Abstract
We have previously isolated (by expression cloning) a human cDNA, termed NFX.1, encoding a nucleic acid-binding protein that interacts with the conserved X1 box cis-element first discovered in class II major histocompatibility complex (MHC) genes. Functional studies involving expression of NFX.1 and assessment of expression from class II reporter constructs and endogenous class II MHC genes indicated that the factor could repress transcription of class II MHC genes. Subsequent studies have extended the biological significance of the factor, indicating that it plays an important role in neuronal development. Indeed, the reiterated RING finger motifs in the central domain of the polypeptide strongly suggest that NF-XI is a probable E3 ubiquitin protein ligase, indicating that the protein may have multiple activities. Here we report the cloning of the mouse homologue of the human NfX.1 cDNA: m-Nfx.1. Comparison of the deduced primary sequence of mouse and human NFX.1 proteins shows very high homology and confirms that m-NFX.1 contains the conserved cysteine-rich DNA-binding motif first described in human NFX.1 (95% homology). Expression of MHC class II genes is substantially reduced following expression of m-NFX.1, which confirms that we have isolated the functional murine homologue of human NfX.1 cDNA. Further evidence comes from the mapping of m-Nfx.1 gene to the proximal region of mouse chromosome 4, a region syntenic to the location of human Nfx.1 (short arm of chromosome 9). Expression profiling shows that m-NFX.1 is expressed ubiquitously in both adult tissues and during development, supporting the hypothesis that it may have yet-undescribed roles in distinct biological processes.
Introduction
Major histocompatibility complex (MHC) class II molecules present fragments of foreign antigens to CD4+ helper T cells and thus play a central role in the immune response (reviewed in refs 1–3). Some immune processes that are dependent upon class II MHC molecules include: thymic selection of T lymphocytes; tolerance induction; antibody production (via T-cell help); direct T-cell-mediated immune responses; and inflammation.
Constitutive expression of MHC class II molecules is normally restricted to selected cell types that act as professional antigen-presenting cells, which include macrophages, dendritic cells and B cells. However, expression of MHC class II genes can also be induced in a variety of MHC class II-negative cells upon binding of cytokines such as interferon-γ (IFN-γ), interleukin (IL)-4 and tumour necrosis factor-α (TNF-α) to their respective receptors.3,4 In humans, lack of both constitutive and induced expression of MHC class II genes results in a serious, often lethal, immunodeficiency disorder known as type II bare lymphocyte syndrome (BLS).5,6 Aberrant expression of MHC class II genes is often observed in target organs of several autoimmune diseases (i.e. in multiple sclerosis, rheumatoid arthritis and insulin-dependent diabetes mellitus).
As different allelic variants of MHC class II molecules are capable of binding exogenous peptides with different efficiency, these highly polymorphic molecules directly determine the ability of each individual to respond appropriately to different antigens. Given their central role in initiating the immune response, great effort has been focused on elucidating the molecular mechanisms controlling proper tissue- and stage-specific expression of MHC class II molecules. Although post-transcriptional mechanisms also contribute to class II MHC gene regulation, this is predominantly controlled at the transcriptional level (reviewed in refs 7–9).
The transcriptional regulation of MHC class II genes is complex and the result of multiple levels of control over the activity of their promoters. At a first level, promoter occupancy by activators or repressors of transcription directly affects gene expression. Detailed analysis of the promoter region of MHC class II genes has shown the presence of three conserved DNA-binding elements, named S, X (divided into X1 and X2) and Y boxes, found within 160 base pairs (bp) upstream of the transcription initiation site. Several conserved transcription factors have been identified that specifically bind to each of the three ‘boxes’ in a co-operative manner. These include, among others, Y box-binding proteins like NF-Y and YB-1, as well as several proteins of the RFX family, that bind to the X boxes (reviewed in refs 7, 8 and 10).
The integrity of all three ‘boxes’ [as well as the octamer motif in the human leucocyte antigen (HLA)-DRA promoter] is necessary for proper transcription of MHC class II genes.3 This is supported by the demonstration that changes in the stereospecific alignment of the three elements results in impaired transcription11 and that X- and Y-box factors bind to their sites in a co-operative manner.12,13 Aside from the X- and Y box-binding proteins, two additional categories of factors have been found to contribute to the regulation of class II transcription. Somatic cell-fusion experiments, using BLS patient-derived cells to define complementation groups, allowed the identification of a master regulator of class II expression: the class II transactivator (CIITA).14 CIITA does not bind to DNA, but interacts with promoter-bound proteins at both X and Y boxes and with the basal transcription machinery to co-ordinate the formation of high-order protein complexes. The final result is the formation of an enhanceosome for class II transcription (reviewed in ref. 15). Finally, we have previously shown that ‘architectural’ proteins such as HMGI(Y) affect the transcription of MHC class II genes by changing the local conformation of DNA at the promoter in order to facilitate the binding of activators.16
Given the role of MHC class II molecules in normal and deviant immune responses, it is critical to broaden our knowledge of factors that control transcription of these genes. Moreover, a better understanding of the molecular events associated with the activity of each of these factors is required for any attempt at manipulating MHC class II expression with a therapeutic goal. As part of our effort to identify transcription factors that control the expression of MHC class II genes, we previously reported the identification of an X1 box-binding protein that functions as a repressor of class II genes in transfected cells. The repressor activity of NFX.1 was demonstrated by its ability to downregulate expression of co-transfected class II reporter constructs and endogenous class II MHC genes (monitored by RNAse I protection analysis). We named this novel protein NFX.1 as it specifically represses class II expression via binding to the X1 region within the X box.17 Here we report the identification and functional characterization of the first mouse homologue of human Nfx.1. Availability of the murine cDNA paves the way for in vivo studies in mice through the creation of transgenic models to better understand the role of NFX.1 in the immune (and perhaps other) systems.
Materials and Methods
Northern blot analysis
WEHI-3 cells were grown to 60% confluence and induced with recombinant human IFN-γ (250 U/ml) (PharMingen, San Diego, CA) for 48 hr at 37° before harvest. Poly A+ RNAs was extracted using a FastTrack 2·0 mRNA isolation kit (Invitrogen, Carlsbad, CA) following the instructions of the manufacturer. Approximately 3 µg of mRNA was electrophoresed in a 1% formaldehyde-denaturing agarose gel, alongside a sample of total RNA to establish the size of the transcript(s). RNA was transferred onto Hybond-N+ membranes (Amersham, Arlington Heights, IL) and UV cross-linked. A PstI–PstI DNA fragment spanning nucleotide (nt) +2018 to nt +2514 (relative to the first ATG) of the m-Nfx.1 coding region was used as a probe and labelled with [α-32P]dCTP (ICN, Costa Mesa, CA). Hybridization was carried out in Quick-Hyb Hybridization solution (Stratagene, St Louis, MO) at 65° with 40 µg/ml of sonicated salmon-sperm DNA (Sigma, St Louis, MO) and 10 ng/ml of probe. The membrane was stringently washed at 65° with 0·1× saline sodium citrate (SSC) containing 0·1% sodium dodecyl sulphate (SDS). Autoradiography was performed using Fujii Rx films (Sigma).
Library screening
A RAPID-SCREEN cDNA library (OriGene Technologies, Rockville, MD) was used to clone the mouse m-NFX.1 cDNA. The library was screened by polymerase chain reaction (PCR) amplification following the instructions of the manufacturer. Two PCR primers: h-NFX.1-5′ (5′-TGCCATGGGGGTCAGTGC-3′) and h-NFX.1-3′ (5′-GGTCCACAGTCTCCTTCATGGCA-3′), were designed on the human Nfx.1 cDNA sequence and used to amplify, by reverse transcription–PCR (RT–PCR), a 300-bp DNA fragment from fetal mouse RNA (E19). The same source of RNA was used to construct the library. For RT–PCR, 200 U of Superscript II reverse transcriptase (Gibco BRL, Gaithersburg, MD) were used following standard protocols. The amplified fragment was sequenced and used to derive a second pair of mouse-specific primers: m-NFX.1-5′ (5′-AGCCTTGTCGGATCATACTG-3′) and m-NFX.1-3′ (5′-GCTTTTCACAGGTGTGGATG-3′), that were used to screen the library. Nfx.1 clone no. 61 was automatically sequenced on both strands using several internal primers on the available sequence. All the primers are indicated in Fig. 1b. Clone Nfx.1 no. 23 was obtained by screening the library with a second set of primers: m-NFX.1-vec (5′-ATGGGCGGTAGGCGTGTACGGTG-3′, on the backbone vector used to build the library) and m-NFX.1-E (5′-CTGAAGGCCATGGTTCTTGAGT-3′, spanning nt +297 to nt +273 on the m-NFX.1 coding region). The entire sequence of clone m-Nfx.1 no. 61 was aligned to the human Nfx.1 sequence and, using the GeneWorks software (IntelliGenetics, Mountain View, CA), identified an open reading frame (ORF). The murine amino acid sequence and hydrophobic profile were also derived from the DNA sequence using the same computer software.
Figure 1 (b).
Deduced amino acid sequence of the murine NFX.1 protein. The complete deduced amino acid sequence of the full-length open reading frame (ORF) from m-Nfx.1 clone 61 is shown here. No other significant ORFs were found in either strand and the size of the transcript is in agreement with the size of the most abundant transcript (≈ 3.9 kb) observed by Northern blot. The seven repeated cysteine-rich motifs are boxed. Arrows show the position and length of some of the primers used to sequence the clone. These sequence data are available from the European Molecular Biology Laboratory (EMBL) GenBank database under accession number AF223576.
Interspecific mouse backcross mapping
Interspecific backcross progeny were generated by mating (C57BL/6J×Mus spretus) F1 females and C57BL/6J males as described previously.18 A total of 205 N2 mice were used to map the Nfx.1 locus (see below for details). DNA isolation, restriction enzyme digestion, agarose-gel electrophoresis, Southern blot transfer and hybridization were performed essentially as described previously.19 All blots were prepared with Hybond-N+ nylon membrane (Amersham). The probe, a 280-bp SmaI–EcoRI fragment of mouse cDNA, was labelled with [α-32P]dCTP (ICN) using a random-primed labelling kit (Stratagene); washing was performed to a final stringency of 1× SSCP, 0·1% SDS, at 65°. Fragments of 21·0, 7·9, 5·2, 2·5, and 1·7 kb were detected in TaqI-digested C57BL.6J DNA and fragments of 14·0, 7·5, 4·4, 3·6, 3·1, and 2·5 kb were detected in TaqI-digested M. spretus DNA. The presence or absence of the 14·0, 7·5, 4·4, 3·6, and 3·1 kb TaqI M. spretus-specific fragments, which co-segregated, was followed in backcross mice. A description of the probes and restriction fragment length polymorphisms (RFLPs) for loci linked to Nfx.1, including Cga, Cntfr and Tgfbr1, has been reported previously.20 Recombination distances were calculated using Map manager, version 2.6.5. Gene order was determined by minimizing the number of recombination events required to explain the allele distribution patterns.
Cell culture
Mouse WEIH-3 cells were maintained in Isocove's Modified Dulbecco's Medium (IMDM) supplemented with 10% fetal calf serum (FCS), 1% l-glutamine, 1% penicillin–streptomycin, 1% sodium pyruvate, 1% non-essential amino acids and 0·05 mm 2-mercaptoethanol. Mouse B-cell lymphoma A20 cells were cultured in RPMI-1640 supplemented with 10% FCS, 1% l-glutamine, 1% penicillin–streptomycin and 0·05 mm 2-mercaptoethanol. All tissue culture reagents were obtained from Gibco BRL.
Transfections, and CAT and β-galactosidase assays
Each plasmid DNA for transfection was prepared by using the Qiagen Plasmid MAXI kit (Qiagen, Chatsworth, CA). The (−4·1 kb) CAT construct (bearing 4·1 kb of the promoter of the gene) was a gift from Dr L. H. Glimcher (Harvard Medical School, Boston, MA).21 pCAT 3-Control and pCAT 3-Basic were purchased from Promega (Madison, WI). pCH110 (a β-galactosidase-expressing vector) was from Pharmacia (Arlington Heights, IL). Cells were cultured in polystyrene flasks (Corning, Cambridge, MA) and pelleted on the day of transfection. A20 cells (5×106) were resuspended in 300 µl of media and transfected using an Electroporator II (Invitrogen) set at 300 V, in 4-mm gap cuvettes (BTX-Genetronix, San Diego, CA). A total amount of 25 µg of plasmid DNA was used for each transfection and cells were incubated on ice for 10 min before and after shocking. After transfection, cells were cultured in six-well plates with fresh media for 48 hr at 37° before harvesting. Each transfection experiment was carried out in duplicate and repeated three times with two independent batches of DNA. For CAT and β-galactosidase assays, cells were washed twice with cold phosphate-buffered saline (PBS) and then lysed in a 200-µl volume of 1× reporter lysis buffer (Promega) for 20 min at room temperature. The lysate was then frozen, thawed and vortexed. A clear supernatant was collected by centrifugation at 9000 g for 5 min at 4°. For β-galactosidase activity, 50 µl of the lysate was used with the β-galactosidase enzyme assay system (Promega) following the protocol of the manufacturer. For CAT activity measurements, 140 µl of lysate was mixed with 360 µl of sample buffer provided with the CAT enzyme-linked immunosorbent assay (ELISA) (Boehringer-Mannheim, Mannheim, Germany) and 200 µl of this solution was used to measure CAT activity, following the instructions included in the kit. Each sample was tested in duplicate.
Expression analysis
A Mouse RAPID-SCAN cDNA panel (OriGene Technologies) was used to define the pattern of expression of m-Nfx.1 in different tissues. PCR-based screening was performed using m-NFX.1-3′ and m-NFX.1-5′ as primers, and samples were loaded on a 1·5% agarose gel stained with GelStar (FMC Bioproducts, Rockland, ME).
In situ hybridization
In situ hybridization was performed as described previously.22 In brief, CD-1 mouse embryos were collected in sterile conditions from pregnant females at embryonic days E12 and E14 and fixed in 4% paraformaldehyde (EMS, Fort Washington, PA) solution in PBS for 16 hr at 4°. Embryos were treated with 30% sucrose solution in PBS for 12 hr before embedding them in OCT (Tissue-Tek, Miles Inc., Elkhart, IN). Two regions of the m-Nfx.1 cDNA – spanning nt −56 to nt +757 (NotI–EcoRV) and nt +2018 to nt +2514 (PstI–PstI) – were subcloned into vector pBluescript II KS+ (Stratagene) and used as templates to generate sense and antisense radiolabelled ([α-[35S]thio-UTP) RNA probes by T3 and T7 polymerase, respectively. The Riboprobe Gemini Core System II transcription kit (Promega) was used following the instructions of the manufacturer. Hybridization was carried out at 52° for 14 hr. Slides were washed at high stringency at 65° for 60 min in 50% formamide, 2× SSC, 20 mm dithiothreitol (DTT) and rinsed twice with 0·5 m NaCl, 10 mm Tris–HCl, pH 7·5, 5 mm EDTA, pH 8, before digesting with RNase A for 30 min at 37°. The high-stringency wash was repeated for an additional 30 min at 65°, followed by two washes in 2× SSC, 1 mm DTT, and 0·1× SSC, 1 mm DTT, respectively. Autoradiography was performed at 4° for 7–15 days. The specificity of the signal was established by using a sense probe and by hybridizing with two different antisense probes spanning independent regions of the m-Nfx.1 cDNA.
Results
Isolation of the murine NFX.1 cDNA clone
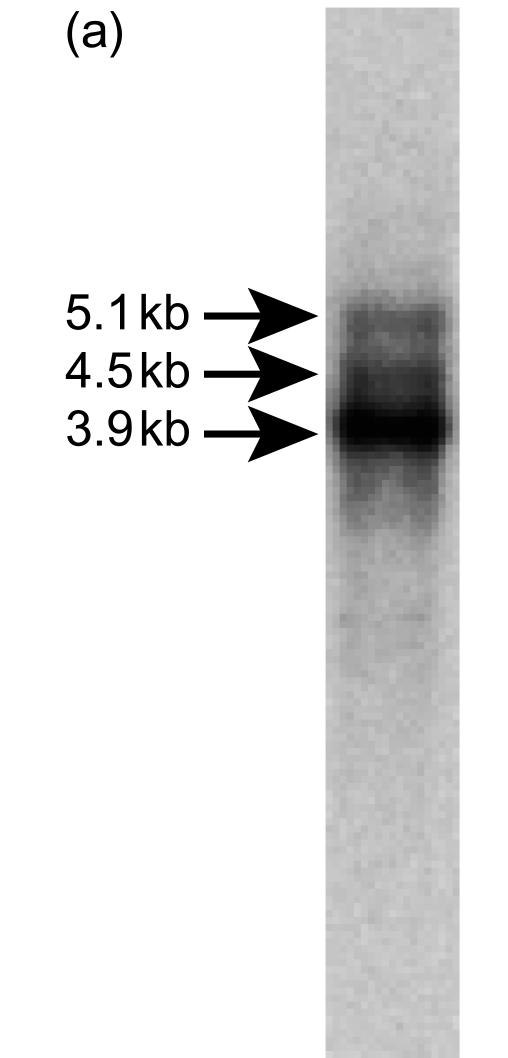
The full-length coding sequence of the murine m-Nfx.1 gene was isolated by screening an E19-fetal mouse cDNA library by PCR using two primers originally derived from the human Nfx.1 cDNA sequence (see the Materials and methods). One of the clones isolated from this screening, clone 61, encodes a full-length copy of the murine m-Nfx.1 cDNA and corresponds to the most abundant mRNA transcript (of ≈ 3·9 kb) observed in Northern blot analysis (Fig. 1a). Two other mRNA transcripts of ≈ 4·5 kb and ≈ 5·1 kb were detected by Northern blotting using RNA from IFN-γ-induced WEHI-3 cells, suggesting the existence of multiple start and/or termination sites of transcription, or alternative splicing of the primary transcript. IFN-γ-induced WEHI-3 cells were used to detect the transcripts as we have previously observed IFN-γ-inducible expression of Nfx.1 in HeLa cells.
Figure 1 (a).
Northern blotting analysis of m-Nfx.1 mRNA. One abundant transcript of ≈ 3.9 kb and two larger, but less abundant, transcripts of ≈ 4.5 kb and ≈ 5.1 kb are shown.
To isolate additional sequence upstream of the short 5′ untranslated region (UTR) of clone 61, we screened the same library with a second set of primers located on the backbone vector used to build the library (forward primer) and within the first 300 bp of the cloned m-Nfx.1 sequence (reverse primer). A partial m-Nfx.1 clone, clone 23, was isolated, which includes a 600-bp 5′-UTR including 500 additional nt upstream of the short 5′-UTR of clone 61 (data not shown). This longer 5′-UTR could result from the existence of a second start site of transcription and may explain the second most abundant transcript (of ≈4·5 kb) detected by Northern blotting.
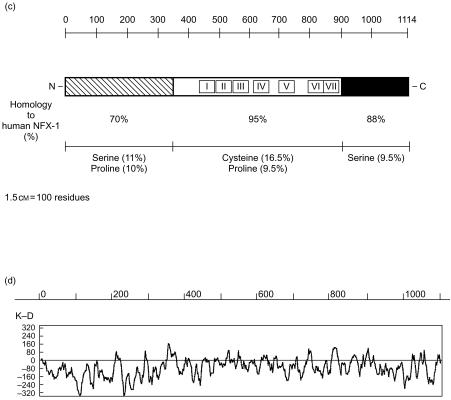
Clone 61 was entirely sequenced on both strands using primers on the backbone vector and multiple internal primers (Fig. 1b). It contains a 3342-base long ORF immediately downstream of a short 5′-UTR, and a 330-base long 3′-UTR. The ORF encodes a 1114 amino acid protein. A comparison of the deduced amino acid sequences of the human and murine Nfx.1 ORFs reveals that they are highly homologous proteins. Similarly to the human NFX.1, murine NFX.1 shows a large central region containing seven conserved cysteine-rich domains of the consensus sequence Cx3C3LxCGx1−5HxCx3CHxGxC (Fig. 1b, 1c). These domains have been shown to be necessary and sufficient for human NFX.1 to bind to the X1 box of the HLA-DRA gene and mediate transcriptional repression.17 The presence of these domains also makes m-NFX.1 the murine homologue of the Drosophila shuttlecraft gene product, STC, which has been shown to play a central role during Drosophila neurogenesis in development.23 As shown in Fig. 1(c), the homology between human and murine NFX.1 is highest (95% homology) in the central part of the protein spanning the DNA-binding, cysteine-rich domain, which supports the idea that specific, high-affinity binding to target sites on DNA is critical for proper function of NFX.1.17 In addition, human and murine NFX.1 are highly homologous at their C-terminus (88% homology), while their homology decreases to 70% at the N-terminus. Interestingly, a similar distribution of homologous residues was also observed in the comparison between human NFX.1 and Drosophila STC proteins, supporting the idea that the DNA-binding domains and the C-terminal portion of these proteins might be conserved as they have critical functions.
Figure 1 (c).
Primary structure analysis of m-NFX.1 and homology to human NFX.1 protein. The seven cysteine-rich DNA-binding domains are indicated with roman numerals in the centre of m-NFX.1 protein and show the highest level of homology to human NFX.1. (d) Kyte-Doolittle hydropathy plot of m-NFX.1 (GeneWorks software; Intelligenetics).
Chromosomal localization of murine Nfx.1
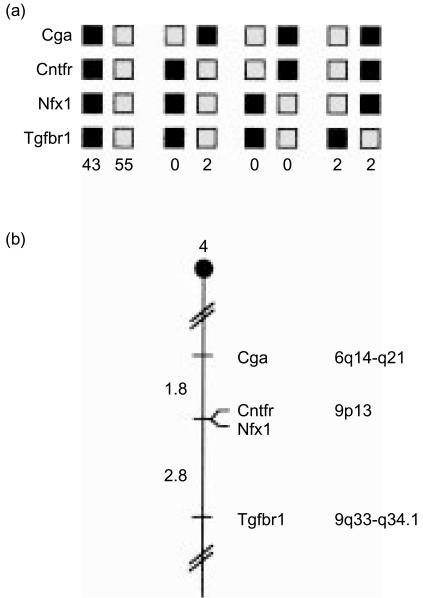
The chromosomal location of Nfx.1 in the mouse genome was determined by interspecific backcross analysis using progeny derived from matings of [(C57BL/6J×Mus spretus)F1×C57BL/6J] mice. This interspecific backcross mapping panel has been typed for over 3000 loci that are well distributed among all autosomes as well as the X chromosome.18 C57BL/6J and M. spretus DNAs were digested with several enzymes and analysed by Southern blot hybridization for informative RFLPs using a mouse cDNA m-Nfx.1 probe. The 14·0, 7·5, 4·4, 3·6, and 3·1 kb TaqI M. spretus RFLPs (see the Materials and methods) were used to follow the segregation of the m-Nfx.1 locus in backcross mice. The mapping results indicated that m-Nfx.1 is located in the proximal region of mouse chromosome 4 linked to Cga, Cntfr and Tgfbr1. Although 104 mice were analysed for every marker and are shown in the segregation analysis (Fig. 2a), up to 181 mice were typed for some pairs of markers. Each locus was analysed in pairwise combinations for recombination frequencies using the additional data. The ratios of the total number of mice exhibiting recombinant chromosomes to the total number of mice analysed for each pair of loci and the most probable gene order are: centromere–Cga–2/111–Cntfr–0/177–m-Nfx.1–5/181–Tgfbr1. The recombination frequencies [expressed as genetic distances in centiMorgans (cm) ± the standard error] are: centromere–Cga–1·8 ± 1·3–[Cntfr, m-Nfx.1]–2·8 ± 1·3. No recombinants were detected between Cntfr and m-Nfx.1 in 177 animals typed in common, suggesting that the two loci are within 1·7 cm of each other (upper 95% confidence limit).
Figure 2.
(a) m-Nfx.1 maps in the proximal region of mouse chromosome 4. m-Nfx.1 was placed on mouse chromosome 4 by interspecific backcross analysis. The segregation patterns of m-Nfx.1 and flanking genes in 104 backcross animals that were typed for all loci are shown. For individual pairs of loci, more than 104 animals were typed (see text). Each column represents the chromosome identified in the backcross progeny that was inherited from the (C57BL/6J×Mus spretus)F1 parent. The shaded boxes represent the presence of a C57BL/6J allele and the white boxes represent the presence of an M. spretus allele. The number of offspring inheriting each type of chromosome is listed at the bottom of each column. (b) Partial chromosome 4 linkage map showing the location of murine m-Nfx.1 in relation to linked genes. Recombination distances between loci [in centimorgans (cM)] are shown to the left of the chromosome and the positions of loci in human chromosomes, where known, are shown to the right. References for the human map positions of loci cited in this study can be obtained from Genome Database (GDB), a computerized database of human linkage information maintained by the William H. Welch Medical Library of The Johns Hopkins University (Baltimore, MD). (c) Schematic of mouse chromosome 4 showing the location of m-Nfx.1 (arrow) relative to a larger number of known genes. Corresponding locations for each gene in human chromosomes are indicated to the left.
We compared our interspecific map of chromosome 4 with a composite mouse linkage map that reports the map location of many uncloned mouse mutations (provided from the Mouse Genome Database, a computerized database maintained at the Jackson Laboratory, Bar Harbor, ME). m-Nfx.1 maps near the location of mouse mutations with interesting phenotypes. Mutation in one gene (vc or vacillans) results in neurological or neuromuscular disorders. Homozygosity at this locus results in a phenotype readily detected at 14 days of age, consisting of violent tremors when walking and swaying of the hindquarters. This is assumed to be caused by a neurological disorder, but there are no gross anatomical abnormalities in the central nervous system (CNS).24 The second mouse mutation mapping near m-NFx.1 affects the wi, or whirler, gene. This is also a neurological defect, resulting in the ‘shaker’ syndrome: trembling, deafness and hyperactivity. As mutations in the Drosophila stc gene (a Nfx.1 homologue) affect neurogenesis, and class II-deficient mice exhibit a ‘shiverer’ phenotype (once again owing to neurological defects), we will investigate whether mutations in m-Nfx.1 may be responsible for these phenotypes.23,25
The proximal region of mouse chromosome 4 shares regions of homology with human chromosomes 6q and 9 (summarized in Fig. 2b). In particular, Cntfr has been assigned to 9p13 in humans. The tight linkage between Cntfr and m-Nfx.1 in mouse suggests that the human homologue of m-Nfx.1 should also map to 9p13. Data from the recently completed human genome sequencing project, as well as results from our own mapping experiments (data not shown), confirm this prediction, and map human Nfx.1 to the short arm of human chromosome 9.26
m-NFX.1 is a repressor of MHC class II gene expression
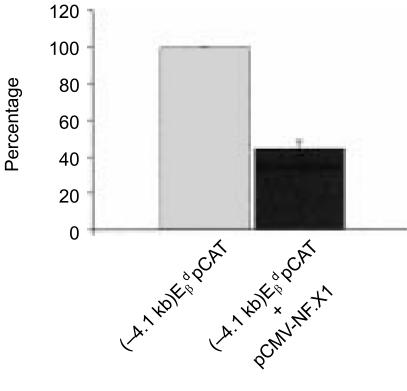
Although our previous demonstration, that human NFX.1 could repress class II MHC gene expression, predicted that a murine homologue would also repress murine class II genes, we wished to test directly whether this was the case in transfection assays. To confirm the functional authenticity of the isolated m-Nfx.1 cDNA, we tested its ability to repress transcription of MHC class II genes in co-transfection experiments. Clone 61 [expressing the full-length murine Nfx.1 coding region under the control of a cytomegalovirus (CMV) promoter] was co-transfected with an -promoter reporter construct [(− 4·1 kb) -pCAT] into the MHC class II-positive cell line A-20. As shown in Fig. 3, m-NFX.1 can decrease expression from the gene promoter to almost 40% of its wild-type activity. This result provides additional indication that we have isolated a full-length, functional murine Nfx.1 clone.
Figure 3.
m-NFX.1 is a repressor of major histocompatibility complex (MHC) class II gene expression. Histogram showing the reduction in CAT activity from the -promoter reporter construct (−4·1 kb), -pCAT, when co-transfected with the pCMV-NFX.1 vector driving expression of full-length m-NFX.1. A reduction of the CAT activity to as little as 40% of wild-type levels is observed. Wild-type activity was set at 100%.
m-NFX.1 is ubiquitously expressed in adult and embryonic tissues
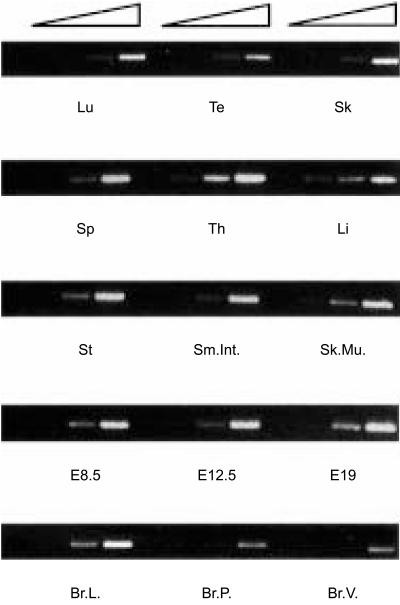
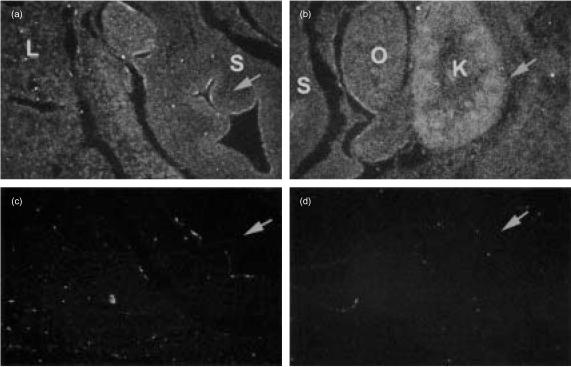
The presence of X1-related sequences on promoters from several genes25 other than MHC class II genes suggests that NFX.1 may also have functions outside the immune system. This is supported by the observation that the Drosophila homologue of murine and human NFX.1, STC, is important for proper development of the CNS during embryogenesis.23 For this reason we have screened both adult and embryonic tissues to assess the distribution of m-Nfx.1 transcripts. m-Nfx.1 is expressed in all tissues tested, with the thymus expressing the highest levels, as detected by RT–PCR analysis (Fig. 4). In situ hybridization analysis shows that m-Nfx.1 is ubiquitously expressed in both E12 and E14 embryos (Fig. 5 and data not shown). The ubiquitous expression profile of m-Nfx.1, as well as the presence of X-box-related consensus sequences on a variety of promoters, support the idea that NFX.1 may have additional undiscovered roles in vivo and suggests that the creation of transgenic/knockout mouse models involving m-Nfx are necessary to probe additional functions of m-Nfx.1.
Figure 4.
m-NfX.1 is ubiquitously expressed in adult tissues. Semiquantitative reverse transcription–polymerase chain reaction (RT–PCR) showing expression of m-NfX.1 in all tissues tested. Lu, lung; Te, testis; Sk, skin; Sp, spleen; Th, thymus; Li, liver; St, stomach; Sm.Int., small intestine; Sk.Mu., skeletal muscle; E8·5, 8·5 d.p.c. whole embryo; E12·5, E12·5 d.p.c whole embryo; E19, E19 d.p.c whole embryo; Br.L., breast lactating; Br.P., breast pregnant; Br.V., breast virgin. All tissues tested show detectable levels of m-NfX.1 mRNA. Four crescent concentration of cDNA were used for each tissue tested.
Figure 5.
m-NfX.1 is ubiquitously expressed during development. In situ hybridization analysis on E14 embryos. m-NfX.1 is expressed ubiquitously through all the body of the developing embryo (this figure and data not shown). (a) Close-up picture showing the embryonic stomach. The Arrow is pointing at the epithelium lining the lumen of the stomach where slightly higher levels of expression are observed. (b) Close-up picture of the embryonic kidney (arrow), showing higher levels of expression of m-NfX.1. (c) and (d) Sense-probe negative controls of (a) and (b), respectively. Arrows are pointing at corresponding regions in (a) and (b). L, liver; S, stomach; O, ovary; K, kidney.
Discussion
The central role of MHC class II molecules in initiating the immune response to foreign antigens has prompted active investigation of the molecular switches that control expression of their genes. In particular, given the fact that MHC class II genes are mainly regulated at the transcriptional level, great attention has been devoted to the identification and characterization of transcription factors that interact with their promoters and affect transcription. Here we report the cloning and an initial characterization of the mouse homologue of the human MHC class II transcriptional repressor, NFX.1.17 m-NfX.1 cDNA contains a 3342-base long ORF that encodes a predicted polypeptide of 1114 amino acids. The predicted m-NFX.1 protein shows very high levels of homology to human NFX.1 and to the Drosophila stc gene product23 and together these proteins define a novel family of transcription factors characterized by the presence of a distinctive cysteine-rich DNA-binding domain. In addition, recent studies indicate that the NFX.1 family of proteins may have additional roles mediated by protein–protein interactions. Indeed, the reiterated RING finger motifs in the central domain of the polypeptide strongly suggest that NF-XI is a probable E3 ubiquitin protein ligase.
Consistent with our findings with human NFX.1, co-transfection experiments show that m-NfX.1 is also able to repress MHC class II transcription. This is of potential therapeutic interest as it suggests that NFX.1 may be used as a target gene for the treatment of disease states where altered MHC class II expression is hypothesized to play a part in disease pathogenesis (such as inflammation and autoimmunity).
The isolation of m-NfX.1 should also allow us to carry out biochemical studies to probe:
occupancy of the X1 box by NFX.1; and
how NFX.1 might influence the binding of the well-characterized X1 and X2 boxes (as well as the Y box and octamer motifs) by factors such as RFXV, CREB, NF-Y and HMG I/Y.7,8,10,16
We hope that the isolation of m-NfX.1 will also permit scientists to probe the range of biological roles that NfX.1 may play within and outside the immune system. The mapping of the gene near known disease loci indicates that the NfX.1 gene should be examined directly in wild-type and mutant mouse strains to determine whether mutation of NfX.1 may directly result in specific diseases. The elegant study by Stroumbakis and colleagues has shown that STC, the Drosophila homologue of NFX.1, seems to have a role in guiding motor-neuron axons to their proper target muscle fields during neurogenesis in the embryo. These data, together with the recent observation that NFX.1 resembles a E3 ubiquitin protein ligase, suggests that NFX.1 will probably have yet undiscovered functional roles. We hypothesize that NFX.1 may participate in biological processes either via its nucleic acid-binding activity, or via protein–protein interactions. This possibility appears feasible owing to our discovery of fairly ubiquitous expression of murine NFX.1 in both adult and embryonic tissues.
Given the role of NFX.1 as a potent repressor of MHC class II expression and its potential involvement in other biological processes (i.e. neurogenesis) the availability of the murine NfX.1 cDNA is of immediate utility for the creation of transgenic and knockout mouse models. These models will be required to ultimately determine the role of NFX.1 in vivo as a prerequisite to any attempt to use it as target molecule in treatment of important immunological conditions.
Acknowledgments
We thank Gail Burke for assisting with the preparation of this manuscript, Dr David Chau for his advice during the course of the experiments and Deborah Householder for her excellent technical assistance with the chromosome mapping studies. We thank Dr Gilbert Jay and Origene Technologies, Inc., for generous donations of reagents used in these experiments. We would also like to thank the Telethon fund, Italy, for supporting Paola Arlotta. This research was supported by the National Institute of Health and, in part, by the National Cancer Institute, Department of Health and Human Services.
References
- 1.Chapman HA. Endosomal proteolysis and MHC class II function. Curr Opin Immunol. 1998;10:93–102. doi: 10.1016/s0952-7915(98)80038-1. [DOI] [PubMed] [Google Scholar]
- 2.McDevitt HO. The role of MHC class II molecules in susceptibility and resistance to autoimmunity. Curr Opin Immunol. 1998;10:677–81. doi: 10.1016/s0952-7915(98)80088-5. [DOI] [PubMed] [Google Scholar]
- 3.Glimcher LH, Kara CJ. Sequences and factors: a guide to MHC class-II transcription. Annu Rev Immunol. 1992;10:13–49. doi: 10.1146/annurev.iy.10.040192.000305. [DOI] [PubMed] [Google Scholar]
- 4.King DP, Jones PP. Induction of Ia and H-2 antigens on a macrophage cell line by immune interferon. J Immunol. 1983;131:315–8. [PubMed] [Google Scholar]
- 5.Hume CR, Lee JS. Congenital immunodeficiencies associated with absence of HLA class II antigens on lymphocytes result from distinct mutations in trans-acting factors. Hum Immunol. 1989;26:288–309. doi: 10.1016/0198-8859(89)90007-4. [DOI] [PubMed] [Google Scholar]
- 6.Benichou B, Strominger JL. Class II-antigen-negative patient and mutant B-cell lines represent at least three, and probably four, distinct genetic defects defined by complementation analysis. Proc Natl Acad Sci USA. 1991;88:4285–8. doi: 10.1073/pnas.88.10.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Abdulkadir SA, Ono SJ. How are class II MHC genes turned on or off? FASEB J. 1995;9:1429–35. doi: 10.1096/fasebj.9.14.7589984. [DOI] [PubMed] [Google Scholar]
- 8.Harton JA, Ting JP. Class II transactivator: mastering the art of major histocompatibility complex expression. Mol Cell Biol. 2000;20:6185–94. doi: 10.1128/mcb.20.17.6185-6194.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Masternak K, Barras E, Zufferey M, et al. A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat Genet. 1998;20:273–7. doi: 10.1038/3081. [DOI] [PubMed] [Google Scholar]
- 10.Tai AKF, Zhou G, Chau KY, Ono SJ. Cis-element dependance and occupancy of the human invariant chain promoter in CIITA-dependant and -independant function. Mol Immunol. 1999;36:447–60. doi: 10.1016/s0161-5890(99)00061-9. [DOI] [PubMed] [Google Scholar]
- 11.Ting JP. Current concepts in DRA gene regulation. Immunol Res. 1993;12:65–77. doi: 10.1007/BF02918369. [DOI] [PubMed] [Google Scholar]
- 12.Durand B, Kobr M, Reith W, Mach B. Functional complementation of major histocompatibility complex class II regulatory mutants by the purified X-box-binding protein RFX. Mol Cell Biol. 1994;14:6839–47. doi: 10.1128/mcb.14.10.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright KL, Vilen BJ, Itoh-Lindstrom Y, et al. CCAAT box binding protein NF-Y facilitates in vivo recruitment of upstream DNA binding transcription factors. EMBO J. 1994;13:4042–53. doi: 10.1002/j.1460-2075.1994.tb06721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–46. [PubMed] [Google Scholar]
- 15.Harton JA, Ting JP. Class II transactivator: mastering the art of major histocompatibility complex expression. Mol Cell Biol. 2000;20:6185–94. doi: 10.1128/mcb.20.17.6185-6194.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abdulkadir SA, Krishna S, Thanos D, Maniatis T, Strominger JL, Ono SJ. Functional roles of the transcription factor Oct-2A and the high mobility group protein I/Y in HLA-DRA gene expression. J Exp Med. 1995;182:487–500. doi: 10.1084/jem.182.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song Z, Krishna S, Thanos D, Strominger JL, Ono SJ. A novel cysteine-rich sequence-specific DNA-binding protein interacts with the conserved X-box motif of the human major histocompatibility complex class II genes via a repeated Cys-His domain and function as a transcriptional repressor. J Exp Med. 1994;180:1763–74. doi: 10.1084/jem.180.5.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Copeland NG, Jenkins NA. Development and application of a molecular genetic linkage map of the mouse genome. Trends Genet. 1991;7:113–8. doi: 10.1016/0168-9525(91)90455-y. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins NA, Copeland NG, Taylor BA, Lee BK. Organization, distribution, and sability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J Virol. 1982;43:26–36. doi: 10.1128/jvi.43.1.26-36.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morales J, Homey B, Vicari AP, et al. CTACK, a skin-associated chemokine that preferentially attracts skin-homing memory T cells. Proc Natl Acad Sci USA. 1999;96:14470–5. doi: 10.1073/pnas.96.25.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Douhan J, III, Brown JA, Gleit ZL, David CS, Glimcher LH. Regulation of the E beta gene in vivo: lessons from E beta d transgenic mice. Int Immunol. 1996;8:255–65. doi: 10.1093/intimm/8.2.255. III. [DOI] [PubMed] [Google Scholar]
- 22.Wert SE, Glasser SW, Korfhagen TR, Whitsett JA. Transcriptional elements from the human SP-C gene direct expression in the primordial respiratory epithelium of transgenic mice. Dev Biol. 1993;156:426–43. doi: 10.1006/dbio.1993.1090. [DOI] [PubMed] [Google Scholar]
- 23.Stroumbakis ND, Li Z, Tolias PP. A homolog of human transcription factor NF-X1 encoded by the Drosophila shuttle craft gene is required in the embryonic central nervous system. Mol Cell Biol. 1996;16:192–201. doi: 10.1128/mcb.16.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marbois BN, Xia YR, Lusis AJ, Clarke CF. Ubiquinone biosynthesis in eukaryotic cells: tissue distribution of mRNA encoding 3,4-dihydroxy-5-polyprenylbenzoate methyltransferase in the rat and mapping of the COQ3 gene to mouse chromosome 4. Arch Biochem Biophys. 1994;15:83–8. doi: 10.1006/abbi.1994.1362. [DOI] [PubMed] [Google Scholar]
- 25.Matsushima GK, Taniike M, Glimcher LH, Grusby MJ, Frelinger JA, Suzuki K, Ting JP. Absence of MHC class II molecules reduces CNS demyelination, microglia/macrophage infiltration, and twitching in murine globoid cell leukodystrophy. Cell. 1994;78:645–56. doi: 10.1016/0092-8674(94)90529-0. [DOI] [PubMed] [Google Scholar]
- 26.Lander ES, Linton LM, Birren B, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. 10.1038/35057062. [DOI] [PubMed] [Google Scholar]