Abstract
CD1 molecules are cell-surface glycoproteins with strong structural similarities to major histocompatibility complex (MHC) class I molecules, and studies in humans and mice have demonstrated that CD1 proteins perform the unique role of presenting lipid antigens to T lymphocytes. Our previous studies have shown that guinea-pigs, unlike the muroid rodents, have an extended family of group 1 CD1 genes. In the current study, we raised monoclonal anibodies (mAbs) against guinea-pig CD1 proteins and generated transfected cell lines expressing individual members of the guinea-pig CD1 family. Our results indicated that multiple members of the guinea-pig CD1 family, including members that are homologous to the human CD1b and CD1c proteins, are expressed at the protein level in transfected cells and in specialized antigen-presenting cells such as monocyte-derived dendritic cells. In addition, CD1 proteins, especially guinea-pig CD1b3, were expressed on a large number of B cells in the guinea-pig, and CD1 expression appeared to be regulated by B-cell maturation or differentiation. Interestingly, three different patterns of intracellular localization were observed for the various guinea-pig CD1 isoforms, a finding that is reminiscent of the distinct patterns of intracellular localization that have been previously demonstrated for human CD1a, CD1b and CD1c. Taken together, these results provide further evidence for substantial similarities between the guinea-pig and human CD1 systems, thus supporting the possibility that the guinea-pig may offer significant advantages as an animal model for the study of the in vivo role of CD1 proteins in infectious and autoimmune diseases.
Introduction
CD1 proteins are antigen-presenting molecules that are required for the recognition of specific lipid and glycolipid antigens by T cells.1 In humans, the CD1 family consists of five non-polymorphic genes that map to chromosome 1.2 These genes encode the transmembrane-anchored heavy chains of five distinct β2-microglobulin (β2m)-associated glycoproteins (designated CD1a, CD1b, CD1c, CD1d and CD1e) that bear striking structural similarities to major histocompatibility complex (MHC) class I antigen-presenting molecules.3,4 A substantial body of data has revealed that three of the human CD1 proteins (CD1a, CD1b and CD1c) function as antigen-presenting molecules for a subset of T cells that responds to specific lipids and glycolipids found in the cell walls of mycobacteria and other bacterial pathogens, and also for T cells that respond to self-glycolipid antigens such as gangliosides.5–8 The ability of CD1 molecules to perform this role is probably a result of their abilities to act as lipid-binding proteins, which trap the hydrophobic alkyl portions within a deep hydrophobic pocket formed by the two membrane distal domains of the protein.4 This leads to the antigenic lipid being displayed in such a way that its hydrophilic or polar head group is accessible for direct interactions with the T-cell receptors (TCRs) of specific CD1-restricted T cells.1,5,9
Sequence similarity and functional analyses have suggested that the various human CD1 isoforms should be classified into two groups.5 Thus, CD1a, -b and -c are designated as group 1 CD1 proteins, whereas the more divergent CD1d protein is designated as group 2 CD1. This separation of human CD1 proteins was first proposed based on evolutionary considerations from comparison of sequences from different species, and has subsequently been reinforced by studies of CD1 expression and function.3,10 The group 1 CD1 proteins show a relatively restricted pattern of expression on cells of haematopoietic lineage. They are highly induced on many myeloid lineage dendritic cells and are expressed on cortical thymocytes and a subset of B cells (CD1c only), but are absent from most other bone marrow-derived cells.3 Functional studies suggest that the major role of group 1 CD1 proteins may be to present specific foreign lipid and glycolipid antigens for the stimulation of specific T-cell responses.5 In contrast, the group 2 CD1d protein is widely and constitutively expressed at low levels on most cells of haematopoietic origin, and has also been identified on several epithelial cell types.11–13 So far, CD1d has not been convincingly shown to present lipid antigens from relevant infectious agents to T cells, although it is known to control the development and functions of a specific subset of T cells known as natural killer (NK) T cells, which appear to mediate considerable immunoregulatory effects on a variety of immune responses.14 Thus, it appears probable that group 1 (CD1a, CD1b and CD1c) and group 2 (CD1d) CD1 proteins have distinct roles in host defence.
In order to understand the role of group 1 CD1 proteins in host immunity, it is necessary to assess the contribution of these molecules during immune responses in vivo using an appropriate animal model. Unfortunately, the muroid rodents (including all strains of mice and rats studied to date) express only group 2 CD1 proteins and appear to have deleted the genes for all known group 1 CD1 isoforms from their genomes. Therefore, while the mouse is an excellent animal model system for studies of the function of group 2 CD1 in vivo, it is useless for evaluation of the importance of group 1 CD1 proteins. In contrast, many other mammals appear to express group 1 CD1 homologues, suggesting that other animal models could potentially be developed to allow the in vivo assessment of these proteins.15,16 We have previously reported that guinea-pigs, unlike the muroid rodents, have an extended family of CD1 genes that includes at least seven genes encoding what are probably evolutionary homologues of two human group 1 CD1 proteins, CD1b and CD1c.17 In the current study, we developed monoclonal antibodies (mAbs) against guinea-pig CD1 (gpCD1) proteins and provided evidence that all seven of the apparently intact group 1 CD1 genes of these animals that we previously identified by cDNA cloning can be expressed as cell-surface proteins. Importantly, we found that multiple gpCD1 proteins are highly expressed on specialized antigen-presenting cells (APC) such as bone marrow-derived dendritic cells (BM-DC), where they may serve a function in antigen presentation similar to that found for the human group 1 CD1 proteins. We also demonstrated strikingly different patterns of subcellular localization for different gpCD1 proteins. This provided a further parallel with the human group 1 CD1 family, and gave further support to the hypothesis that these proteins are individually specialized to survey different intracellular compartments for their lipid antigen content. Our results should provide a useful foundation for the further development of the guinea-pig as a uniquely relevant small animal model for in vivo studies of the functions of group 1 CD1 proteins in infectious and autoimmune diseases.
Materials and methods
Animals
Female Hartley strain guinea-pigs weighing 200–300 g were purchased from Charles River Laboratories (Wilmington, MA). Strain 2 guinea-pigs were obtained from the National Cancer Institute (Frederick, MD) and established as a breeding colony at our animal facilities. BALB/c mice were purchased from Charles River. Animals were housed under specific pathogen-free conditions and all animal experiments were performed according to our institutional guidelines on animal welfare and humane treatment of laboratory animals.
Generation of gpCD1 transfectants
Cell lines K.BALB (mouse-transformed fibroblast line), CHO-K1 (Chinese hamster ovary cell line), 104C1 (guinea-pig fetal carcass-derived fibroblast-like line) and GPC-16 (guinea-pig colonic adenocarcinoma line) were obtained from the American Type Culture Collection (ATCC, Manassas, VA). Complementary DNA inserts encoding the various gpCD1 proteins were cloned from Hartley and Strain 2 guinea-pig thymocyte cDNA, as previously reported.17 Full-length CD1 cDNAs were directionally subcloned into the eukaryotic expression vector, pcDNA3.1, (Invitrogen, Carlsbad, CA) and transfected into K.BALB, 104C1 and GPC-16 cell lines using Lipofectamine Plus lipofection reagents (Invitrogen), as described previously.17 Growing cells obtained after selection with G418 (Geneticin; Invitrogen) were screened by flow cytometry analysis using the guinea-pig cross-reactive anti-human CD1b mAb, BCD1b3 (subclone BCD1b3.2, which produces a mAb identical to that produced by previously described subclone BCD1b3.1).18 Transfectant cells expressing gpCD1 were then isolated by sterile sorting using a FACSort flow cytometer (Becton-Dickinson Immunocytometry Systems, San Jose, CA) or magnetic bead positive selection using anti-gpCD1 mAbs and goat anti-mouse immunoglobulin G (IgG)-coated magnetic beads (Dynabeads M450; Dynal, Inc. Lake Success, NY). Subclones of gpCD1-expressing transfectants were obtained by limiting dilution of sorted bulk transfectant lines.
Production of soluble gpCD1–mouse IgG fusion proteins
Purified gpCD1–mouse immunoglobulin Fc fusion proteins were used to immunize mice for anti-CD1 mAb production and as the solid-phase antigen in a screening enzyme-linked immunosorbent assay (ELISA) for detection of anti-gpCD1 mAb-secreting hybridomas. Soluble gpCD1–mouse IgG1 Fc fusion protein cDNA constructs were produced by fusing the sequence encoding the extracellular α1+α2+α3 domains of the gpCD1b3 or gpCD1b4 proteins to a sequence encoding the CH2 and CH3 domains of mouse IgG1 using standard polymerase chain reaction (PCR) and molecular cloning techniques. To prevent protein aggregation via a free cysteine normally directed to the immunoglobulin light chain, the immunoglobulin hinge region was designed to contain the HA tag peptide and omit this cysteine. The gpCD1–Fc fusion constructs were subcloned into expression vector pcDNA3.1 and co-transfected (using lipofection) into CHO-K1 cells together with the guinea-pig β2m cDNA in the expression vector pZeo (Invitrogen). Transfected CHO-K1 cells were grown in AlphaMEM medium (JRH Biosciences, Lenexa, KS) containing G418 and Zeocin (Invitrogen) for selection of stably co-transfected cells. Secretion of gpCD1–Fc fusion proteins was detected by performing a standard capture ELISA on supernatant samples using reagents specific for mouse IgG. Positive bulk transfectant lines were subcloned by limiting dilution, and subclones secreting relatively high levels of gpCD1–immunoglobulin Fc fusion proteins were selected and expanded. Culture supernatants containing high levels of CD1 fusion proteins were analysed by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) to ascertain the molecular weight of the products. The fusion proteins were purified by MAPS II protein A affinity resin (Bio-Rad, Hercules, CA). The purified fusion proteins were concentrated by centrifugation in a 10 K molecular weight cut-off Centricon-10 concentrator (Millipore, Bedford, MA) and then dialysed extensively against phosphate-buffered saline (PBS).
Generation of anti-gpCD1 mAbs
To generate mAbs against gpCD1, two different immunization strategies were successfully employed. Three anti-gpCD1 hybridoma lines (CD1F2/5E3, CD1F2/6B5 and CD1F2/1B12) were produced from BALB/c mice that were immunized intraperitoneally with irradiated K.BALB transfectant cell lines expressing either gpCD1b3 or gpCD1b4 suspended in PBS without adjuvant. The mice received three such immunizations with transfected K.BALB cells at 3-week intervals, and then were rested for 5 weeks following which an intravenous (i.v.) boost with 100 µg of gpCD1b3–IgG1 fusion protein in saline was given. One other anti-gpCD1 hybridoma line (CD1.4-1D12) was produced from pooled spleen cells of mice that were immunized three times intraperitoneally (i.p.) and intradermally (i.d.) at biweekly intervals with 2·5 µg total of purified gpCD1b1–murine (m)IgG1 fusion protein, plus RIBI adjuvant (Corixa, Seattle, WA) and finally boosted i.v. with 100 µg of gpCD1b1–mIgG1 in saline. Mice were killed 48 hr after i.v. boosting, and their spleen cells were fused with NSO myeloma cells and plated in 96-well plates according to standard methods for hybridoma generation.18 Supernatants of the hybridomas were screened by fluorescence-activated cell sorter (FACS) analysis of guinea-pig CD1b3, -b4 and mock transfectant cells or guinea-pig thymocytes. CD1.4-1D12 was screened by ELISA and then by FACS analysis of gpCD1 transfectants or thymocytes. Limiting-dilution cloning of positive hybridomas was performed at least twice to obtain stable lines secreting mAbs against gpCD1. Anti-gpCD1 mAbs were purified from culture supernatants of hybridomas grown in medium supplemented with ultra-low IgG fetal bovine serum (Hyclone, Logan, UT) by protein G–affinity column chromatography (Amersham Pharmacia Biotech, Inc., Piscataway, NJ).
Antibodies and FACS analysis
In addition to our newly generated mAbs against gpCD1 proteins (CD1F2/5E3, CD1F2/6B5, CD1F2/1B12 and CD1.4-1D12), the following mAbs were obtained from Serotec Ltd (Raleigh, NC): CT6 (anti-guinea-pig CD8), CT7 (anti-guinea-pig CD4), Msgp4 (anti-guinea-pig MHC class I), Msgp3 (anti-guinea-pig IgG), Msgp9 (anti-guinea-pig pan-B-cell marker; demonstrated in this study to be reactive with gpCD1b3) and Cl.13.1 (anti-guinea-pig MHC class II). A hybridoma line producing mAb IVA12 (anti-human MHC class II monomorphic determinant, cross-reactive with guinea-pig MHC class II) was obtained from the ATCC. The anti-human CD1b mAb BCD1b3 (subclone BCD1b3.1 or BCD1b3.2, cross-reactive with gpCD1 proteins) has been described previously17 and anti-human CD1b mAbs BCD1b1, BCD1b2 and BCD1b4 (all mouse IgG1/κ-isotype), were produced using the same methods used to generate BCD1b3.18 The anti-human CD1d mAb CD1d69 (subclone CD1d69.4, mouse IgG2a/κ) was produced in our laboratory from the same fusion that generated a series of previously published anti-CD1d mAbs.19 Other previously described anti-CD1 mAbs used in the current study included 7C4 (kindly provided by Drs O. Madjic and W. Knapp, University of Vienna, Vienna, Austria;20), WM-25 (gift of Dr E. Favaloro, Westmead Hospital, Westmead, Australia;21), 20–27 (obtained from The University of Melbourne, Center for Animal Biotechnology, Victoria, Australia;22), CC-20 (kindly provided by C. J. Howard, Institute for Animal Health, Compton, Newbury, Berkshire, UK)23,24 and Fe5.5C1 (kindly provided by Dr P. F. Moore, University of California, Davis, CA, USA). FACS analysis was performed as previously described.17 Briefly, primary antibodies were added to single-cell suspensions (1 µg/106 cells) for 30 min on ice, washed twice with FACS buffer [2% fetal calf serum (FCS), 0·01% sodium azide in PBS] and then incubated with fluorescein isothiocyanate (FITC)-conjugated goat F(ab′)2 anti-mouse IgG/IgM (BioSource International, Camarillo, CA) for 30 min on ice. After staining, cells were analysed with a fluorescence-activated cell sorter (FACScan; Becton-Dickinson). Dead cells were excluded by propidium iodide (Sigma Chemical Co., St Louis, MO) gating.
Generation of guinea-pig BM-DC
Hartley or Strain 2 guinea-pig femurs and tibias were isolated aseptically and flushed by syringe with RPMI-1640 (Gibco-BRL, Grand Island, NY), and the mononuclear cells in the suspension were isolated by centrifugation at 400 g on a layer of density-gradient medium formulated to a density appropriate for guinea-pig cells (density 1·107), made by combining 2·5 volumes of Histopaque 1119 (Sigma) and 1 volume of Ficoll Paque Plus (Amersham Pharmacia Biotech). The isolated mononuclear cells were cultured at a density of 1×106/ml in a 150-cm2 flask in 30 ml of RPMI-1640 supplemented with human granulocyte–macrophage colony-stimulating factor (GM-CSF) (300 U/ml; PeproTech, Rocky Hill, NJ) and 15% conditioned medium from the guinea-pig JH4 fibroblast cell line (obtained from the ATCC). After 24 hr, non-adherent cells were transferred to a new flask. Cultures were fed every second day by removing 50% of the medium and replacing it with fresh medium containing GM-CSF. This procedure allowed depletion of the non-adherent lymphocytes and other nucleated cells from the starting marrow preparation. By day 5, aggregates of round adherent cells became visible, which continued to increase in number and size but remained firmly adherent until days 8–10, at which point they became more loosely adherent and could be easily detached from the surface of the flask. These non-adherent and loosely adherent cells, representing a population highly enriched for BM-DC, were collected between days 10 and 14 of culture and used for in vitro studies or cryopreserved.
Isolation of guinea-pig splenic DC (SP-DC)
Spleens from Hartley guinea-pigs or Strain 2 guinea-pigs were aseptically removed and perfused in a Petri dish with 20 ml of collagenase D solution (100 Mandl U/ml; Roche, Indianapolis, IN) using a syringe. The collagenase-perfused spleens were cut into fragments that were gently ground between the frosted ends of two microscope slides to produce a single-cell suspension. The connective tissue fragments of the ground spleen were then redigested with 400 Mandl U/ml of collagenase D (4·5 ml for one spleen) for 30 min at 37°, and the cells released after this more vigorous collagenase treatment and moderate agitation (pipetting) were combined with the first cell suspension. The pooled cell suspension was then spun down and the cell pellet resuspended in 60% Percoll (Amersham Pharmacia Biotech) in PBS and overlaid on 30% Percoll/PBS, and then centrifuged at 400 g for 20 min at room temperature. The low density cells from the interface were collected, washed and resuspended in complete RPMI-1640 containing 10% heat-inactivated FCS (Hyclone), 2 mm HEPES, 5×10−5 m 2-mercaptoethanol (2-ME), 100 U/ml penicillin G, 100 µg/ml streptomycin and 2×10−3 m l-glutamine, and cultured in a 75-cm2 flask for 2 hr at 37° after which non-adherent cells were removed and discarded. The adherent fraction was then cultured in complete RPMI-1640 containing 10% FCS and 300 U/ml of human GM-CSF. After 48 hr, the SP-DC in these cultures became non-adherent and were removed with gentle agitation of the culture flask.
Reverse transcription–polymerase chain reaction (RT–PCR) of gpCD1 genes and restriction digest analysis of PCR products
RNA was extracted from BM-DC or SP-DC using the S.N.A.P.™ Total RNA isolation kit (Invitrogen), and first-strand cDNA was synthesized using Oligo(dT) and SuperScript II reverse transcriptase (Invitrogen). To amplify transcripts from all known guinea-pig group 1 CD1 genes, primers were selected that were complementary to completely identical regions among all seven group 1 CD1 genes [5′ primer gp1: 5′-AACTTCAGCAATGA-3′ (base pairs 223–236 in the α1 domain); 3′ primer gp2 : 5′-AGGCTTTGGGTAGAA-3′ (bp 686–699 in the α3 domain)]. The samples were first denatured at 95° for 3 min and then subjected to PCR amplification for 35 cycles of 30 seconds at 95°, 1 min at 54° and 1 min at 72°, and a final cycle of 7 min at 72°. PCR products were purified with the QIAquick PCR purification kit (Qiagen, Valencia, CA) and digested with different restriction enzymes, as detailed in the text. All restriction enzymes were purchased from New England BioLabs (Beverly, MA). Patterns obtained by restriction enzyme digestion of PCR products were visualized on 1·8–2·4% agarose gels following staining with ethidium bromide and UV transillumination.
Immunohistochemistry
Tissue samples were mounted in optimal cutting temperature (OCT) compound (Tissue-Tek, Torrance, CA), frozen in liquid nitrogen, and stored at −80°. Frozen tissue sections (5-µm thick) were fixed in acetone for 10 min, air-dried and stained by an indirect immunoperoxidase method using avidin–biotin–peroxidase complex (Vector Laboratories, Burlingame, CA) and 3-amino-9-ethylcarbazole (Sigma) as the chromogen.
Immunofluoresence microscopy
104C1 cell transfectants expressing different gpCD1 isoforms were grown for 48 hr on glass coverslips to allow attachment and spreading, and then fixed with 3·7% paraformaldehyde and permeabilized with 0·2% saponin/5% donkey serum (Jackson Immunoresearch Laboratories, West Grove, PA) in PBS (permeabilization buffer). The coverslips were then incubated with primary mAbs (anti-gpCD1 or non-specific control mouse IgG antibodies), followed by FITC-labelled donkey anti-mouse IgG in permeabilization buffer. Cover slips were mounted for observation on glass slides in Vectashield mounting medium (Vector Laboratories) as an anti-fading agent.
Results
Generation of mAbs against gpCD1 proteins and gpCD1 transfectant cells
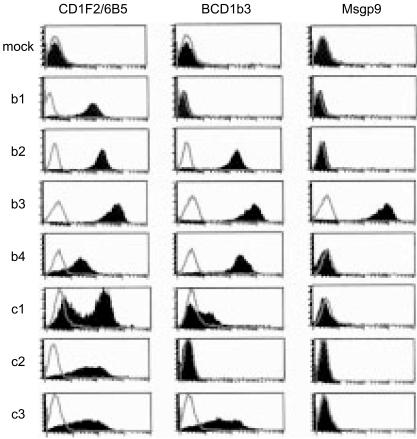
To raise a panel of mAbs that would enable us to detect all of the known group 1 CD1 isoforms expressed in the guinea-pig, we first generated gpCD1 transfectant cells in K.BALB and soluble gpCD1–Fc fusion proteins. We were able to detect the gpCD1b3- and gpCD1b4-transfected cells by flow cytometry using anti-human CD1b mAb BCD1b3, which we previously showed (by FACS and immunoprecipitation) to be cross-reactive with a subset of CD1 proteins expressed on guinea-pig thymocytes.17 Using the K.BALB murine cell transfectants and gpCD1b3–Fc and gpCD1b4–Fc fusion proteins constructed with the mouse IgG1 constant-region domains, we were able to develop effective strategies for immunization of BALB/c mice that facilitated the selective production of antibodies against gpCD1 proteins. Fusions of spleen cells from these immunized mice with non-secreting NSO myeloma cells led to the isolation of four new hybridoma cell lines that secreted mAbs reactive with gpCD1 proteins (CD1F2/5E3, CD1F2/6B5, CD1F2/1B12, CD1.4-1D12). Using these new mAbs, we were then able to identify the expression of each of the known forms of group 1 gpCD1 proteins following the transfection of expression constructs containing cDNAs encoding these proteins into guinea-pig cell lines 104C1 and GPC16, including several gpCD1 proteins that could not be clearly detected using mAb BCD1b3. As shown in Fig. 1, mAb CD1F2/6B5 proved to be reactive with transfectants expressing all of the known group 1 gpCD1 family genes, whereas mAb BCD1b3 detected only gpCD1b2, -b3, -c3, -b4 and, very weakly, gpCD1c1. Thus, based on the reactivity of mAb CD1F2/6B5 with our panel of transfected 104C1 cells, we concluded that all of the previously cloned guinea-pig group 1 CD1 sequences, excluding those known to have obvious pseudogene features,17 can be expressed as proteins at the cell surface.
Figure 1.
Fluorescence-activated cell sorter (FACS) analysis of guinea-pig CD1 (gpCD1) transfectant cells. Guinea-pig cell line 104C1 was transfected with cDNAs encoding each of the indicated isoforms of gpCD1 in vector pcDNA3.1, or with an empty vector alone (mock). Transfectant cells were first stained with CD1F2/6B5 [anti-pan gpCD1 monoclonal antibody (mAb)], BCD1b3 (anti-human CD1b), Msgp9 (anti-gp-pan B-cell mAb, specific for gpCD1b3) or an isotype-matched non-binding control mAb [P3, murine immunoglobulin G1 (mIgG1)]. Subsequently, the cells were stained with an anti-mouse IgG/IgM conjugated to fluorescein isothiocyanate (FITC) as a secondary reagent. Control mAb P3 staining is shown as unfilled histograms, and the filled histograms represent staining with the indicated specific mAbs.
Using the collection of transfected cell lines expressing the various group 1 gpCD1 proteins, we determined the pattern of reactivity of each of the newly derived anti-gpCD1 mAbs and also for a collection of previously published antibodies with known specificity for CD1 proteins of humans or other mammals that were found to cross-react with gpCD1 proteins. As summarized in Table 1, three general patterns of reactivity were observed. As mentioned above, mAb CD1F2/6B5 was unique in that it was reactive with all of the gpCD1 proteins tested (‘pan-gpCD1’). In contrast, four mAbs were found to recognize only a single form of gpCD1. These were: CD1.4-1D12, which recognized gpCD1b1; CD1d69, which recognized gpCD1b4; and Msgp9 and WM-25, which were both specific for gpCD1b3 (Table 1 and Fig. 1). The remainder of the mAbs reacted with two or more, but not all, of the group 1 gpCD1 proteins, in some cases showing a preference for CD1b-like (e.g. BCD1b2, BCD1b4) or CD1c-like (e.g. Fe5.5C1) isoforms. We also tested the cross-reactivity of the four newly derived anti-gpCD1 mAbs against human CD1a, -b, -c and -d using transfected C1R B-lymphoblastoid cells expressing each of these proteins (data not shown). CD1F2/5E3 detected human CD1a weakly, bound strongly to human CD1b and CD1c, but showed no reactivity with human CD1d. CD1F2/1B12 could only detect human CD1b, and surprisingly, CD1F2/6B5 (which detected all forms of gpCD1 tested) did not bind detectably to any human CD1 isoforms.
Table 1. Specificity of monoclonal antibodies (mAbs) against guinea-pig CD1 proteins.
| Guinea-pig CD1 isoforms | ||||||||
|---|---|---|---|---|---|---|---|---|
| mAbs | Derived against | b1 | b2 | b3 | b4 | c1 | c2 | c3 |
| CD1F2/5E3 | Guinea-pig CD1 | − | ± | ++ | + | ++ | ++ | + |
| CD1F2/6B5 | Guinea-pig CD1 | + | ++ | ++ | + | ++ | ++ | + |
| CD1F2/1B12 | Guinea-pig CD1 | − | ++ | ++ | ++ | − | − | + |
| CD1·4-1D12 | Guinea-pig CD1 | + | − | − | − | − | − | − |
| Msgp9 | Guinea-pig CD1 | − | − | ++ | − | − | − | − |
| 7C4 | Human CD1b | − | + | + | − | − | − | − |
| WM25 | Human CD1b | − | − | ++ | − | − | − | − |
| BCD1b1 | Human CD1b | − | − | ++ | + | − | − | − |
| BCD1b2 | Human CD1b | − | ++ | ++ | ++ | − | − | + |
| BCD1b3 | Human CD1b | − | ++ | ++ | ++ | ± | − | + |
| BCD1b4 | Human CD1b | − | ++ | ++ | − | − | − | − |
| CD1d69 | Human CD1d | − | − | − | ±∼ +* | − | − | − |
| 20–27 | Sheep CD1 | − | − | ++ | ++ | ± | − | − |
| CC-20 | Cow CD1 | − | ++ | + | ++ | − | − | − |
| Fe5·5C1 | Feline CD1 | − | − | ++ | ± | ++ | ++ | + |
New mAbs generated in this study and mAbs against CD1 of human or other species were tested for their reactivity with guinea-pig CD1 (gpCD1) proteins using transfectants of guinea-pig cell lines 104C1 and GPC-16 expressing individual isoforms of gpCD1. All mAbs were mouse immunoglobulin G (IgG) and were detected with anti-mouse IgG/IgM-conjugated to fluorescein isothiocyanate (FITC) as a secondary reagent. Symbols signify the following: ‘−’, no reactivity above the level of isotype-matched non-binding control antibody; ‘±’, weak but significant shift from isotype-matched control antibody; ‘+’, positive staining with a mean fluorescence intensity (MFI) of 10–100; ‘++’, strong staining with a MFI of >100.
gpCD1b4 expression detected by CD1d69 was ‘+’ in GPC-16 cells transfected with gpCD1b4 and ‘±’ in 104C1 transfectant cells.
Tissue distribution of gpCD1 proteins
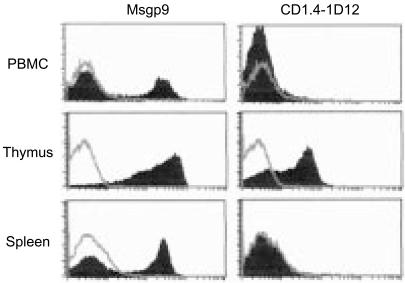
The availability of a large panel of mAbs with defined specificities for various gpCD1 proteins allowed us to examine the patterns of cellular expression and tissue distribution of these proteins. Flow cytometry of guinea-pig thymocytes revealed that all of the antibodies reacted with gpCD1 proteins expressed on the surface of these cells (Fig. 2, and additional data not shown). The gpCD1 proteins expressed by thymocytes probably include gpCD1b1 and gpCD1b3, based on staining and FACS analysis with mAbs CD1.4-1D12 and Msgp9, respectively (Fig. 2). The mean fluorescence intensity values for thymocytes stained with mAb CD1.4-1D12 were lower compared to those observed with CD1F2/6B5 or Msgp9, suggesting that the cell-surface expression level of gpCD1b1 on these cells may be relatively low compared to other gpCD1 isoforms. Staining of peripheral blood mononuclear cells (PBMC) and splenocytes with mAb Msgp9 showed expression by a subset of cells that was consistent with the proportion of B lymphocytes present in those cell suspensions (Fig. 2), supporting previous suggestions that the molecule recognized by Msgp9 is expressed on most or all B cells. The percentage of Msgp9-positive cells in spleen cell suspensions was almost identical to the percentage of cells positive for the pan-gpCD1-reactive mAb, CD1F2/6B5. Using the CD1.4-1D12 mAb, gpCD1b1 expression was not detected by FACS of splenocytes, suggesting that this gpCD1 isoform may be excluded from B cells. CD1d69 stained thymocytes but did not detectably stain either PBMC or spleen cells, indicating the absence or low expression of gpCD1b4 in these latter two cell populations (data not shown).
Figure 2.
Fluorescence-activated cell sorter (FACS) analysis of CD1 expression on guinea-pig leucocyte suspensions. Peripheral blood mononuclear cells (PBMC), thymocytes and spleen cell suspensions obtained from normal adult Strain 2 guinea-pigs were stained with Msgp9 [anti-guinea-pig (gp)CD1b3], CD1.4-1D12 (anti-gpCD1b1) or isotype-matched control monoclonal antibody (mAb) [P3, murine immunoglobulin G1 (mIgG1)], and detected with an anti-mouse IgG/IgM conjugated to fluorescein isothiocyanate (FITC). Control mAb P3 staining is shown as unfilled histograms, and the filled histograms represent staining with the indicated specific mAbs.
Expression of gpCD1 proteins by dendritic cells
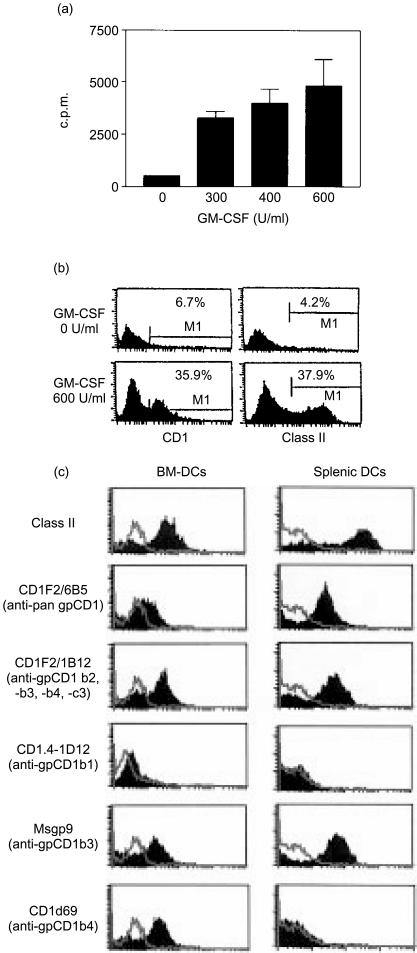
In humans, group 1 CD1 proteins are highly expressed by myeloid DCs in a variety of different tissues, including dermal DCs, interdigitating cells in lymphoid tissues and epidermal Langerhans' cells.3 This prominent expression by specialized APC is consistent with the view that group 1 CD1 proteins are involved in the stimulation of antigen-specific T cells for the generation of adaptive immune responses and immunological memory against lipid antigens of microbial pathogens. It was therefore of interest to determine whether the guinea-pig group 1 CD1 proteins would also show this pattern of strong upregulation on specialized APC such as BM-DC and SP-DCs. This was initially evaluated by isolating DCs and DC precursors from bone marrow or spleen and culturing these in the presence of recombinant GM-CSF, a cytokine that has been found to be central to the growth and differentiation of myeloid DCs in both humans25 and mice.26 As guinea-pig GM-CSF has not yet been characterized and cannot be obtained in purified form, we first tested whether human recombinant GM-CSF could stimulate myeloid cell precursors from guinea-pig bone marrow. As shown in Fig. 3(a), we found that human GM-CSF stimulated proliferation of cultured guinea-pig bone marrow cells, as measured by tritiated thymidine incorporation. In addition, GM-CSF significantly upregulated both CD1 and MHC class II expression in these cultures, indicating that human recombinant GM-CSF can cross-react with GM-CSF receptors expressed by guinea-pig bone marrow cells and induce their differentiation into DCs in a manner similar to that described for humans and mice.
Figure 3.
Expression of guinea-pig CD1 (gpCD1) proteins on bone marrow-derived dendritic cells (BM-DCs). (a) Bone marrow cell cultures from normal adult guinea-pigs were harvested on day 4 and reseeded in 96-well plate wells (at 1×105 cells/well) in medium containing different doses of human granulocyte–macrophage colony-stimulating factor (GM-CSF) and cultured for a further 3 days. The proliferative responses were determined by incorporation of [3H]thymidine. (b) Bone marrow cells were cultured in the presence or absence of human GM-CSF for 3 days in 24-well plates and the effect of GM-CSF on the induction of CD1 and major histocompatibility complex (MHC) class II expression was determined by fluorescence-activated cell sorter (FACS) analysis using anti-gpCD1 monoclonal antibody (mAb) or anti-class II mAbs (Cl.13.1). (c) FACS analysis of BM-DCs and splenic DCs from normal adult guinea-pigs. BM-DCs harvested on day 14 or splenic DCs harvested 48 hr after isolation were stained with the indicated mAbs against guinea-pig MHC class II or gpCD1 proteins. Control mAb P3 staining is shown as unfilled histograms, and the filled histograms represent staining with the indicated specific mAbs.
Based on these results and the methods described in the literature for derivation of DCs from bone marrow of mice26 and rat,27 we established a culture system for the production of myeloid-lineage DCs from guinea-pig bone marrow. These in vitro-cultured BM-DCs showed a morphology characteristic of cultured murine or human DCs (data not shown) and expressed high levels of MHC class II and CD1 molecules on their surfaces (Fig. 3c). Based on staining with the panel of anti-CD1 mAbs, it appeared that gpCD1b3 and gpCD1b4 were strongly expressed on cultured guinea-pig BM-DCs, while gpCD1b1 expression was low or undetectable. Expression of other isoforms of gpCD1 could not be determined because of the lack of monospecific mAbs against these proteins. To confirm and extend these results obtained with BM-DCs, we also analysed DCs isolated from the guinea-pig spleen by collagenase digestion followed by Percoll gradient separation and short-term culture in medium supplemented with GM-CSF.28 Similar, high expression of CD1 molecules and MHC class II were found on SP-DCs. Interestingly, the patterns of group 1 CD1 expression observed for BM-DCs and SP-DCs, although very similar, were not identical as staining with mAb CD1d69 (anti-gpCD1b4) was clearly observed on BM-DCs but not on SP-DCs. This observation provides the first evidence to our knowledge of differential regulation of group 1 CD1 isoforms on subsets of DCs of different tissue origin in the guinea-pig, a feature that has been previously documented for human group 1 CD1 expression.3
Because of the limitations of FACS analysis using the currently available panel of anti-gpCD1 mAbs to identify individual gpCD1 isoforms, we also developed a method based on RT–PCR amplification of CD1 transcripts coupled with restriction endonuclease digestion to analyse the expression of specific CD1 isoforms. CD1 transcripts were amplified by RT–PCR using a pair of universal primers complementary to all known group 1 gpCD1 transcripts. After purifying the RT–PCR products (477 bp), we performed restriction digests, using a panel of restriction enzymes, to generate patterns of DNA fragments that could be correlated with the expression of individual group 1 gpCD1 isoforms. As summarized in Table 2, digestion of the 477-bp product with a panel of nine restriction endonucleases was predicted to generate distinct patterns of DNA fragments for each of the seven group 1 CD1 isoforms. In most cases, these patterns were sufficiently different to be distinguished using agarose-gel electrophoresis and ethidium bromide staining of the digested PCR products.
Table 2. Restriction enzyme digestion patterns predicted for reverse transcription–polymerase chain reaction (RT–PCR) products for each guinea-pig CD1 (gpCD1) isoform.
| Guinea-pig CD1 isoforms | |||||||
|---|---|---|---|---|---|---|---|
| Restriction endonuclease | b1 | b2 | b3 | b4 | c1 | c2 | c3 |
| None | 477 | 477 | 477 | 477 | 477 | 477 | 477 |
| SalI | – | – | – | – | 430/47 | 430/47 | 430/47 |
| StyI | 247/230 | – | 391/86 | – | – | 292/185 | – |
| StuI | – | 394/83 | – | – | – | 259/218 | – |
| XbaI | – | – | – | 246/231 | – | – | – |
| XhoI | – | – | – | – | 420/57 | 420/57 | – |
| DraI | – | – | – | – | – | – | 295/182 |
| KpnI | – | 243/234 | – | – | – | – | – |
| EcoRV | – | 334/143 | 288/189 | – | – | – | – |
| HindIII | – | 313/164 | 313/164 | – | – | 313/164 | – |
Transcripts corresponding to all isoforms of gpCD1 genes could be amplified by PCR with equal efficiency using the universal gpCD1 primers to yield PCR products of 477 bp. From the nucleotide sequences of each isoform, digestion patterns for PCR products corresponding to each isoform of gpCD1were predicted as shown (numbers indicate fragment length in base pairs). For example, DraI digests only the PCR product from gpCD1c3 (295 and 182 bp fragments), and will not digest the PCR product corresponding to other known gpCD1 isoforms. These predicted restriction enzyme digestion patterns were each confirmed by actual digestion of PCR products generated by using plasmids containing cDNA inserts for each isoform of all seven known group 1 gpCD1 genes as templates.
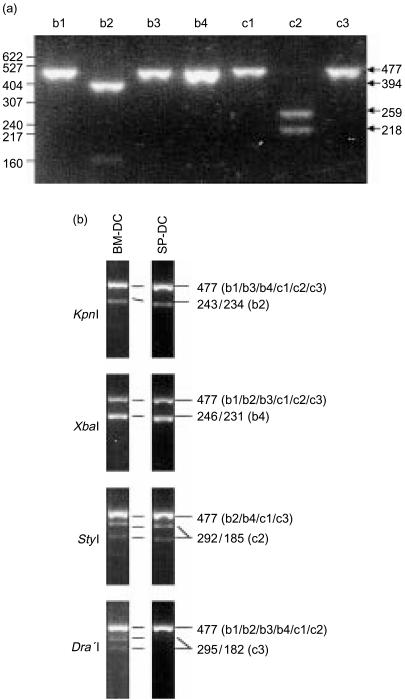
To validate this approach experimentally, we first confirmed that the universal primers amplified all the isoforms of gpCD1 genes equally by using plasmids containing the cloned gpCD1 cDNAs in vector pcDNA3 as templates. This produced strong PCR products consistent with the predicted size of 477 bp in all cases (data not shown). These PCR products were then digested with the nine restriction enzymes listed in Table 2 and subsequently analysed by agarose-gel electrophoresis. Figure 4(a) shows a representative example of the restriction digestion patterns obtained using the enzyme StuI, which cleaved only gpCD1b2 and gpCD1c2 products, yielding unique patterns of DNA fragments for these two CD1 isoforms. Similar preliminary studies confirmed that each of the restriction enzymes listed in Table 2 generated DNA fragments of sizes consistent with the patterns predicted on the basis of the known sequence information (data not shown).
Figure 4.
Detection of guinea-pig CD1 (gpCD1) gene usage in splenic dendritic cells (SP-DC) and bone marrow dendritic cells (BM-DC) by reverse transcription–polymerase chain reaction (RT–PCR) and restriction enzyme digestion pattern analysis. (a) A representative example of a restriction enzyme digestion that distinguishes different isoforms of gpCD1. Polymerase chain reaction (PCR) products produced using pcDNA3 plasmid constructs containing each isoform of the gpCD1 gene were digested with StuI and run on a 2·4% agarose gel. As predicted in Table 2, StuI cut only the gpCD1b2 and gpCD1c2 PCR products to yield fragments of the predicted sizes. Numbers on the left indicate molecular weight (MW) standards in base pairs (bp), and numbers on the right indicate the sizes of the various bands visualized on the gel. The 477-bp band corresponds to the undigested PCR products. (b) Restriction enzyme digestion patterns of RT–PCR CD1 gene products from BM-DCs and SP-DCs. RNA was extracted from bone BM-DCs and SP-DCs, and RT–PCR analyses were performed using the universal gpCD1 primers. The resulting gpCD1 PCR products were purified and digested with the different restriction enzymes indicated, and the restriction digests were resolved on 2·4% agarose gels with ethidium bromide staining. The undigested PCR products have a size of 477 bp, and smaller fragments indicated cleavage by the restriction endonuclease. Sizes of the restriction fragments and the gpCD1 isoforms that these correspond to are indicated on the right.
We applied this method for distinguishing transcripts of specific gpCD1 genes to the analysis of mRNA extracted from guinea-pig BM-DCs or SP-DCs. The RT–PCR reactions using universal gpCD1 primers generated PCR products of expected size (477 bp) from cDNA templates prepared from RNA extracted from both types of DCs. As shown in Fig. 4(b), restriction enzyme digestion of these products allowed the specific detection of transcripts corresponding to four different gpCD1 isoforms. Thus, KpnI digestion of PCR products from BM-DCs and SP-DCs showed two bands – one of 477 bp (undigested) and one of ≈230–240 bp (consistent with a doublet of 243/234 bp) – specifically indicating the presence of transcripts encoding gpCD1b2. Similarly, digestion patterns obtained using XbaI, StyI and DraI provided evidence for expression of gpCD1b4, gpCD1c2 by both types of DCs and expression of gpCD1c3 by BM-DCs. In contrast, bands indicating the presence of gpCD1b1 were not detected in the StyI digest, consistent with the extremely low or absent expression of this isoform by DCs that was observed by FACS (Fig. 3c).
A summary of the detection of specific isoforms of gpCD1 by the FACS staining and RT–PCR methods is shown in Table 3. For the three gpCD1 isoforms that could be individually detected with both of these methods (gpCD1b1, gpCD1b3 and gpCD1b4), there was only partial concordance in the expression at the RNA versus protein levels. Thus, SP-DCs were negative for CD1d69 (anti-gpCD1b4), but showed an RT–PCR/restriction digest pattern consistent with gpCD1b4 mRNA expression. In addition, RT–PCR/restriction digest pattern analysis did not detect gpCD1b3 mRNA from either DC preparation, but both were stained with mAb Msgp9, which our initial studies indicated to be monospecific for gpCD1b3 (Table 1). These findings suggest that mRNA and protein levels may not always be tightly correlated for certain gpCD1 proteins, although further investigations are being undertaken to determine the precise explanations for these discrepancies. Nevertheless, the combined analysis provides a strong indication that multiple specific isoforms of gpCD1b and gpCD1c proteins are probably expressed by various DC subsets in the guinea-pig.
Table 3. Summary of guinea-pig CD1 (gpCD1) isoforms detected by reverse transcription–polymerase chain reaction (RT–PCR) and fluorescence-activated cell sorter (FACS) analysis on bone marrow-derived dendritic cells (BM-DC) and splenic dendritic cells (SP-DC).
| Guinea-pig CD1 isoforms | ||||||||
|---|---|---|---|---|---|---|---|---|
| Cell type | Method of detection | b1 | b2 | b3 | b4 | c1 | c2 | c3 |
| BM-DC | RT–PCR | ND | + | ND | + | ? | + | + |
| FACS | ± | ? | + | + | ? | ? | ? | |
| SP-DC | RT–PCR | ND | + | ND | + | ? | + | ND |
| FACS | ND | ? | + | ND | ? | ? | ? | |
‘+’ indicates specific detection of the indicated isoform (‘±’ for FACS indicates only weak detection on a minority of cells in the sample); ‘ND’ indicates ‘not detected’ (i.e. below the level of detection for the method); and ‘?’ signifies the lack of a method to specifically detect the indicated isoform.
In situ tissue expression of gpCD1 proteins on dendritic cells and B lymphocytes
A subset of anti-gpCD1 mAbs was found to stain specifically when used for immunohistochemical analysis of frozen tissue sections by the immunoperoxidase method, and this approach was used to study the distribution of group 1 gpCD1 protein expression in tissues in situ. Prominent expression was revealed of gpCD1 proteins on dermal DCs, including CD1F2/6B5 (anti-pan-gpCD1) staining of trunk skin (Fig. 5a) and CD1d69 (anti-gpCD1b4) staining of the superficial dermal layers in the ear (Fig. 5b). These results indicate that dermal DCs in the guinea-pig express gpCD1b4 and potentially other group 1 gpCD1 isoforms. Together with our analysis of in vitro-cultured DCs from bone marrow and spleen, these results indicated that guinea-pig group 1 CD1 proteins are highly expressed on specialized APC, suggesting a similar pattern of expression and potential role in antigen presentation, as previously established for the human group 1 CD1 family.
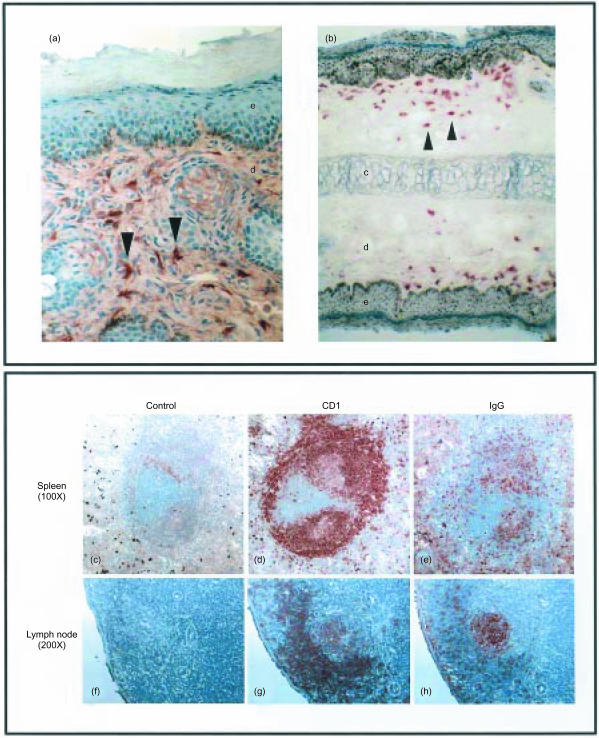
Figure 5.
Immunohistochemical analysis of guinea-pig CD1 (gpCD1) proteins in tissue. (A) and (B) Frozen sections of trunk skin (A, ×400 magnification) and ear (B, ×200 magnification) were stained with anti-pan gpCD1 monoclonal antibody (mAb) (CD1F2/6B5) or CD1d69 (anti-gpCD1b4), respectively. e, epidermis; d, dermis; c, cartilage; arrowheads show representative gpCD1-positive dendritic cells (DCs) that are abundant in the dermal layer. (C) to (H) Frozen serial sections of spleen (C), (D) and (E) or lymph node (F), (G) and (H) were stained with anti-pan gpCD1 mAb (CD1F2/6B5) or anti-gp IgG (Msgp3).
We also used immunohistochemistry to assess the pattern of gpCD1 expression in situ in the spleen and lymph node. As shown in Fig. 5(c) to Fig. 5(e), serial frozen section of spleen stained with anti-pan-gpCD1 (CD1F2/6B5, Fig. 5d) or anti-guinea-pig IgG (Msgp3, Fig. 5e) indicated prominent expression of gpCD1 proteins on splenic B cells. Guinea-pig CD1 proteins were highly expressed on B cells in the peripheral corona of the B-cell areas and the marginal zone, and weakly expressed on germinal centre B cells. This was further suggested by discrepancies in the staining patterns of lymph node obtained with anti-pan-gpCD1 and anti-guinea-pig IgG, as the latter stained most strongly in the lymph node germinal centres where anti-gpCD1 staining was relatively weak (Fig. 5g, 5h). Similarly, the antigen recognized by mAb Msgp9 (probably gpCD1b3) has been reported to be expressed highly on B cells in the B-cell corona but weakly on germinal centre B cells,29 suggesting that expression of gpCD1b3 and potentially other group 1 gpCD1 proteins may be regulated on B cells by their state of differentiation or activation. A similar mechanism has been proposed in humans and mice, based on the selective expression of human CD1c by mantle zone B cells30 or increased CD1d expression on murine marginal zone B cells.31,32
Intracellular distribution of gpCD1 proteins
The cytoplasmic tails of several CD1 molecules, including human CD1b and CD1c, and both mouse and human CD1d, have been shown to contain a tyrosine-based targeting motif (YXXZ, where Y=tyrosine, X=any amino acid and Z=an amino acid with a bulky hydrophobic side-chain) that is important for their endosomal targeting.33,34 Human CD1a lacks this YXXZ-targeting motif and remains primarily surface localized, whereas CD1b possesses the YXXZ motif and shows strong steady-state localization to endosomes.35,36 Recent investigations on CD1c revealed that it is localized mainly to early and late endosomes, whereas CD1b is also directed to lysosomal compartments and MHC class II-containing compartments (MIICs) in APCs.37 The different patterns of subcellular localization of the various CD1 isoforms have led to the hypothesis that each member of the CD1 family may be specialized for sampling of lipid antigens that accumulate in different intracellular compartments.5,38 This has been proposed as one of the factors that may explain the diversification of CD1 proteins into a multigene family of distinct isoforms.
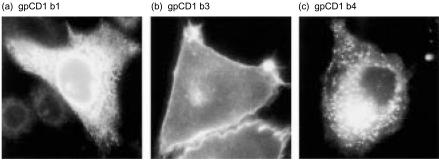
As previously reported by us, the gpCD1b3 cytoplasmic domain, like human CD1a, lacks the YXXZ targeting motif, while all other group 1 gpCD1 proteins possess this endosomal targeting motif.17 This suggested that the different group 1 gpCD1 isoforms may show different steady-state localizations in cells, perhaps reflecting the specialized functions of the various isoforms. To assess this possibility, we analysed the intracellular localization of each gpCD1 protein in transfected cell lines that each expressed a single isoform. A survey of all seven of the known group 1 gpCD1 proteins by immunofluorescence microscopy following staining of permeabilized 104C1 transfectant cells with anti-gpCD1 mAb (CD1F2/6B5, CD1F2/1B12 and CD1F2/5E3), revealed three distinct patterns of intracellular localization (Fig. 6). As predicted, gpCD1b3, which lacks a YXXZ targeting motif, showed a strong cell-surface staining pattern which closely resembled that previously demonstrated for human CD1a.35 In marked contrast, gpCD1b4, -b2, -c2, and -c3, all of which contain YXXZ sequences in their cytoplasmic tails, showed weak cell-surface but striking punctate intracellular staining, consistent with a prominent accumulation in endosomes, as previously found for CD1 proteins that possess YXXZ targeting motifs (Fig. 6, and data not shown). Surprisingly, gpCD1b1 showed a clear reticular staining pattern that is typical for proteins which are mainly localized to the endoplasmic reticulum (ER), despite the fact that this molecule contains a cytoplasmic tail YXXZ motif and does not appear to have an identifiable ER retention or retrieval motif. These results suggest that the various members of the gpCD1 family, as for the human CD1 family, have evolved to have differences in subcelluar localization and intracellular trafficking that may allow the lipid antigen content of different compartments to be sampled and subsequently presented to T cells.
Figure 6.
Intracellular localization of guinea-pig CD1 (gpCD1) proteins. Transfectants of guinea-pig cell line 104C1 expressing gpCD1b1, -b3 or -b4 were grown on cover slips, fixed, permeabilized and stained with anti-gpCD1 monoclonal antibody (mAb) (gpCD1b1, -b3: mAb CD1F2/6B5, gpCD1b4: mAb CD1F2/1B12). Binding of the anti-gpCD1 mAb was detected with fluorescein isothiocyanate (FITC)-conjugated donkey anti-mouse immunoglobulin G (IgG).
Discussion
In this study we have provided several lines of evidence to demonstrate that the multiple group 1 CD1 proteins encoded in the genome of the guinea-pig are expressed as proteins in a cell type-specific manner consistent with their proposed role as antigen-presenting molecules. Our results showed that various gpCD1b and gpCD1c isoforms are expressed by specialized APCs. Thus, in addition to the prominent expression of at least one gpCD1 protein on B lymphocytes (gpCD1b3), we obtained evidence confirming that multiple gpCD1 isoforms are expressed by DCs cultured from spleen or bone marrow in vitro, and probably also on DCs that are normally present in tissues such as the skin. These results are consistent with the proposed immunological functions of these proteins in T-cell recognition of foreign lipid antigens and provide a foundation for further studies to develop the guinea-pig as a small animal model for studies of the in vivo functions of group 1 CD1 molecules.
Although some isoforms of gpCD1, such as gpCD1b1 and -b3, could be detected by mAbs that appeared to be monospecific for these isoforms, we could not unequivocally determine the gpCD1 gene usage by the various cell populations that we examined by analysis with combinations of mAbs alone. Thus, we developed a strategy using RT–PCR combined with restriction enzyme digestion as an additional approach to study the pattern of gpCD1 gene usage by particular cell populations of interest, such as BM-DCs or SP-DCs. This allowed us to gain further insight into the expression pattern of the complex family of group 1 CD1 genes in the guinea-pig and provided evidence for expression, at the transcriptional level, of at least two CD1b-like (gpCD1b2 and -b4) and two CD1c-like (gpCD1c2 and -c3) molecules on guinea-pig bone marrow and spleen-derived DCs.
One hypothesis that has been proposed to explain the presence of multiple isoforms of CD1 in humans is that these related, but distinct, molecules are each specialized to efficiently sample the lipid antigen content of different compartments within the APC.34 This concept has recently gained support from studies carried out on human group 1 CD1 proteins in vitro. The human CD1b protein, for example, has been shown to traffic to late endosomes, co-localizing with MHC class II and HLA-DM molecules.36 This was shown to be dependent upon the presence of an intact YXXZ endosomal-targeting motif. In contrast, human CD1a lacks the YXXZ motif and shows substantially lower steady-state accumulation in endosomes, suggesting that human CD1a and CD1b are directed to different compartments for antigen loading and mediate distinct pathways for antigen presentation.35 A similar argument has been put forth for human CD1c, which appears to localize in a broad subset of endosomes that overlaps partially, but not completely, with the compartments in which CD1b accumulates.37
In the current study, we found similar indications that the gpCD1 proteins may have diverged from each other to some extent with regard to their intracellular trafficking and localization properties. Six of the seven known group 1 gpCD1 proteins also possess a YXXZ endosomal-targeting motif in their cytoplasmic tails, and examination of the intracellular localization of five of these showed that all had a prominent intracellular steady-state accumulation. For four proteins (gpCD1b2, -b4, -c2 and -c3), the pattern of intracellular accumulation was similar to that observed previously for human CD1b, and was clearly consistent with prominent endosomal localization. Whether or not these four endosomally targeted isoforms display subtle differences in the particular endosomal compartments to which they traffic, as found to be the case for human CD1b and CD1c, will require further studies with detailed characterization of co-localizing endosomal markers. An unexpected finding of these studies was the observation that gpCD1b1, which has a typical YXXZ motif and no other intracellular targeting signal that we could identify, showed a unique pattern of intracellular localization that was consistent with marked retention in the ER. This preferential accumulation of a CD1 protein in the ER, if it is confirmed to be a consistent feature of gpCD1b1 when expressed in normal cells, is to our knowledge unprecedented. Although direct studies of T-cell recognition of antigens presented by this molecule will be needed to determine whether this unusual intracellular localization is relevant to the function of gpCD1b1, it is interesting to speculate that such a pattern of localization could reflect a major role for this CD1 molecule in surveying the lipid content of the ER.
The gpCD1b3 protein was also unique among the group 1 gpCD1 family in that this protein lacks the critical YXXZ endosomal-targeting motif at the C-terminus. Examination of the subcellular localization of gpCD1b3 revealed prominent expression on the cell surface with relatively little intracellular accumulation. This pattern is essentially identical to human CD1a, which also lacks the YXXZ motif. We speculate that, as shown for human CD1a in antigen presentation studies in vitro, the gpCD1b3 molecule may be specialized for presentation of lipid antigens that are found at the plasma membrane or in the early recycling endosomal compartments.35,39 Overall, our findings on the subcellular localization of the gpCD1 isoforms suggest that the multiple gpCD1 isoforms may have evolved for the purpose of providing antigen-presenting molecules that are specialized to efficiently sample the lipid antigen content of different intracellular compartments, as has been suggested to be the case for the human CD1 family. Therefore, the multiple CD1b and CD1c isoforms in the guinea-pig may provide an excellent model in which to further study both in vitro and in vivo this proposed functional specialization of the individual group 1 CD1 isoforms.
One of the notable findings in this study was the high level of CD1 expression by a large proportion of B cells. Through our analysis of gpCD1-transfected cells, we found that the anti-pan guinea-pig B-cell mAb, Msgp9, actually recognized the gpCD1b3 protein. Subsequent detailed analysis revealed that most B cells in the spleen or blood were also positive for staining with our anti-pan gpCD1 mAb, CD1F2/6B5. Immunohistochemistry showed that gpCD1 proteins were expressed highly in the B-cell areas of the spleen and lymph nodes, in addition to scattered staining suggestive of expression by interfollicular DCs. In humans, CD1c is expressed on some B-cell populations, such as circulating B cells (especially in newborns and infants),40 mantle zone B cells of the tonsil and lymph nodes, and the marginal zone B cells of the spleen.30,41 Similarly, expression of gpCD1 proteins is differentially regulated on different B-cell subsets, as we found gpCD1 to be highly expressed on mantle zone and splenic marginal zone B cells, but only weakly expressed on germinal centre B cells. Our studies thus far could not identify which specific isoforms of gpCD1 might be expressed on B cells in addition to gpCD1b3. The resolution of this issue will require the development of additional reagents or methodologies that will allow the more accurate identification of specific isoforms of gpCD1. However, even with the current level of information, it is reasonable to speculate that the prominent expression of one or more members of the CD1 family on guinea-pig B cells will probably be linked to interactions between B cells and CD1-restricted T cells.
It is also of interest that we could not find mAbs which selectively recognized the complete set of gpCD1b isoforms or gpCD1c isoforms according to our current classification of these proteins. One mAb that came close to having this property was Fe5.5C1, which recognized the three gpCD1c isoforms but also bound strongly to gpCD1b3. This pattern of reactivity raises the possibility that these four isoforms could belong to one group which may be expressed preferentially on B cells, considering the fact that human CD1c proteins are expressed on a subset of B cells and gpCD1b3 is strongly expressed by guinea-pig B cells. Thus, while the current classification that we have proposed provides a useful working model, it seems clear that further studies on the expression patterns and functions of each of the many gpCD1 isoforms will be needed to fully establish the most appropriate grouping and classification of these proteins.
It should be noted that a previous study showed significant upregulation of staining with mAb Msgp9 in demyelinating lesions present in the brain and spinal cord tissues of guinea-pigs with experimental allergic encephalomyelitis (EAE), a model of multiple sclerosis (MS) or other autoimmune demyelinating diseases.29 Given our evidence from this study that mAb Msgp9 is highly specific for gpCD1b3, it thus appears probable that one or possibly more isoforms of gpCD1 can be upregulated at sites of autoimmune-mediated tissue damage. In fact, we have recently applied the methods described in the current study to the analysis of brain and spinal cord samples from guinea-pigs with active EAE, and have found prominent upregulation of gpCD1 proteins on several cell types in areas of active central nervous system (CNS) disease (B. Cipriani, K. Hiromatsu, S. Porcelli, C. Brosnan, manuscript in preparation). Along with previous reports suggesting a possible link between the human CD1b molecule and MS,42,43 our current results provide further impetus to more fully evaluate the possible involvement of the CD1 system in autoimmune demyelinating diseases and also suggest that guinea-pigs can be used as an animal model to test this hypothesis.
A significant accomplishment of this study was the development of systems for the culture of DCs from bone marrow and spleens of guinea-pigs. We demonstrated that these DCs expressed high levels of multiple group 1 gpCD1 proteins on their surfaces and transcribed multiple gpCD1 genes. These findings should be valuable in future efforts to study the role of gpCD1 molecules on specialized APCs. In fact, we have recently found that guinea-pig BM-DCs, produced according to the methods described in the current study, can stimulate lipid antigen-specific T-cell responses after exposure to foreign lipid antigens derived from mycobacteria (K. Hiromatsu, S. Porcelli, manuscript in preparation). Thus, the reagents and methods produced and characterized in the current study should provide an excellent opportunity for testing the in vivo relevance of the group 1 CD1 system to host defence against microbial pathogens or autoimmune diseases using the guinea-pig as an animal model system.
Acknowledgments
S.P. was supported by a grant from the Irene Diamond Foundation, and by grants from the NIH/NIAID (AI48933 and AI45889). K.H. was supported in part by a fellowship from the Medical Foundation. Part of this work was supported by NIH SBIR AI-40798 (to K.L. and C.G.B.). We thank Penny Cloutier-Lyons, Cindy Greer and Maryann Giel for preparation of mAb CD1.4-1D12.
Abbreviations
- β2m
β2-microglobulin
- BM-DC
bone marrow-derived dendritic cells
- SP-DC
splenic dendritic cells
- gpCD1
guinea-pig CD1
References
- 1.Porcelli SA, Segelke BW, Sugita M, Wilson IA, Brenner MB. The CD1 family of lipid antigen presenting molecules. Immunol Today. 1998;19:362–8. doi: 10.1016/s0167-5699(98)01289-4. [DOI] [PubMed] [Google Scholar]
- 2.Albertson DG, Fishpool R, Sherrington P, Nacheva E, Milstein C. Sensitive and high resolution in situ hybridization to human chromosomes using biotin labelled probes: assignment of the human thymocyte CD1 antigen genes to chromosome 1. EMBO J. 1988;7:2801–5. doi: 10.1002/j.1460-2075.1988.tb03135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Porcelli SA. The CD1 family: a third lineage of antigen-presenting molecules. Adv Immunol. 1995;59:1–98. doi: 10.1016/s0065-2776(08)60629-x. [DOI] [PubMed] [Google Scholar]
- 4.Zeng Z, Castano AR, Segelke BW, Stura EA, Peterson PA, Wilson IA. Crystal structure of mouse CD1: an MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339–45. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 5.Porcelli SA, Modlin RL. The CD1 system: antigen-presenting molecules for T cell recognition of lipids and glycolipids. Annu Rev Immunol. 1999;17:297–329. doi: 10.1146/annurev.immunol.17.1.297. [DOI] [PubMed] [Google Scholar]
- 6.Moody DB, Ulrichs T, Muhlecker W, et al. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 2000;404:884–8. doi: 10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- 7.Rosat JP, Grant EP, Beckman EM, et al. CD1-restricted microbial lipid antigen-specific recognition found in the CD8+ αβ T cell pool. J Immunol. 1999;162:366–71. [PubMed] [Google Scholar]
- 8.Beckman EM, Melian A, Behar SM, et al. CD1c restricts responses of mycobacteria-specific T cells. Evidence for antigen presentation by a second member of the human CD1 family. J Immunol. 1996;157:2795–803. [PubMed] [Google Scholar]
- 9.Brenner M, Porcelli S. Antigen presentation: a balanced diet. Science. 1997;277:332. doi: 10.1126/science.277.5324.332. [DOI] [PubMed] [Google Scholar]
- 10.Calabi F, Jarvis JM, Martin L, Milstein C. Two classes of CD1 genes. Eur J Immunol. 1989;19:285–92. doi: 10.1002/eji.1830190211. [DOI] [PubMed] [Google Scholar]
- 11.Brossay L, Jullien D, Cardell S, Sydora BC, Burdin N, Modlin RL, Kronenberg M. Mouse CD1 is mainly expressed on hemopoietic-derived cells. J Immunol. 1997;159:1216–24. [PubMed] [Google Scholar]
- 12.Bonish B, Jullien D, Dutronc Y, Huang BB, Modlin R, Spada FM, Porcelli SA, Nickoloff BJ. Overexpression of CD1d by keratinocytes in psoriasis and CD1d-dependent IFN-gamma production by NK-T cells. J Immunol. 2000;165:4076–85. doi: 10.4049/jimmunol.165.7.4076. [DOI] [PubMed] [Google Scholar]
- 13.Balk SP, Burke S, Polischuk JE, Frantz ME, Yang L, Porcelli S, Colgan SP, Blumberg RS. β2-microglobulin-independent MHC class Ib molecule expressed by human intestinal epithelium. Science. 1994;265:259–62. doi: 10.1126/science.7517575. [DOI] [PubMed] [Google Scholar]
- 14.Godfrey DI, Hammond KJ, Poulton LD, Smyth MJ, Baxter AG. NKT cells: facts, functions and fallacies. Immunol Today. 2000;21:573–83. doi: 10.1016/s0167-5699(00)01735-7. [DOI] [PubMed] [Google Scholar]
- 15.Hayes SM, Knight KL. Group 1 CD1 genes in rabbit. J Immunol. 2001;166:403–10. doi: 10.4049/jimmunol.166.1.403. [DOI] [PubMed] [Google Scholar]
- 16.Rhind SM, Hopkins J, Grant ES. Differential expression of ovine CD1. Immunology. 2000;101:452–7. doi: 10.1046/j.1365-2567.2000.00134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dascher CC, Hiromatsu K, Naylor JW, et al. Conservation of a CD1 multigene family in the guinea pig. J Immunol. 1999;163:5478–88. [PubMed] [Google Scholar]
- 18.Behar SM, Porcelli SA, Beckman EM, Brenner MB. A pathway of costimulation that prevents anergy in. J Exp Med. 1995;182:2007–18. doi: 10.1084/jem.182.6.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spada FM, Borriello F, Sugita M, Watts GF, Koezuka Y, Porcelli SA. Low expression level but potent antigen-presenting function of CD1d on monocyte lineage cells. Eur J Immunol. 2000;30:3468–77. doi: 10.1002/1521-4141(2000012)30:12<3468::AID-IMMU3468>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 20.Amiot M, Bernard A, Raynal B, Knapp W, Deschildre C, Boumsell L. Heterogeneity of the first cluster of differentiation. Characterization and epitopic mapping of three CD1 molecules on normal human thymus cells. J Immunol. 1986;136:1752–8. [PubMed] [Google Scholar]
- 21.Favaloro EJ, Bradstock KF, Kamath S, Dowden G, Gillis D, George V. Characterization of a p43 human thymocyte antigen. Dis Markers. 1986;4:261–70. [PubMed] [Google Scholar]
- 22.Mackay CR, Maddox JF, Gogolin-Ewens KJ, Brandon MR. Characterization of two sheep lymphocyte differentiation antigens, SBU-T1 and SBU-T6. Immunology. 1985;55:729–37. [PMC free article] [PubMed] [Google Scholar]
- 23.MacHugh ND, Bensaid A, Davis WC, Howard CJ, Parsons KR, Jones B, Kaushal A. Characterization of a bovine thymic differentiation antigen analogous to CD1 in the human. Scand J Immunol. 1988;27:541–7. doi: 10.1111/j.1365-3083.1988.tb02381.x. [DOI] [PubMed] [Google Scholar]
- 24.Howard CJ, Sopp P, Bembridge G, Young J, Parsons KR. Comparison of CD1 monoclonal antibodies on bovine cells and tissues. Vet Immunol Immunopathol. 1993;39:77–83. doi: 10.1016/0165-2427(93)90166-2. [DOI] [PubMed] [Google Scholar]
- 25.Caux C, Dezutter-Dambuyant C, Schmitt D, Banchereau J. GM-CSF and TNF-alpha cooperate in the generation of dendritic Langerhans' cells. Nature. 1992;360:258–61. doi: 10.1038/360258a0. [DOI] [PubMed] [Google Scholar]
- 26.Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, Muramatsu S, Steinman RM. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med. 1992;176:1693–702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Talmor M, Mirza A, Turley S, Mellman I, Hoffman LA, Steinman RM. Generation or large numbers of immature and mature dendritic cells from rat bone marrow cultures. Eur J Immunol. 1998;28:811–7. doi: 10.1002/(SICI)1521-4141(199803)28:03<811::AID-IMMU811>3.0.CO;2-S. 10.1002/(sici)1521-4141(199803)28:03<811::aid-immu811>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 28.Girolomoni G, Simon JC, Bergstresser PR, Cruz PD., Jr Freshly isolated spleen dendritic cells and epidermal Langerhans' cells undergo similar phenotypic and functional changes during short-term culture. J Immunol. 1990;145:2820–6. [PubMed] [Google Scholar]
- 29.Butter C, Healey DG, Baker D, Turk JL. A quantitative immunocytochemical study of the infiltrating lymphocytes in the spinal cord of guinea pigs with chronic relapsing experimental allergic encephalomyelitis. J Neuroimmunol. 1989;25:169–76. doi: 10.1016/0165-5728(89)90134-3. [DOI] [PubMed] [Google Scholar]
- 30.Smith ME, Thomas JA, Bodmer WF. CD1c antigens are present in normal and neoplastic B cells. J Pathol. 1988;156:169–77. doi: 10.1002/path.1711560212. [DOI] [PubMed] [Google Scholar]
- 31.Roark JH, Park SH, Jayawardena J, Kavita U, Shannon M, Bendelac A. CD1.1 expression by mouse antigen-presenting cells and marginal zone B cells. J Immunol. 1998;160:3121–7. [PubMed] [Google Scholar]
- 32.Amano M, Baumgarth N, Dick MD, Brossay L, Kronenberg M, Herzenberg LA, Strober S. CD1 expression defines subsets of follicular and marginal zone B cells in the spleen. Beta 2-microglobulin-dependent and independent forms. J Immunol. 1998;161:1710–7. [PubMed] [Google Scholar]
- 33.Robinson MS, Bonifacino JS. Adaptor related proteins. Curr Opin Cell Biol. 2001;13:444–53. doi: 10.1016/s0955-0674(00)00235-0. [DOI] [PubMed] [Google Scholar]
- 34.Briken V, Moody DB, Porcelli SA. Diversification of CD1 proteins: sampling the lipid content of different cellular compartments. Semin Immunol. 2000;12:517–25. doi: 10.1006/smim.2000.0274. [DOI] [PubMed] [Google Scholar]
- 35.Sugita M, Grant EP, van Donselaar E, Hsu VW, Rogers RA, Peters PJ, Brenner MB. Separate pathways for antigen presentation by CD1 molecules. Immunity. 1999;11:743–52. doi: 10.1016/s1074-7613(00)80148-x. [DOI] [PubMed] [Google Scholar]
- 36.Sugita M, Jackman RM, van Donselaar E, Behar SM, Rogers RA, Peters PJ, Brenner MB, Porcelli SA. Cytoplasmic tail-dependent localization of CD1b antigen-presenting molecules to MIICs. Science. 1996;273:349–52. doi: 10.1126/science.273.5273.349. [DOI] [PubMed] [Google Scholar]
- 37.Briken V, Jackman RM, Watts GF, Rogers RA, Porcelli SA. Human CD1b and CD1c isoforms survey different intracellular compartments for the presentation of microbial lipid antigens. J Exp Med. 2000;192:281–8. doi: 10.1084/jem.192.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaible UE, Hagens K, Fischer K, Collins HL, Kaufmann SH. Intersection of group I CD1 molecules and mycobacteria in different intracellular compartments of dendritic cells. J Immunol. 2000;164:4843–52. doi: 10.4049/jimmunol.164.9.4843. [DOI] [PubMed] [Google Scholar]
- 39.Ulrichs T, Porcelli SA. CD1 proteins: targets of T cell recognition in innate and adaptive immunity. Rev Immunogenet. 2000;2:416–32. [PubMed] [Google Scholar]
- 40.Durandy A, Thuillier L, Forveille M, Fischer A. Phenotypic and functional characteristics of human newborns' B lymphocytes. J Immunol. 1990;144:60–5. [PubMed] [Google Scholar]
- 41.Delia D, Cattoretti G, Polli N, Fontanella E, Aiello A, Giardini R, Rilke F, Della PG. CD1c but neither CD1a nor CD1b molecules are expressed on normal, activated, and malignant human B cells: identification of a new B-cell subset. Blood. 1988;72:241–7. [PubMed] [Google Scholar]
- 42.Battistini L, Fischer FR, Raine CS, Brosnan CF. CD1b is expressed in multiple sclerosis lesions. J Neuroimmunol. 1996;67:145–51. doi: 10.1016/0165-5728(96)00045-8. [DOI] [PubMed] [Google Scholar]
- 43.Shamshiev A, Donda A, Carena I, Mori L, Kappos L, de Libero G. Self glycolipids as T-cell autoantigens. Eur J Immunol. 1999;29:1667–75. doi: 10.1002/(SICI)1521-4141(199905)29:05<1667::AID-IMMU1667>3.0.CO;2-U. 10.1002/(sici)1521-4141(199905)29:05<1667::aid-immu1667>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]