Abstract
Retinoic acid (RA) has been shown to regulate cellular growth and differentiation of a variety of cell types, including cells of the myelomonocytic lineage. We used the monocytic leukaemia cell line THP-1, which differentiates to macrophages in response to phorbol 12-myristate 13-acetate (PMA), to investigate the regulation by RA of genes in the scavenger receptor type B family (CD36) in human monocyte/macrophages. Reverse transcription–polymerase chain reaction and flow cytometry demonstrated that, like PMA and the natural peroxisome-proliferator-activated receptor-γ (PPARγ) ligand 15d-PGJ2, RA induced CD36 gene expression in these cells. Moreover, RA plus 15d-PGJ2 further enhanced CD36 protein and mRNA levels over that seen with the RA or PPARγ compounds alone. The PPARγ antagonist GW9662 was shown to block completely PPARγ-ligand induction of CD36 gene expression, but had little effect on the action of RA. Our data indicated that RXR- and RAR-specific ligands (LG153 and TTNPB, respectively) were each alone able to increase CD36 mRNA and surface protein levels. By using calphostin C, a specific protein kinase C (PKC) inhibitor, we demonstrated that induction of CD36 by PMA, as well as by PPARγ and RXR ligands were dependent upon PKC activation. In contrast, activation of CD36 through the RAR pathway was not affected by inhibition of PKC activity. Taken together, these data demonstrate that RA can up-regulate CD36 expression in human monocytes/macrophages. This regulation appears to be predominantly mediated through the RAR/RXR pathway of action and, unlike previously described methods of CD36 modulation, is independent of PPARγ and PKC signalling. This study suggests a possible role for RA in physiological processes involving the scavenger receptor function in cells of the monocyte/macrophage lineage.
Introduction
Scavenger receptor type B (SR-B) is a structurally heterogeneous family of proteins that includes CD36, the receptor for selective cholesteryl ester uptake, scavenger receptor class B type I (SR-BI), and lysosomal integral membrane protein II (LIMP-II).1,2 In cells of the monocyte/macrophage lineage, the scavenger receptor CD36 has been implicated in the phagocytosis of apoptotic cells and in the formation of foam cells during atherogenesis. Regulation of CD36 expression is a complex process resulting from the co-ordinated interplay between multiple soluble factors and cell surface adhesion molecules.3,4 Expression of CD36 has been shown to be regulated through the peroxisome-proliferator-activated-receptor γ (PPARγ) pathway and to be directly correlated with the maturational stage of the cells. Its expression is dramatically increased on monocytes upon their interaction with activated endothelium and by treatment of monocytes with macrophage colony-stimulating factor or interleukin-4 (IL-4). Furthermore, it has been shown that a variety of specific PPARγ ligands can, by themselves, up-regulate CD36, indicating that activation of PPARγ is sufficient to induce its expression.5–7 A common feature that appears to accompany CD36 up-regulation on macrophages induced by different methodologies (e.g. interaction with activated endothelium or treatment with macrophage colony-stimulating factor) is the differentiation of the cells as reflected by increased adherence and a variety of morphological criteria.1,8
We now demonstrate a mechanism of CD36 induction that involves a retinoid-mediated signalling pathway. In a recent report, Wuttge et al.9 showed that retinoic acid (RA) can up-regulate the expression of CD36, that this effect is mediated by RA nuclear receptors (RARs), and that RA treatment can enhance the ability of specific PPARγ ligands to up-regulate CD36. The present studies confirm and extend these observations. Our results indicate that up-regulation of CD36 by RA is independent of PPARγ signalling and is not associated with monocyte adherence or differentiation. Furthermore, unlike previously characterized mechanisms of CD36 regulation, we have determined that induction by RA is unique in not being dependent on protein kinase C (PKC) activity. A thorough understanding of this novel mechanism of action may lead to the discovery of a new class of drugs that can be utilized for clinical applications of selective CD36 modulation and increased scavenger activity in cells of the monocyte/macrophage lineage.
Materials and methods
Cell culture and chemicals
Human THP-1 cells were obtained from the American Type Culture Collection (Rockville, MD). Cells were grown in RPMI-1640 medium (Cellgro, Heradon, VA) supplemented with 10% heat-inactivated fetal bovine serum (FBS), HEPES buffer, 50 IU/ml penicillin/streptomycin, and 1 µg amphotericin (complete medium) as previously described.10 All-trans-retinoic acid (RA) and phorbol 12-myristate 13-acetate (PMA), purchased from Sigma (St Louis, MO), were dissolved in dimethylsulphoxide (0·2% of final volume). 15-deoxy-delta prostaglandin J2 (15d-PGJ2) was purchased from Alexis Biochemical (San Diego, CA). The synthetic PPARγ antagonist GW966211,12 was synthesized by the Medicinal Chemistry Department at Glaxo Wellcome Research and Development and was a generous gift from Dr T. M. Willson at same Institute (Glaxo Wellcome Co, Research Triangle Park, NC). LG100153 (LG153) and (E)-4-[2-(5,5,8,8-tetramethyl-5,6,7,8-tetrahydro-2-naphthalenyl)-1-propenyl] benzoic acid (TTNPB) was obtained from The Department of Medicinal Chemistry, Ligand Pharmaceuticals, Inc. (San Diego, CA). Calphostin C (CAL) was purchased from Calbiochem (La Jolla, CA). All reverse transcription–polymerase chain reaction (RT-PCR) kit components were from Perkin Elmer Co. (Foster City, CA). All other chemicals were purchased from Sigma (St. Louis, MO). Unless otherwise indicated, THP-1 cells were treated with RA (5 µm), PMA (100 nm), 15d-PGJ2 (10 µm), LG153 (5 µm), TTNPB (5 µm), CAL (0·5 µm) and GW9662 (20 µm) for 24 or 48 hr before analysis by flow cytometry or semiquantitative RT-PCR as described below.
RT-PCR
Total RNA was prepared from cultured THP-1 cells using TRIzol Reagent (Gibco BRL, Rockville, MD). To amplify 474 bp PPARγ, 389 bp CD36, and 200 bp GAPDH cDNA fragments, the sequences of PCR primers were: for PPARγ sense (5′-TCTCTCCGTAATGGAAGACC-3′), antisense (5′-GCATTATGAGACATCCCCAC-3′); for CD36 sense (5′-GAGAACTGTTATGGGGCTAT-3′), antisense (5′-TTCAACTGGAGAGGCAAAGG-3′); and for GAPDH sense (5′-CCATGGAGAAGGCTGGGG-3′), antisense (5′-CAAAGTTGTCATGGATGACC-3′), according to the published data.10,13,14 The RT-PCR was carried out as described.10 The ratio of primer concentrations for PPARγ and CD36 to GAPDH was about 1·3 : 0·7 if run in the same reaction tubes; otherwise they were run separately in order to get satisfactory results. The samples were first denatured at 95° for 30 seconds, followed by 32 PCR cycles, each with temperature variations as follows: 95° for 30 seconds, 60° for 30 seconds, and 72° for 30 seconds. The last cycle was followed by an additional extension incubation of 7 min at 72°. Analysis of amplicons was visualized on 1% agarose gel containing 0·2 µg/μl ethidium bromide and visualized under an ultraviolet transilluminator. The densitometric analysis of PCR products was performed using computer software (Bio-Rad Quantity One) and a GS-700 Imaging Densitometer (Bio-Rad, Hercules, CA), and was standardized by the GAPDH product. The ratios of PPARγ : GAPDH or CD36 : GAPDH density bands in control groups were considered as 100%. Values of treatment group PPARγ : GAPDH or CD36 : GAPDH ratios are given as percentage of controls. A 100-bp ladder (Gibco BRL, Rockville, MD) was used as a size standard.
Flow cytometry analysis
THP-1 cells were decanted from the culture flask if they were growing in suspension or were harvested by scraping or trypsin–ethylenediaminetetraacetic acid detachment if they were adherent as a result of some culture conditions (e.g. PMA treatment) and centrifuged. Aggregates were disrupted by vigorous pipetting. Cells (2 × 105–106) were washed twice with phosphate-buffered saline (PBS) and resuspended in 100 µl PBS containing 1% bovine serum albumin, incubated at 4° for 1 hr with appropriate dilutions of either purified R-phycoerythrin (R-PE)-conjugated mouse anti-human (CD36) monoclonal antibody or mouse immunoglobulin G (IgG), kappa isotype control (Pharmingen, San Diego, CA). The cells were washed and then analysed on a FAC-Scan cytofluorograph (Becton Dickinson, San Jose, CA). For each experiment, 10 000 events were collected and analysed using the CellQuest program based on their forward and side light-scatter profiles. The data were expressed as per cent positive and mean fluorescence intensity (MFI; arbitrary units). The ‘M1’ positive gates were set from isotype controls for each treatment condition such that these controls showed less than 2% positive individual cells. The isotype control histograms are not shown in order to simplify the figures unless otherwise indicated.
Statistical methods
All RT-PCR data were expressed as a mean±SE. Statistical significance was determined with Student's t-test (two-tailed) comparison between two groups of data sets. Asterisks shown in the figures indicate significant differences of experimental groups in comparison with the corresponding control condition (see figure legends).
Results
Induction of CD36 protein and mRNA in human monocytes
THP-1 cells were used as a model for investigating regulation of CD36 expression on cells of the monocyte/macrophage lineage. These cells can be induced to differentiate into macrophages by a variety of stimuli including PMA, granulocyte–macrophage colony-stimulating factor (GM-CSF), IL-415,16 Analysis of CD36 by flow cytometry (Fig. 1a) illustrates that untreated THP-1 cells show little, if any, surface expression of CD36. When these cells are cultured in the presence of PMA (100 nm), they undergo major differentiation changes, beginning at 12 hr of treatment which corresponds to the cells taking on the characteristics of mature macrophages as assessed by their morphological, antigenic and adherence properties.17 Figure 1(a) demonstrates that these cells also showed a dramatic increase in surface CD36 expression that correlated with this differentiation process. Thus, THP-1 cells appear to possess the same characteristics as normal human monocytes of increased CD36 expression as they differentiate into macrophages.18,19
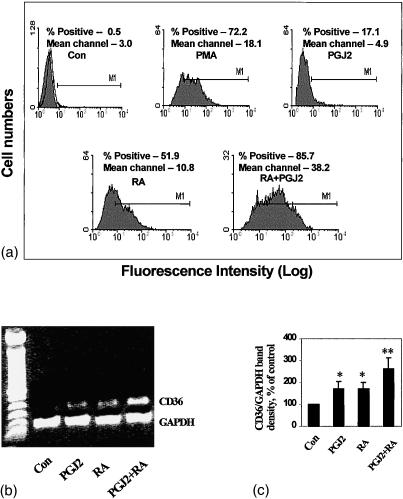
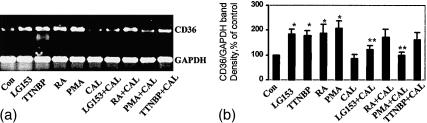
Figure 1.
Induction of CD36 protein and mRNA in THP-1 cells. Cells were cultured for 48 hr in the compounds as indicated or solvent control (Con). CD36 surface protein was measured by flow cytometry (a). In control cells (a, top left), the filled area indicates isotope control immunoglobulin as fluorescence background; the unfilled area represents specific staining with anti-CD36 monoclonal antibody. Cell surface staining of CD36 for all other treatment conditions is represented graphically by the filled areas. There were no significant differences in isotype control staining between treatment conditions. CD36 mRNA levels were assessed by RT-PCR (b) using CD36 and GAPDH primers. GAPDH mRNA expression was used as an internal control for normalization purposes. (c) represents the mean±SE CD36/GAPDH mRNA levels of at least three independent experiments; * indicates significant differences as compared to the solvent control while ** indicates significance between combination treatment and treatment with RA or 15d-PGJ2 alone (P < 0·05).
Studies from a number of laboratories have indicated that CD36 expression is regulated through the PPARγ pathway and that its expression can be induced by ligands to this nuclear receptor.5,7 Both normal monoctyes and THP-1 cells have been shown to express PPARγ.5,20 As expected, Fig. 1(a) demonstrates that the natural PPARγ ligand 15d-PGJ2 can significantly induce surface expression of CD36. The data show that RA is also a potent inducer of CD36 surface protein expression. To determine whether the observed RA-induced increase in CD36 surface protein on THP-1 cells reflects regulation at the transcriptional level, mRNA levels were assessed by semiquantitative RT-PCR.10,21 Positive controls for these experiments included cells treated with PMA and 15d-PGJ2. As shown in Fig. 1(b), a 2-day treatment with 5 µm RA, as well as with 10 µm 15d-PGJ2, caused a marked increase in steady-state levels of CD36 mRNA. Changes induced by RA were found to be rapid; THP-1 cells exposed to RA for less than 24 hr showed a significant increase in the levels of CD36 mRNA while maximum stimulation occurred after 2 days of treatment (data not shown). As with surface protein expression, the highest levels of CD36 mRNA were obtained after combination treatment with RA and 15d-PGJ2 (Fig. 1b,c).
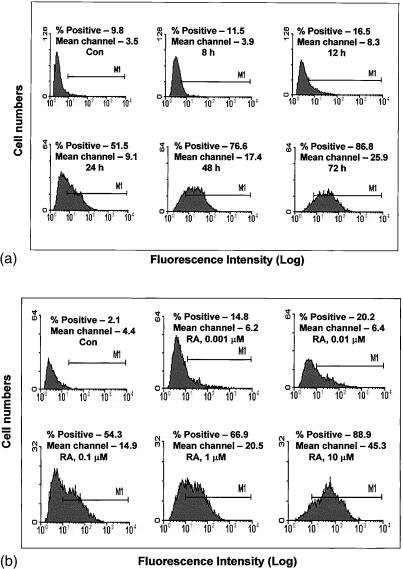
Figure 2(a) illustrates the time-dependent nature of this RA effect; culturing THP-1 cells with 5 µm RA resulted in an induction of CD36 protein beginning at 12 hr of treatment, and reached maximum levels after 48–72 hr. No further increase of CD36 was seen after 72 hr of treatment. As shown in Fig. 2(b), a significant increase in protein expression of CD36 was first observed at a RA concentration of 0·001 µm and a maximal increase was seen at 5–10 µm RA. No further increase in the presence of higher concentrations of RA was found. Unlike cultures with PMA where the cells became substrate adherent and took on the morphological features of macrophages (flattened morphology with multiple pseudopod formation), treatment with RA produced no apparent differentiation changes and cells remained non-adherant (data not show). These results support the finding of Zhu et al. who observed that 9-cis RA also did not induce differentiation of THP-1 cells.22
Figure 2.
Time and dose dependency of RA effect on CD36 surface protein level. THP-1 cells were cultured for various periods of time as shown in the presence of 5 µm RA (a), or for 48 hr in various concentrations of RA as indicated (b), then the CD36 surface protein levels were assessed by flow cytometry. All filled areas indicate cell surface staining of CD36 with anti-CD36 monoclonal antibody. Isotype control as fluorescence background is not shown.
Expression and up-regulation of PPARγ
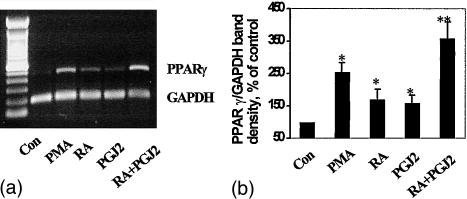
Expression of PPARγ mRNA in THP-1 cells following different treatment protocols was assessed by RT-PCR. Figure 3 demonstrates that cells treated for 1 day with 15d-PGJ2 (10 µm) as well as with RA (5 µm) showed up-regulation of PPARγ mRNA levels. A further increase of PPARγ expression was observed when RA was combined with 15d-PGJ2. These results extend previous reports demonstrating the ability of 9-cis RA and 15d-PGJ2 to up-regulate PPARγ in THP-1 cells.22
Figure 3.
Effect of RA on PPARγ mRNA levels. Total RNA was isolated from THP-1 cells cultured for 24 hr with PMA (100 nm), 15d-PGJ2 (10 µm), RA (5 µm), 15d-PGJ2 (10 µm) + RA (5 µm), or solvent control, then subjected to RT-PCR analysis using PPARγ and GAPDH primers (a). GAPDH mRNA expression was used as an internal control for normalization purposes. The bar graph (b) represents the mean±SE PPARγ/GAPDH mRNA levels of at least three independent experiments. * indicates significant differences as compared to the solvent control while ** indicates significance between combination treatment and single treatment values (P < 0·05).
RA induction of CD36 is not mediated through the PPARγ-signalling pathway
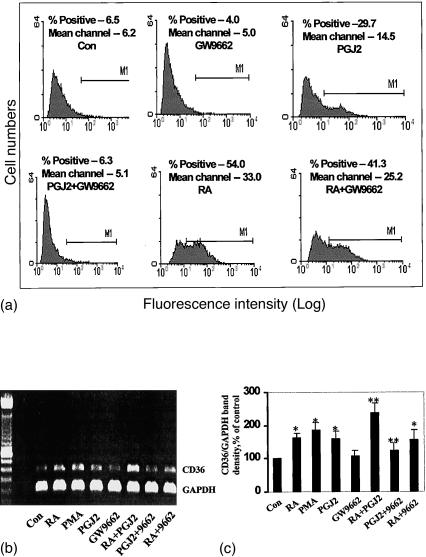
As shown above, RA induces both CD36 and PPARγ expression in THP-1 cells. Since CD36 is known to be regulated through PPARγ activation,23 this finding supported the contention that induction of CD36 by RA is caused by up-regulation of PPARγ. We addressed this possibility in a series of experiments utilizing specific PPARγ antagonists and other culture protocols that would prevent PPARγ signalling. As expected, while the PPARγ antagonist GW9662 alone (20 µm) had no effect on CD36 protein or mRNA levels in THP-1 cells (Fig. 4), it completely blocked its induction as assessed by both these criteria when the cells were treated with the PPARγ ligand 15d-PGJ2. In contrast, GW9662 only showed a slight inhibition of RA-induced CD36 expression; about 23% inhibition in the case of surface expression as assayed by flow cytometry and 25% inhibition in the case of mRNA levels as assessed by semiquantitative RT-PCR.
Figure 4.
The effect of PPARγ antagonist on CD36 protein and mRNA levels. The cells were cultured for 48 hr in the compounds as indicated. CD36 surface protein was measured by flow cytometry (a), with the filled areas representing specific staining with anti-CD36 monoclonal antibody. Isotype control immunoglobulin as fluorescence background is not shown. CD36 mRNA levels were assessed by RT-PCR (b) using CD36 and GAPDH primers. GAPDH mRNA expression was used as an internal control for normalization purposes. The bar graph (c) represents the mean±SE CD36/GAPDH mRNA levels of at least three independent experiments. * indicates significant differences as compared to the solvent control while ** indicates significance between combination treatment and treatment with RA or 15d-PGJ2 alone (P < 0·05).
The culture medium of normally passaged THP-1 cells and in the experiments described above included RPMI-1640 plus 10% fetal calf serum. Both FBS and RPMI-1640 contain ligands for PPARγ, for example, low-density lipoprotein (LDL) and fatty acids in the former and para-aminobenzoic acid in the latter. Since it has been shown that PPARγ-mediated gene activation requires liganded receptors,6,23 we determined whether up-regulation of CD36 by RA (5 µm) could be accomplished in Hanks' balanced salt solution (i.e. whether RA induction could be accomplished in the complete absence of any possible PPARγ ligands). Our results demonstrated that RA markedly increased CD36 surface protein (mean fluorescence intensity) and CD36 mRNA levels as compared to untreated controls under these conditions (data not show).
RA induction of CD36 is mediated through both RAR and RXR and is not dependent on PKC activation
Recent studies have shown that multiple methods of inducing CD36 in human monocytes, such as treatment with oxidized LDL, 15d-PGJ2 and IL-4, utilize a common signalling pathway dependent on both PKC and PPARγ. To determine whether PKC activity is required for RA-induced expression of CD36 mRNA, we evaluated expression of CD36 mRNA in THP-1 cells after treatment with RA (5 µm) alone, or in combination with the PKC inhibitor CAL.16 In these experiments, the cells were preincubated with CAL for 20 min prior to the addition of RA. Cells treated with PMA were used as positive controls for showing the effectiveness of blocking PKC-dependent activation of CD36 by this PKC inhibitor. Figure 5 shows a marked reduction in the levels of CD36 induced by PMA when the cells were co-treated with CAL (0·5 µm) for 24 hr. In contrast, RA up-regulation of CD36 expression was only marginally affected by CAL.
Figure 5.
RAR, RXR ligand regulation of CD36 mRNA levels in the presence of calphostin C. Total RNA was isolated from cells cultured for 48 hr with the indicated compounds or solvent control, then subjected to RT-PCR analysis using CD36 and GAPDH primers (a). GAPDH mRNA expression was used as an internal control for normalization purposes. The bar graphic (b) represents the mean±SE CD36/GAPDH mRNA levels of at least three independent experiments. * indicates significant differences as compared to the solvent control while ** indicates significance between combination treatment and treatment with PMA or LG153 alone (P < 0·05).
RA can isomerize in vitro to 9-cis RA, the natural ligand for RXR which is the heterodimeric partner of PPARγ.24 Work by Zhu et al.22 and others25 has demonstrated that 9-cis RA can synergize with PPARγ ligands in the activation of PPARγ–RXR heterodimers for induction of PPARγ-sensitive genes. Thus, it is possible that treatment with RA could interact with PPARγ signalling through cross-talk mediated by 9-cis RA-liganded RXR. In order to delineate RAR versus possible PPARγ/RXR-mediated effects of RA on CD36 expression and inhibition by CAL, cells were treated with the synthetic RAR and RXR ligands, TTNPB and LG153, respectively.26 As shown in Fig. 5, both TTNPB and LG153 effectively up-regulated CD36 mRNA levels in THP-1 cells. However, CD36 induction by these two ligands was differentially affected by the PKC inhibitor; while CAL had no significant effect on the ability of TTNPB to increase CD36 (21·5% inhibition), LG153-induced increases were inhibited by more than 60%. Similar effects by TTNPB and LG153 were seen on CD36 surface protein levels on THP-1 cells (data not shown).
Discussion
Despite the existence of a substantial amount of data documenting CD36 expression in a variety of cell types, only scant information is available on the mechanisms underlying its regulation. Unlike the LDL receptor, CD36 is not subject to negative regulation by high levels of intracellular cholesterol. On the contrary, binding by at least one of its ligands, oxLDL, can induce the expression of this receptor.5,7 The mechanism of this induction has been shown to be mediated by PPARγ. It has been demonstrated that induction of CD36 by IL-4 and PKC activators (e.g. diacylglycerol) are also PPARγ dependent.16 Recent findings indicated that down-regulation of CD36 by transforming growth factor-β1 (TGF-β1) and TGF-β2 was also mediated through the PPARγ-signalling pathway by alteration of PPARγ phosphorylation.6 Finally, it has been demonstrated that a variety of specific PPARγ ligands can, by themselves, up-regulate CD36 indicating that activation of PPARγ is sufficient for inducing surface protein expression and mRNA levels of this scavenger receptor.6,16 Thus, it appears that a variety of divergent physiological and pharmacological agents utilize a common pathway involving PPARγ to modulate CD36 expression.
Our studies have confirmed and extended the observations by Wuttge et al.9 that RA can up-regulate CD36 surface expression and mRNA levels in human monocytes. Unlike other methods of CD36 regulation that have been described, our findings indicate that this RA-mediated induction is largely independent of PPARγ signalling: (1) RA remained a potent inducer of CD36 surface protein and mRNA expression in the complete absence of any possible ligands for PPARγ; (2) a PPARγ antagonist (GW9662) only marginally inhibited RA induction of CD36; and (3) RA-mediated up-regulation of CD36 was not dependent on PKC activity as are PPARγ-dependent mechanisms of action.5 Our data indicated that the greatest expression of CD36 occurred following combination treatment with RA plus the natural PPARγ ligand 15d-PGJ2, further supporting a contention of non-coincidental pathways of action for these compounds. Wuttge et al.9 showed similar co-operation between RA and oxLDL in the induction of CD36 on primary peripheral blood monocytes.
Our studies with the RAR- and RXR-specific ligands TTNPB and LG153, respectively, indicated that both RAR- and RXR-mediated pathways can be involved in CD36 regulation. Since ligands to RXR (e.g. 9-cis RA) have been shown to synergize with PPARγ ligands in activating the PPARγ–RXR dimer complex,27 it is possible that LG153 induction of CD36 reflects enhanced signalling through the PPARγ–RXR pathway. This contention is supported by the fact that LG153-induction of CD36 was inhibited by the PKC inhibitor CAL, as was induction by PPARγ ligands. In contrast, induction by the specific RAR ligand TTNPB was not affected by CAL, suggesting that the RAR-mediated signalling pathway is the component of RA induction that is PPARγ and PKC independent.
The molecular mechanism of CD36 transcriptional activation by RA is unknown. Armesilla and Vega28 delineated the structural organization of the CD36 gene, which revealed the presence of a TATA box located 28 bp upstream from the transcription initiation site. In later work, these investigators also defined CD36 gene regions and identified transcription factors that are important for the expression of CD36 in monocyte/macrophage cells.29 Their data demonstrated that the transcription of the CD36 gene in monocytic cell lines is mainly controlled by its proximal promoter region and that transcription factors belonging to the polyomavirus enhancer-binding protein2 (PEBP2) family play a major role in the transcriptional regulation of CD36. Interestingly, it has been shown that members of this PEBP2 family of transcription factors can be up-regulated by RA in F9 mouse teratocarcinoma cells.30 Although similar studies in other cell types have not been reported, these findings suggest that one mechanism of RA induction of CD36 in human monocytes is through up-regulation of PEBP2.
In addition to macrophages, CD36 is found on platelets and microvascular endothelial cells in some organs, such as the liver.1,6,19 Monocytes from humans genetically deficient in CD36 have a decreased capacity to take up oxLDL31 and this scavenger receptor has been shown to be required for atherogenesis in mice.32 These findings incontrovertibly link CD36 to the pathogenesis of atherosclerosis. Because of this link, the induction of CD36 by RA might be considered as evidence that retinoid signalling is involved in atherosclerosis and that treatment with RA could hasten atherogenesis. This argument has been proposed by Wuttge et al.9 and was supported by their demonstration of retinoid ligands and RA receptors in human atherosclerotic plaques. However, atherosclerosis is a complex syndrome with multiple factors that may be affected by CD36 function. This fact is emphasized by the finding that no correlation has been seen between CD36 induction in vivo, and atherogenesis.33 Indeed, induction of CD36 by treatment of mice with PPARγ ligands has shown a net reduction (rather than increase) of atherosclerosis.33 How might this be explained? First of all, pharmacological methods for up-regulating CD36 are not specific. For example, PPARγ has been determined to differentially regulate various macrophage-expressed scavenger receptors and functions.34,35 Thus, PPARγ ligands can suppress the induction of SR-A by phorbol esters while at the same time promoting the expression of CD36.34 Another possibility is that CD36 expression is not ordinarily rate-limiting in the development of foam cells; while CD36 is required for foam cell formation, there is no evidence that increased expression of CD36 can further accelerate this process. It is more probable that levels of LDL cholesterol or the conversion to oxLDL limit the rate of foam cell development. In this regard, it is worth noting that PPARγ ligands induce CD36 in multiple tissues, including fat. Hence, the removal of oxLDL from the circulation into CD36-rich adipose or other tissues may provide a net total benefit on atherogenesis that is not counteracted by elevated CD36 in macrophages. The fact that heterozygous LDL receptor knockout mice are protected from atherosclerosis by overexpression of CD36 in the liver demonstrates the validity of this idea.36 Future studies will determine the ability of retinoids to modulate CD36 expression in multiple cell types and tissues in order to appreciate fully their potential benefits and risks as compounds for altering immunological and atherosclerotic events.
Acknowledgments
This work was supported by National Institutes of Health Research grant P01-HD35276.
References
- 1.Huh HY, Pearce SF, Yesner LM, Schindler JL, Silverstein RL. Regulated expression of CD36 during monocyte-to-macrophage differentiation: potential role of CD36 in foam cell formation. Blood. 1996;87:2020–8. [PubMed] [Google Scholar]
- 2.Febbraio M, Hajjar DP, Silverstein RL. CD36: a class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J Clin Invest. 1901;108:785–91. doi: 10.1172/JCI14006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Silverstein RL, Febbraio M. CD36 and atherosclerosis. Curr Opin Lipidol. 2000;11:483–91. doi: 10.1097/00041433-200010000-00006. 10.1097/00041433-200010000-00006. [DOI] [PubMed] [Google Scholar]
- 4.Janabi M, Yamashita S, Hirano K, et al. Reduced adhesion of monocyte-derived macrophages from CD36-deficient patients to type I collagen. Biochem Biophys Res Commun. 2001;283:26–30. doi: 10.1006/bbrc.2001.4718. [DOI] [PubMed] [Google Scholar]
- 5.Tontonoz P, Nagy L, Alvarez JG, Thomazy VA, Evans RM. PPARgamma promotes monocyte/macrophage differentiation and uptake of oxidized LDL. Cell. 1998;93:241–52. doi: 10.1016/s0092-8674(00)81575-5. [DOI] [PubMed] [Google Scholar]
- 6.Han J, Ajar DP, Tauras JM, Feng J, Gotto AM, Jr, Nicholson AC. Transforming growth factor-beta1 (TGF-beta1) and TGF-beta2 decrease expression of CD36, the type B scavenger receptor, through mitogen-activated protein kinase phosphorylation of peroxisome proliferator-activated receptor-gamma. J Biol Chem. 2000;275:1241–6. doi: 10.1074/jbc.275.2.1241. [DOI] [PubMed] [Google Scholar]
- 7.Nagy L, Tontonoz P, Alvarez JG, Chen H, Evans RM. Oxidized LDL regulates macrophage gene expression through ligand activation of PPARgamma. Cell. 1998;93:229–40. doi: 10.1016/s0092-8674(00)81574-3. [DOI] [PubMed] [Google Scholar]
- 8.Kerkhoff C, Sorg C, Tandon NN, Nacken W. Interaction of S100A8/S100A9-arachidonic acid complexes with the scavenger receptor CD36 may facilitate fatty acid uptake by endothelial cell. Biochemistry. 2001;40:241–8. doi: 10.1021/bi001791k. [DOI] [PubMed] [Google Scholar]
- 9.Wuttge DM, Romer A, Eriksson U, Torma H, Hansson GK, Sirsjo A. Induction of CD36 by all-trans retinoic acid: retinoic acid receptor signaling in the pathogenesis of atherosclerosis. FASEB J. 2001;15:1221–3. doi: 10.1096/fj.00-0488fje. [DOI] [PubMed] [Google Scholar]
- 10.Han SW, Greene ME, Pitts J, Wada RK, Sidell N. Novel expression and function of peroxisome proliferator-activated receptor gamma (PPARgamma) in human neuroblastoma cells. Clin Cancer Res. 2001;7:98–104. [PubMed] [Google Scholar]
- 11.Miyahara T, Schrum L, Rippe R, et al. Peroxisome proliferator-activated receptors and hepatic stellate cell activation. J Biol Chem. 2000;275:35715–22. doi: 10.1074/jbc.M006577200. [DOI] [PubMed] [Google Scholar]
- 12.Han S, Wada RK, Sidell N. Differentiation of human neuroblastoma by phenylacetate is mediated by peroxisome proliferator-activated receptor gamma. Cancer Res. 2001;61:3998–4002. [PubMed] [Google Scholar]
- 13.Pietsch A, Erl W, Lorenz RL. Lovastatin reduces expression of the combined adhesion and scavenger receptor CD36 in human monocytic cells. Biochem Pharmacol. 1996;52:433–9. doi: 10.1016/0006-2952(96)00245-6. [DOI] [PubMed] [Google Scholar]
- 14.Satta MA, Korbonit M, Jacos RA, et al. Expression of menin gene mRNA in pituitary tumours. Eur J Endocrinol. 1999;140:358–61. doi: 10.1530/eje.0.1400358. [DOI] [PubMed] [Google Scholar]
- 15.Yesner LM, Huh H, Pearce SF, Silverstein RL. Regulation of monocyte CD36 and thrombospondin-1 expression by soluble mediators. Arterioscler Thromb Vasc Biol. 1996;16:1019–25. doi: 10.1161/01.atv.16.8.1019. [DOI] [PubMed] [Google Scholar]
- 16.Feng J, Han J, Pearce SF, Silverstein RL, Gotto AM, Jr, Hajjar DP, Nicholson AC. Induction of CD36 expression by oxidized LDL and IL-4 by a common signaling pathway dependent on protein kinase C and PPAR-gamma. J Lipid Res. 2000;41:688–96. [PubMed] [Google Scholar]
- 17.Tsuchiya S, Kobayashi Y, Goto Y, Okumura H, Nakae S, Konno T, Tada K. Induction of maturation in cultured human monocytic leukemia cells by phorbol diester. Cancer Res. 1982;42:1530–6. [PubMed] [Google Scholar]
- 18.Keisari Y, Robin G, Nissimov L, Wang H, Mesika A, Dimri R, Ofek I. Role of cytokines in the maturation and function of macrophages. Effect of GM-CSF and IL-4. Adv Exp Med Biol. 2000;479:73–89. doi: 10.1007/0-306-46831-X_7. [DOI] [PubMed] [Google Scholar]
- 19.Greenwalt DE, Lipsky RH, Ockenhouse CF, Ikeda H, Tandon NN, Jamieson GA. Membrane glycoprotein CD36: a review of its roles in adherence, signal transduction, and transfusion medicine. Blood. 1992;80:1105–15. [PubMed] [Google Scholar]
- 20.Kintscher U, Goetze S, Wakino S, et al. Peroxisome proliferator-activated receptor and retinoid X receptor ligands inhibit monocyte chemotactic protein-1-directed migration of monocytes. Eur J Pharmacol. 2000;401:259–70. doi: 10.1016/s0014-2999(00)00461-1. [DOI] [PubMed] [Google Scholar]
- 21.Goswami PC, Albee LD, Spitz DR, Ridnou LA. A polymerase chain reaction assay for simultaneous detection and quantitation of proto-oncogene and GAPD mRNAs in different cell growth rates. Cell Prolif. 1997;30:271–82. doi: 10.1046/j.1365-2184.1997.00093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu L, Gong B, Bisgaier CL, Aviram M, Newton RS. Induction of PPARgamma1 expression in human THP-1 monocytic leukemia cells by 9-cis-retinoic acid is associated with cellular growth suppression. Biochem Biophys Res Commun. 1998;251:842–8. doi: 10.1006/bbrc.1998.9567. [DOI] [PubMed] [Google Scholar]
- 23.Tontonoz P, Nagy L. Regulation of macrophage gene expression by peroxisome-proliferator-activated receptor gamma: implications for cardiovascular disease. Curr Opin Lipidol. 1999;10:485–90. doi: 10.1097/00041433-199912000-00002. 10.1097/00041433-199912000-00002. [DOI] [PubMed] [Google Scholar]
- 24.Kliewer SA, Xu HE, Lambert MH, Willson TM. Peroxisome proliferator-activated receptors: from genes to physiology. Recent Prog Horm Res. 1901;56:239–63. doi: 10.1210/rp.56.1.239. [DOI] [PubMed] [Google Scholar]
- 25.Benson S, Padmanabhan S, Kurtz TW, Pershadsingh HA. Ligands for the peroxisome proliferator-activated receptor-gamma and the retinoid X receptor-alpha exert synergistic antiproliferative effects on human coronary artery smooth muscle cells. Mol Cell Biol Res Commun. 2000;3:159–64. doi: 10.1006/mcbr.2000.0209. [DOI] [PubMed] [Google Scholar]
- 26.Rosati R, Ramnath N, Adil MR, Ou X, Ali MA, Heyman RA, Kalemkerian GP. Activity of 9-cis-retinoic acid and receptor-selective retinoids in small cell lung cancer cell lines. Anticancer Res. 1998;18:4071–5. [PubMed] [Google Scholar]
- 27.Schulman IG, Shao G, Heyman RA. Transactivation by retinoid X receptor-peroxisome proliferator-activated receptor gamma (PPARgamma) heterodimers: intermolecular synergy requires only the PPARgamma hormone-dependent activation function. Mol Cell Biol. 1998;18:3483–94. doi: 10.1128/mcb.18.6.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Armesilla AL, Vega MA. Structural organization of the gene for human CD36 glycoprotein. J Biol Chem. 1994;269:18985–91. [PubMed] [Google Scholar]
- 29.Armesilla AL, Calvo D, Vega MA. Structural and functional characterization of the human CD36 gene promoter: identification of a proximal PEBP2/CBF site. J Biol Chem. 1996;271:7781–7. doi: 10.1074/jbc.271.13.7781. [DOI] [PubMed] [Google Scholar]
- 30.Geoffroy V, Ducy P, Karsenty GA. PEBP2 alpha/AML-1-related factor increases osteocalcin promoter activity through its binding to an osteoblast-specific cis-acting element. J Biol Chem. 1995;270:30973–9. doi: 10.1074/jbc.270.52.30973. [DOI] [PubMed] [Google Scholar]
- 31.Nozaki S, Kashiwagi H, Yamashita S, et al. Reduced uptake of oxidized low density lipoproteins in monocyte-derived macrophages from CD36-deficient subjects. J Clin Invest. 1995;96:1859–65. doi: 10.1172/JCI118231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Febbraio M, Podrez EA, Smith JD, Hajjar DP, Hazen SL, Hoff HF, Sharma K, Silverstein RL. Targeted disruption of the class B scavenger receptor CD36 protects against atherosclerotic lesion development in mice. J Clin Invest. 2000;105:1049–56. doi: 10.1172/JCI9259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicholson AC, Febbraio M, Han J, Silverstein RL, Hajjar DP. CD36 in atherosclerosis. The role of a class B macrophage scavenger receptor. Ann N Y Acad Sci. 2000;902:128–31. [PubMed] [Google Scholar]
- 34.Li AC, Brown KK, Silvestre MJ, Willson TM, Palinski W, Glass CK. Peroxisome proliferator-activated receptor gamma ligands inhibit development of atherosclerosis in LDL receptor-deficient mice. J Clin Invest. 2000;106:523–31. doi: 10.1172/JCI10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chinetti G, Gbaguidi FG, Griglio S, et al. CLA-1/SR-BI is expressed in atherosclerotic lesion macrophages and regulated by activators of peroxisome proliferator-activated receptors. Circulation. 2000;101:2411–17. doi: 10.1161/01.cir.101.20.2411. [DOI] [PubMed] [Google Scholar]
- 36.Arai T, Wang N, Bezouevski M, Welch C, Tal AR. Decreased atherosclerosis in heterozygous low density lipoprotein receptor-deficient mice expressing the scavenger receptor BI transgene. J Biol Chem. 1999;274:366–71. doi: 10.1074/jbc.274.4.2366. [DOI] [PubMed] [Google Scholar]