of linkage to the high affinity IgE receptor. Clin Exp Allergy. 1994;24:868–77. doi: 10.1111/j.1365-2222.1994.tb01809.x. [DOI] [PubMed] [Google Scholar]
- 26.Rich SS, Roitman-Johnson B, Greenberg B, Roberts S, Blumenthal MN. Genetic analysis of atopy in three large kindreds: no evidence of linkage to D11S97. Clin Exp Allergy. 1992;22:1070–6. doi: 10.1111/j.1365-2222.1992.tb00132.x. [DOI] [PubMed] [Google Scholar]
- 27.Amelung PJ, Panhuysen CI, Postma DS, Levitt RC, Koeter GH, Francomano CA, Bleecker ER, Meyers DA. Atopy and bronchial hyperresponsiveness: exclusion of linkage to markers on chromosomes 11q and 6p. Clin Exp Allergy. 1992;22:1077–84. doi: 10.1111/j.1365-2222.1992.tb00133.x. [DOI] [PubMed] [Google Scholar]
- 28.Lympany P, Welsh K, MacCochrane G, Kemeny DM, Lee TH. Genetic analysis using DNA polymorphism of the linkage between chromosome 11q13 and atopy and bronchial hyperresponsiveness to methacholine. J Allergy Clin Immunol. 1992;89:619–28. doi: 10.1016/0091-6749(92)90330-5. [DOI] [PubMed] [Google Scholar]
- 29.Hizawa N, Yamaguchi E, Ohe M, Itoh A, Furuya K, Ohnuma N, Kawakami Y. Lack of linkage between atopy and locus 11q13. Clin Exp Allergy. 1992;22:1065–9. doi: 10.1111/j.1365-2222.1992.tb00131.x. [DOI] [PubMed] [Google Scholar]
- 30.Torres-Galvan MJ, Quiralte J, Pestano JJ. IL4–R1 (5q31-q33) and FcepsilonRI-betaca (11q13) markers and atopy: a case/control study in a spanish population. Allergy. 2001;56:159–63. doi: 10.1034/j.1398-9995.2001.056002159.x. 10.1034/j.1398-9995.2001.056002159.x. [DOI] [PubMed] [Google Scholar]
- 31.Postma DS, Bleecker ER, Amelung PJ, Holroyd KJ, Xu J, Panhuysen CI, Meyers DA, Levitt RC. Genetic susceptibility to asthma-bronchial hyperresponsiveness coinherited with a major gene for atopy. N Engl J Med. 1995;333:894–900. doi: 10.1056/NEJM199510053331402. [DOI] [PubMed] [Google Scholar]
- 32.Bleecker ER, Amelung PJ, Levitt RC, Postma DS, Meyers DA. Evidence for linkage of total serum IgE and bronchial hyperresponsiveness to chromosome 5q: a major regulatory locus important in asthma. Clin Exp Allergy. 1995;25:84–8. doi: 10.1111/j.1365-2222.1995.tb00430.x. [DOI] [PubMed] [Google Scholar]
- 33.Noguchi E, Shibasaki M, Arinami T, et al. Evidence for linkage between asthma/atopy in childhood and chromosome 5q31-q33 in a Japanese population. Am J Respir Crit Care Med. 1997;156:1390–3. doi: 10.1164/ajrccm.156.5.9702084. [DOI] [PubMed] [Google Scholar]
- 34.Beyer K, Nickel R, Freidhoff L, et al. Association and linkage of atopic dermatitis with chromosome 13q12–14 and 5q31–33 markers. J Invest Dermatol. 2000;115:906–8. doi: 10.1046/j.1523-1747.2000.00096.x. [DOI] [PubMed] [Google Scholar]
- 35.Shek LP, Tay AH, Chew FT, Goh DL, Lee BW. Genetic susceptibility to asthma and atopy among Chinese in Singapore – linkage to markers on chromosome 5q31–33. Allergy. 2001;56:749–53. doi: 10.1034/j.1398-9995.2001.056008749.x. [DOI] [PubMed] [Google Scholar]
- 36.Blumenthal MN, Wang Z, Weber JL, Rich SS. Absence of linkage between 5q markers and serum IgE levels in four large atopic families. Clin Exp Allergy. 1996;26:892–6. [PubMed] [Google Scholar]
- 37.Kamitani A, Wong ZY, Dickson P, et al. Absence of genetic linkage of chromosome 5q31 with asthma and atopy in the general population. Thorax. 1997;52:816–17. doi: 10.1136/thx.52.9.816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mansur AH, Bishop DT, Markham AF, Britton J, Morrison JF. Association study of asthma and atopy traits and chromosome 5q cytokine cluster markers. Clin Exp Allergy. 1998;28:141–50. doi: 10.1046/j.1365-2222.1998.00229.x. 10.1046/j.1365-2222.1998.00229.x. [DOI] [PubMed] [Google Scholar]
- 39.Holberg CJ, Halonen M, Solomon S, Graves PE, Baldini M, Erickson RP, Martinez FD. Factor analysis of asthma and atopy traits shows 2 major components, one of which is linked to markers on chromosome 5q. J Allergy Clin Immunol. 2001;108:772–80. doi: 10.1067/mai.2001.119158. [DOI] [PubMed] [Google Scholar]
- 40.McIntire JJ, Umetsu SE, Akbari O, et al. Identification of Tapr (an airway hyperreactivity regulatory locus) and the linked Tim gene family. Nat Immunol. 2001;2:1109–16. doi: 10.1038/ni739. 10.1038/ni739. [DOI] [PubMed] [Google Scholar]
- 41.Marsh DG, Neely JD, Breazeale DR, et al. Linkage analysis of IL4 and other chromosome 5q31.1 markers and total serum immunoglobulin E concentrations. Science. 1994;264:1152–6. doi: 10.1126/science.8178175. [DOI] [PubMed] [Google Scholar]
- 42.Song Z, Casolaro V, Chen R, Georas SN, Monos D, Ono SJ. Polymorphic nucleotides within the human IL-4 promoter that mediate overexpression of the gene. J Immunol. 1996;156:424–9. [PubMed] [Google Scholar]
- 43.Hook S, Cheng P, Holloway J, Riley G, Sawyer G, Le Gros G, Beasley R. Analysis of two IL-4 promoter polymorphisms in a cohort of atopic and asthmatic subjects. Exp Clin Immunogenet. 1999;16:33–5. doi: 10.1159/000019094. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki I, Hizawa N, Yamaguchi E, Kawakami Y. Association between a C+33T polymorphism in the IL-4 promoter region and total serum IgE levels. Clin Exp Allergy. 2000;30:1746–9. doi: 10.1046/j.1365-2222.2000.00983.x. 10.1046/j.1365-2222.2000.00983.x. [DOI] [PubMed] [Google Scholar]
- 45.Shirakawa T, Deichmann KA, Izuhara I, Mao I, Adra CN, Hopkin JM. Atopy and asthma: genetic variants of IL-4 and IL-13 signalling. Immunol Today. 2000;21:60–4. doi: 10.1016/s0167-5699(99)01492-9. [DOI] [PubMed] [Google Scholar]
- 46.Rosenwasser LJ, Borish L. Genetics of atopy and asthma: the rationale behind promoter-based candidate gene studies (IL-4 and IL-10) Am J Respir Crit Care Med. 1997;156:S152–5. doi: 10.1164/ajrccm.156.4.12tac-14. [DOI] [PubMed] [Google Scholar]
- 47.Noguchi E, Shibasaki M, Arinami T, et al. Association of asthma and the interleukin-4 promoter gene in Japanese. Clin Exp Allergy. 1998;28:449–53. doi: 10.1046/j.1365-2222.1998.00256.x. 10.1046/j.1365-2222.1998.00256.x. [DOI] [PubMed] [Google Scholar]
- 48.Walley AJ, Cookson WO. Investigation of an interleukin-4 promoter polymorphism for associations with asthma and atopy. J Med Genet. 1996;33:689–92. doi: 10.1136/jmg.33.8.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dizier MH, Sandford A, Walley A, Philippi A, Cookson W, Demenais F. Indication of linkage of serum IgE levels to the interleukin-4 gene and exclusion of the contribution of the (−590 C to T) interleukin-4 promoter polymorphism to IgE variation. Genet Epidemiol. 1999;16:84–94. doi: 10.1002/(SICI)1098-2272(1999)16:1<84::AID-GEPI7>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 50.Hijazi Z, Haider MZ. Interleukin-4 gene promoter polymorphism [C590T] and asthma in Kuwaiti Arabs. Int Arch Allergy Immunol. 2000;122:190–4. doi: 10.1159/000024396. [DOI] [PubMed] [Google Scholar]
- 51.Elliott K, Fitzpatrick E, Hill D, et al. The −590C/T and −34C/T interleukin-4 promoter polymorphisms are not associated with atopic eczema in childhood. J Allergy Clin Immunol. 2001;108:285–7. doi: 10.1067/mai.2001.117180. [DOI] [PubMed] [Google Scholar]
- 52.Grunig G, Warnock M, Wakil AE, et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282:2261–3. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, Donaldson DD. Interleukin-13: central mediator of allergic asthma. Science. 1998;282:2258–61. doi: 10.1126/science.282.5397.2258. 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 54.van der Pouw Kraan TC, van Veen A, Boeije Lc, et al. An IL-13 promoter polymorphism associated with increased risk of allergic asthma. Genes Immun. 1999;1:61–5. doi: 10.1038/sj.gene.6363630. [DOI] [PubMed] [Google Scholar]
- 55.Heinzmann A, Mao XQ, Akaiwa M, et al. Genetic variants of IL-13 signalling and human asthma and atopy. Hum Mol Genet. 2000;9:549–59. doi: 10.1093/hmg/9.4.549. [DOI] [PubMed] [Google Scholar]
- 56.Graves PE, Kabesch M, Halonen M, et al. A cluster of seven tightly linked polymorphisms in the IL-13 gene is associated with total serum IgE levels in three populations of white children. J Allergy Clin Immunol. 2000;105:506–13. doi: 10.1067/mai.2000.104940. [DOI] [PubMed] [Google Scholar]
- 57.Pantelidis P, Jones MG, Welsh KI, Taylor AN, du Bois RM. Identification of four novel interleukin-13 gene polymorphisms. Genes Immun. 2000;1:341–5. doi: 10.1038/sj.gene.6363679. [DOI] [PubMed] [Google Scholar]
- 58.Liu X, Nickel R, Beyer K, et al. An IL13 coding region variant is associated with a high total serum IgE level and atopic dermatitis in the German multicenter atopy study (MAS-90) J Allergy Clin Immunol. 2000;106:167–70. doi: 10.1067/mai.2000.107935. [DOI] [PubMed] [Google Scholar]
- 59.Howard TD, Whittaker PA, Zaiman AL, et al. Identification and association of polymorphisms in the interleukin-13 gene with asthma and atopy in a Dutch population. Am J Respir Cell Mol Biol. 2001;25:377–84. doi: 10.1165/ajrcmb.25.3.4483. [DOI] [PubMed] [Google Scholar]
- 60.Leung TF, Tang NL, Chan IH, Li AM, Ha G, Lam CW. A polymorphism in the coding region of interleukin-13 gene is associated with atopy but not asthma in Chinese children. Clin Exp Allergy. 2001;31:1515–21. doi: 10.1046/j.1365-2222.2001.01212.x. [DOI] [PubMed] [Google Scholar]
- 61.Ober C, Leavitt SA, Tsalenko A, et al. Variation in the interleukin 4-receptor alpha gene confers susceptibility to asthma and atopy in ethnically diverse populations. Am J Hum Genet. 2000;66:517–26. doi: 10.1086/302781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hershey GK, Friedrich MF, Esswein LA, Thomas ML, Chatila TA. The association of atopy with a gain-of-function mutation in the alpha subunit of the interleukin-4 receptor. N Engl J Med. 1997;337:1720–5. doi: 10.1056/NEJM199712113372403. [DOI] [PubMed] [Google Scholar]
- 63.Oiso N, Fukai K, Ishii M. Interleukin 4 receptor alpha chain polymorphism Gln551Arg is associated with adult atopic dermatitis in Japan. Br J Dermatol. 2000;142:1003–6. doi: 10.1046/j.1365-2133.2000.03485.x. [DOI] [PubMed] [Google Scholar]
- 64.Wang HY, Shelburne CP, Zamorano J, Kelly AE, Ryan JJ, Keegan AD. Effects of an allergy-associated mutation in the human IL-4R alpha (Q576R) on human IL-4-induced signal transduction. J Immunol. 1999;162:4385–9. [PubMed] [Google Scholar]
- 65.Grimbacher B, Holland SM, Puck JM. The interleukin-4 receptor variant Q576R in hyper-IgE syndrome. N Engl J Med. 1998;338:1073–4. doi: 10.1056/NEJM199804093381516. [DOI] [PubMed] [Google Scholar]
- 66.Noguchi E, Shibasaki M, Arinami T, et al. Lack of association of atopy/asthma and the interleukin-4 receptor alpha gene in Japanese. Clin Exp Allergy. 1999;29:228–33. doi: 10.1046/j.1365-2222.1999.00458.x. 10.1046/j.1365-2222.1999.00458.x. [DOI] [PubMed] [Google Scholar]
- 67.Mitsuyasu H, Izuhara K, Mao XQ, et al. Ile50Val variant of IL4R alpha upregulates IgE synthesis and associates with atopic asthma. Nat Genet. 1998;19:119–20. doi: 10.1038/472. 10.1038/472. [DOI] [PubMed] [Google Scholar]
- 68.Mitsuyasu H, Yanagihara Y, Mao XQ, et al. Dominant effect of Ile50Val variant of the human IL-4 receptor alpha-chain in IgE synthesis. J Immunol. 1999;162:1227–31. [PubMed] [Google Scholar]
- 69.Kruse S, Japha T, Tedner M, Sparholt SH, Forster J, Kuehr J, Deichmann KA. The polymorphisms S503P and Q576R in the interleukin-4 receptor alpha gene are associated with atopy and influence the signal transduction. Immunology. 1999;96:365–71. doi: 10.1046/j.1365-2567.1999.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Andrews RP, Burrell L, Rosa-Rosa L, Cunningham CM, Brzezinski JL, Bernstein JA, Khurana Hershey GK. Analysis of the Ser786Pro interleukin-4 receptor alpha allelic variant in allergic and nonallergic asthma and its functional consequences. Clin Immunol. 2001;100:298–304. doi: 10.1006/clim.2001.5082. 10.1006/clim.2001.5082. [DOI] [PubMed] [Google Scholar]
- 71.Hizawa N, Freidhoff LR, Chiu YF, et al. Genetic regulation of Dermatophagoides pteronyssinus-specific IgE responsiveness: a genome-wide multipoint linkage analysis in families recruited through 2 asthmatic sibs. Collaborative Study on the Genetics of Asthma (CSGA) J Allergy Clin Immunol. 1998;102:436–42. doi: 10.1016/s0091-6749(98)70132-0. [DOI] [PubMed] [Google Scholar]
- 72.van Neerven RJ. The role of allergen-specific T cells in the allergic immune response: relevance to allergy vaccination. Allergy. 1999;54:552–61. doi: 10.1034/j.1398-9995.1999.t01-1-00092.x. [DOI] [PubMed] [Google Scholar]
- 73.Matsushita S, Muto M, Suemura M, Saito Y, Sasazuki T. HLA-linked nonresponsiveness to Cryptomeria japonica pollen antigen. I. Nonresponsiveness is mediated by antigen-specific suppressor T cell. J Immunol. 1987;138:109–15. [PubMed] [Google Scholar]
- 74.Caraballo L, Marrugo J, Jimenez S, Angelini G, Ferrara GB. Frequency of DPB1*0401 is significantly decreased in patients with allergic asthma in a mulatto population. Hum Immunol. 1991;32:157–61. doi: 10.1016/0198-8859(91)90051-a. [DOI] [PubMed] [Google Scholar]
- 75.Lara-Marquez ML, Yunis JJ, Layrisse Z, et al. Immunogenetics of atopic asthma: association of DRB1*1101 DQA1*0501 DQB1*0301 haplotype with Dermatophagoides spp.-sensitive asthma in a sample of the Venezuelan population. Clin Exp Allergy. 1999;29:60–71. doi: 10.1046/j.1365-2222.1999.00461.x. 10.1046/j.1365-2222.1999.00461.x. [DOI] [PubMed] [Google Scholar]
- 76.Kim YK, Oh HB, Oh SY, Cho SH, Kim YY, Min KU. HLA-DRB1*07 may have a susceptibility and DRB1*04 a protective effect upon the development of a sensitization to house dust mite Dermatophagoides pteronyssinus. Clin Exp Allergy. 2001;31:110–15. [PubMed] [Google Scholar]
- 77.Nickel RG, Casolaro V, Wahn U, et al. Atopic dermatitis is associated with a functional mutation in the promoter of the C-C chemokine RANTES. J Immunol. 2000;164:1612–16. doi: 10.4049/jimmunol.164.3.1612. [DOI] [PubMed] [Google Scholar]
- 78.Fryer AA, Spiteri MA, Bianco A, et al. The −403 G→A promoter polymorphism in the RANTES gene is associated with atopy and asthma. Genes Immun. 2000;1:509–14. doi: 10.1038/sj.gene.6363717. [DOI] [PubMed] [Google Scholar]
- 79.Szalai C, Kozma GT, Nagy A, Bojszko AA, Krikovszky D, Szabo T, Falus A. Polymorphism in the gene regulatory region of MCP-1 is associated with asthma susceptibility and severity. J Allergy Clin Immunol. 2001;108:375–81. doi: 10.1067/mai.2001.117930. [DOI] [PubMed] [Google Scholar]
- 80.Lee YA, Wahn U, Kehrt R, et al. A major susceptibility locus for atopic dermatitis maps to chromosome 3q21. Nat Genet. 2000;26:470–3. doi: 10.1038/82625. 10.1038/82625. [DOI] [PubMed] [Google Scholar]
- 81.Mitchell TJ, Walley AJ, Pease JE, Venables PJ, Wiltshire S, Williams TJ, Cookson WO. Delta 32 deletion of CCR5 gene and association with asthma or atopy. Lancet. 2000;356:1491–2. doi: 10.1016/S0140-6736(00)03144-5. [DOI] [PubMed] [Google Scholar]
- 82.Casolaro V, Keane-Myers AM, Swendeman SL, Steindler C, Zhong F, Sheffery M, Georas SN, Ono SJ. Identification and characterization of a critical CP2-binding element in the human interleukin-4 promoter. J Biol Chem. 2000;275:36605–11. doi: 10.1074/jbc.M007086200. [DOI] [PubMed] [Google Scholar]
- 83.Takeda K, Tanaka T, Shi W, et al. Essential role of Stat6 in IL-4 signaling. Nature. 1996;380:627–30. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 84.Shimoda K, van Deursen J, Sangster MY, et al. Lack of IL-4-induced Th2 response and IgE class switching in mice with disrupted Stat6 gene. Nature. 1996;380:630–3. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 85.Gao PS, Mao XQ, Roberts MH, et al. Variants of STAT6 (signal transducer and activator of transcription 6) in atopic asthma. J Med Genet. 2000;37:380–2. doi: 10.1136/jmg.37.5.380a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–2. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- 87.Adra CN, Gao PS, Mao XQ, Baron BW, Pauker S, Miki T, Shirakawa T, Hopkin JM. Variants of B cell lymphoma 6 (BCL6) and marked atopy. Clin Genet. 1998;54:362–4. doi: 10.1034/j.1399-0004.1998.5440418.x. [DOI] [PubMed] [Google Scholar]
- 88.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–69. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 89.Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J Biol Chem. 1997;272:21597–603. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- 90.Kishikawa H, Sun J, Choi A, Miaw SC, Ho IC. The cell type-specific expression of the murine IL-13 gene is regulated by GATA-3. J Immunol. 2001;167:44414–20. doi: 10.4049/jimmunol.167.8.4414. [DOI] [PubMed] [Google Scholar]
- 91.Finotto S, Neurath MF, Glickman JN, et al. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science. 2002;295:336–8. doi: 10.1126/science.1065544. [DOI] [PubMed] [Google Scholar]
- 92.Nakamura Y, Ghaffar O, Olivenstein R, Taha RA, Soussi-Gounni A, Zhang DH, Ray A, Hamid Q. Gene expression of the GATA-3 transcription factor is increased in atopic asthma. J Allergy Clin Immunol. 1999;103:215–22. doi: 10.1016/s0091-6749(99)70493-8. [DOI] [PubMed] [Google Scholar]
- 93.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 94.Vender JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 95.Brutsche MH, Brutsche IC, Wood P, Mogulkoc N, Custovic A, Egan J, Woodcock A. B-cell isotype control in atopy and asthma assessed with cDNA array technology. Am J Physiol Lung Cell Mol Physiol. 2001;280:L627–37. doi: 10.1152/ajplung.2001.280.4.L627. [DOI] [PubMed] [Google Scholar]
- 96.Brutsche MH, Brutsche IC, Wood P, et al. Apoptosis signals in atopy and asthma measured with cDNA arrays. Clin Exp Immunol. 2001;123:181–7. doi: 10.1046/j.1365-2249.2001.01441.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Karp CL, Grupe A, Schadt E, et al. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat Immunol. 2000;1:221–6. doi: 10.1038/79759. [DOI] [PubMed] [Google Scholar]
- 98.Westergren-Thorsson G, Malmstrom J, Marko-Varga G. Proteomics – the protein expression technology to study connective tissue biology. J Pharm Biomed Anal. 2001;24:815–24. doi: 10.1016/s0731-7085(00)00548-3. [DOI] [PubMed] [Google Scholar]