Abstract
Persistent antigenic stimulation during chronic hepatitis C may alter the T-cell receptor variable chain beta (TCR BV) repertoire as well as the cytokine responses of hepatitis C virus (HCV)-specific T lymphocytes. We analysed the distribution of the TCR BV subsets 2.1, 3.1, 5.1, 6.1, 8, 13.1, 13.6, 14.1, 17.1, 21.3 in relation to intracytoplasmic expression of interleukin-2, interferon-γ, interleukin-4 and interleukin-10. Using flow cytometry, CD45RO+ memory T cells of 27 patients with chronic hepatitis C, eight patients with resolved HCV infection and 16 non-HCV-related controls were studied with and without stimulation by the HCV core, NS3, NS4, NS5a and NS5b proteins. Patients with chronic and resolved hepatitis C differed by larger basal TCR BV2.1+, BV6.1+, BV17.1+ and BV21.3+ subsets in chronic hepatitis C, which were correlated to the numbers of T cells with spontaneous interleukin-2 and interferon-γ production (r=0·51–0·73, P<0·05). Upon HCV-specific stimulation these subsets did not expand, whereas a marked in vitro expansion of TCR BV8+ T cells in response to all HCV proteins was selectively noted in chronic hepatitis C (P<0·05). This expansion of TCR BV8+ memory T cells was significantly correlated to HCV-induced interleukin-10 expression (r=0·58–0·98, P<0·01). Thus, differential involvement of selected TCR BV subsets may be related to the outcome of HCV infection.
Introduction
Hepatitis C virus (HCV)-specific T cells are considered crucial for the outcome of HCV infection. The antigen specificity of a T cell is determined by the structure of its T-cell receptor (TCR), which is a heterodimeric glycoprotein composed of disulphide-linked α- and β-chain variable regions. During T-cell differentiation unique variable region genes are randomly rearranged from multiple variable (V), diversity (D) and joining (J) segments for the β locus and V and J segments for the α locus.1 Furthermore, T-cell subpopulations bearing specific TCR variable β (TCR BV) sequences may be engaged by HCV antigens, and therefore may lead to expansions of cognate TCR BV subsets that may then be followed by clonal exhaustion, anergy, or deletions of TCR BV subsets, as has been implicated in the pathogenesis of other viral diseases.2
Memory T cells represent that part of the T-cell repertoire that has been activated by exposure to antigen in the past.3,4 Compared to naive T cells, these cells are more rapidly dividing, express adhesion molecules and the low-molecular-weight isoform of CD45 (CD45RO).5,6 Thus, analysis of the TCR repertoire of T cells responding to viral antigens may provide important insights in the T-cell subsets participating in the course of disease. Therefore, we have chosen 10 TCR BV families that span a high proportion of the T-cell repertoire and determined TCR BV phenotypes with and without HCV antigen stimulation in the memory T cells of patients with different outcomes of hepatitis C by flow cytometry.7,8 Since T-cell subsets can exert different functions depending on their cytokine profiles,9 we simultaneously analysed the intracytoplasmic expression of the cytokines interleukin-2 (IL-2), interferon-γ (IFN-γ), IL-4 and IL-10. The aim of the study was twofold: first to test whether antigen-specific challenge of T lymphocytes from patients with different outcomes of HCV infection leads to clonal expansion or exhaustion of subsets with distinct TCR BV chains. Second to evaluate differences in antigen-specific cytokine reponse that could correspond to either clearance or persistence of hepatitis C virus infection.
Materials and methods
Patients
Three groups of patients were included in this study. Group 1 consisted of 27 patients with chronic hepatitis C with elevated liver enzymes and viral RNA detectable in the serum. Further demographic and virological data of the patient groups are given in Table 1. Members of group 2 were considered to have recovered from HCV infection. This group consisted of eight carefully selected patients who were anti-HCV positive and had recovered from an episode of acute hepatitis but had consistently normal aminotransferases for more than 2 years without detectable viral RNA in their serum on repeated examination. Group 3, comprising 16 patients with liver diseases not related to hepatitis C, served as a control group to ensure antigen specificity of HCV-induced immune responses (alcoholic liver cirrhosis n=10, hepatitis B n=5, sclerosing cholangitis n=1). There were no significant differences between the two anti-HCV groups with respect to total immunoglobulin levels. All patients were negative for cryoglobulins and autoantibodies.
Table 1. Patient characteristics of the study groups.
| Patient group | n | Sex F/M | Age (years) | ALT (U/l) | Virus load (copies/ml) | HCV typing (genotype) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Group 1 | 27 | 11/16 | 38 median | 48·5 | 62·4×104 median | 1a | 1b | 2a | 2b | 3a | 4c | 1b/3a | 4c/4d | other |
| range 23–75 | range 14–150 | range 25·9×104–84·9×104 | 5 | 10 | 1 | 1 | 3 | 1 | 1 | 1 | 4 | |||
| Group 2 | 8 | 1/7 | 27·5 median | 9 median | ND | serotype | ||||||||
| range 17–67 | range 5–19 | type 1, n=5, no type specific reactivity, n=3 | ||||||||||||
| Controls | 16 | 7/9 | 43 median | 27·5 median | ND | ND | ||||||||
| range 21–68 | range 8–196 | |||||||||||||
Group 1, chronic hepatitis C; Group 2, self-limited HCV infection; Controls, non-HCV disease.
ND, not determined.
The nature and purpose of this study were explained to all subjects and informed consent was obtained. The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the local ethics committee.
Serological tests for anti-HCV
HCV antibodies were determined with a microparticle enzyme immunoassay (MEIA) according to the instructions of the manufacturer (Abbott, Wiesbaden, Germany). Seroreactivities in all individuals were confirmed using a dot immunoassay obtained from the same manufacturer to dissect HCV antibodies with respect to their specificity for the HCV proteins core, NS3, NS4, and NS5.
Detection of HCV RNA
HCV RNA was detected after isolation of RNA from 140 µl serum with a nucleic acid purification kit (Viral Kit, Qiagen, Hilden, Germany) by reverse transcription and nested polymerase chain reaction as described by Imberti and co-authors.10 Briefly, RNA was reverse-transcribed with the Moloney murine leukaemia virus (MMuLV) reverse transcriptase (Boehringer, Mannheim, Germany) in the presence of the antisense (5′-GATGCACGGTCTACGAGACCTC-3′) primer. The cDNA was amplified by thermostable DNA-polymerase (Expand High Fidelity, Boehringer Mannheim) with 5′-GATGCACGGTCTACGAGACCTC-3′ (antisense) and 5′-AACTACTGTCTTCACGCAGAA-3′ (sense) as first primers and with 5′-GCGACCCAACACTACTCGGCT-3′ (antisense) and 5′-ATGGCGTTAGTATGAGTG-3′ (sense) as second primers. All primer sets were chosen from the 5′-noncoding region of the HCV genome. Quantitative determination of HCV RNA copies was carried out using the branched DNA technology of Chiron (Emeryville, CA).
With regard to the infecting HCV subtype, all subjects from Group 1 were characterized by the INNO-LiPA HCV II test according to the instructions of the manufacturer (Innogenetics, Zwijndrecht, Belgium). Patients in Group 2 were characterized by serotype-specific antibodies (Murex, Abbott Wiesbaden, Germany).11
HCV-specific immune analysis
HCV proteins
The purified recombinant proteins derived from the HCV-1 prototype sequence were purchased from Mikrogen (Munich, Germany). HCV proteins were expressed in Escherichia coli, purified by ion exchange chromatography followed by preparative gel electrophoresis and were supplied in Tris–glycine buffer.12 The recombinant proteins corresponded to the following regions of the HCV polyprotein: r-core (truncated): amino acids (aa) 1–115, r-NS3: aa 1007–1534, r-NS4: aa 1616–1862, r-NS5a: aa 2007–2268, r-NS5b: aa 2622–2868. The bacterial lipopolysaccharide content of the proteins had been checked by Limulus assay (Max-Pettenkofer-Institut, Munich, Germany) and found to be between 4·0 and 20 pg/μg recombinant protein, which is in the range assumed to be acceptable for cell culture experiments (<50 pg/μg).13
Antibodies and reagents
The following fluorescein isothiocyanate (FITC)-labelled TCR BV antibodies were purchased from Immunotech (Hamburg, Germany): BV2.1 [mouse immunoglobulin M (IgM), clone E22E7.2], BV3.1 (mouse IgG2a, clone LE-89), BV5.1 (mouse IgG2a, clone Immu157), BV6.1 (mouse IgM, clone, CRI 304.3), BV8 (8.1 and 8.2) (mouse IgG2a, clone 56C5.2), BV13.1 (mouse IgG2b, clone Immu222), BV13.6 (mouse IgG1, clone JU-74.3), BV14 (mouse IgG1, clone CAS 1.13), BV17.1 (mouse IgG1, clone E 17.5F3), BV21.3 (mouse IgG2a, clone IG125). Appropriate isotype controls were also purchased from Immunotech. Anti-IL-2 (mouse IgG1, clone DMS-1), anti-IL-4 (mouse IgG1, clone M1), anti IFN-γ (mouse IgG2a, clone H21) monoclonal antibodies (mAbs) were purchased from Genzyme (Ismaning, Germany). Anti-IL-10 (rat IgG1, clone JES5-2A5) antibody was purchased from Pharmingen (Hamburg, Germany). The following phycoerythrin (PE)-labelled mAbs were purchased from Becton Dickinson (Heidelberg, Germany): anti-CD3 (mouse IgG1, Leu-4 clone SK7), anti-CD4 (mouse IgG1, Leu 3a and Leu 3b, clones SK3 and SK4), anti-CD8 (mouse IgG2a, Leu 2a, clone SK2), anti-HLA DR (mouse IgG2a, clone L243), anti-CD45RO (mouse IgG2a, clone UCHL-1). Appropriate isotype controls were also purchased from Becton Dickinson (Heidelberg, Germany). Goat anti-mouse IgG F(ab′)2; goat anti-mouse and goat anti-rat second step antibodies [IgG F(ab′)2 Texas Red-labelled] (all preabsorbed to human proteins) were obtained from Dianova (Hamburg, Germany). Recombinant cytokines were purchased from Genzyme (rIL-2, rIL-4, rIFN-γ) and R & D Systems (rIL-10; Wiesbaden, Germany).
Cells and cell culture
Peripheral blood mononuclear cells (PBMC) were isolated from freshly heparinized blood by Ficoll (Biochrom, Berlin, Germany) density-gradient centrifugation. Unfractionated PBMC (1×106/ml) were resuspended in RPMI-1640 (Biochrom) containing 10% autologous human serum, 100 units/ml penicillin, 100 units/ml streptomycin, and incubated at 37° with 5% CO2 in 96-well microtitre plates (Nunc, Wiesbaden, Germany) in the presence of recombinant HCV proteins for 60 hr (final concentration 10 µg/ml). In preceding experiments the effects of tissue culture on TCR BV subset composition and relevant toxicity could be excluded. Based on previously published data optimal exposure time to monensin was determined.14–17 Adding monensin (Sigma, Munich Germany) (3·0 µm) 12 hr prior to harvesting was found to be optimal for enhancing the signal to noise ratio by inhibiting vesicular traffic of the cells.18
Three-colour flow cytometry for immunophenotyping and intracytoplasmic staining of cytokines
The surface markers CD45RO, CD4, CD8, human leucocyte antigen (HLA) DR and TCR BV chains were detected via direct immunofluorescence; then free IgG epitopes were blocked by an incubation step with goat anti-mouse IgG. Intracytoplasmic staining for cytokines was performed as indirect immunofluorescence (30 min incubation at room temperature) according to the modified paraformaldehyde (PFA)-saponin procedure described by Sander et al. using 4% PFA as fixative and 0·1% saponin for permeabilization.19 In brief, cultured cells were washed twice in Hanks' balanced salt solution (HBSS; Gibco, Eggenstein, Germany) and stained for the surface markers CD45RO and TCR BV chains (20 min incubation at 4° in the dark). After one further wash in HBSS free IgG epitopes were blocked by an incubation step with goat anti-mouse IgG (final concentration 100 µg/ml, 20 min incubation at 4° in the dark). After one further wash the cells were fixed in ice-cold HBSS containing 4% PFA for 5 min and washed again. Cells were resuspended in HBSS containing 0·1% saponin (saponin buffer) and subsequently washed twice in saponin buffer. Then cytokine-specific antibodies diluted in saponin buffer were added at a concentration of 2·5–5·0 µg/ml and incubated for 30 min at room temperature in the dark. Cells were washed twice in saponin buffer and subsequently incubated with appropriate second-step antibodies for 30 min at room temperature in the dark. Finally, cells were washed in HBSS.
Flow cytometric analysis of surface markers and cytokines was performed by triple-colour immunofluorescence cytometry on a FACSort flow cytometer (Becton Dickinson). Control experiments were performed with CD3, CD4 and CD8 antibodies to check whether the antigen-specific immune response was confined to T helper cells. To ensure specificity of the staining procedure, the binding of each mAb was blocked with a molar excess of recombinant cytokine (rIL-2, rIL-4, rIFN-γ and rIL-10). Forward and side scatter and gating for CD45RO+ cells were used to exclude non-T cells from the analysis. Gating was routinely counterchecked by staining for CD3. The analysis for CD45RO, TCR BV expression and intracytoplasmic cytokines was performed with the CellQuest™ software (Becton Dickinson) after counting 10 000 CD45RO+ cells. All experiments were performed in duplicate.
Statistics
Differences in TCR and cytokine expression as well as induction of positive cells with and without stimulation were tested by Kruskal–Wallis analysis of variance. Having established a significant difference between the three groups, the Mann–Whitney U-test was used to determine which groups differed significantly from each other.
All calculations were performed on a personal computer with Statview 4.5 software (Abacus Concepts inc., Berkeley, CA). P-values <0·05 were regarded as statistically significant.
Results
Spontaneous TCR BV distribution in the peripheral blood of patients with different outcomes of HCV infection
Flow cytometric analysis of unstimulated T-lymphocyte subsets after 60-hr culture indicated significantly greater fractions of peripheral blood T cells with TCR BV2.1+, 6.1+, 13.6+, 17.1+ and 21.3+ T cells in the peripheral blood of patients with chronic hepatitis C (Table 2) than in patients with self-limited HCV infection (P<0·05). Compared to non-HCV disease controls these differences were significant only for TCR BV13.6+ (P<0·05). No significant differences existed between the non-HCV disease controls and patients with previous self-limited disease.
Table 2. TCR BV chain distribution (%) in unstimulated memory (CD45RO+) T cells.
| Chronic hepatitis C (n=27) | Self-limited hepatitis C (n=8) | Non-HCV disease controls (n=16) | |
|---|---|---|---|
| BV2.1 | 17·4* (11·4–39·4) | 5·2 (3·1–18·2) | 11·8 (1·6–37·3) |
| BV3.1 | 10·1 (0·5–43·8) | 3·5 (0·3–22·7) | 6·1 (3·1–45·1) |
| BV5.1 | 15·1 (1·0–50·3) | 10·3 (0·4–29·5) | 8·9 (3·6–39·4) |
| BV6.1 | 15·6* (3·0–44·2) | 1·4 (0·4–16·8) | 7·9 (4·1–39·5) |
| BV8 | 10·1 (0·5–40·9) | 8·5 (1·8–22·7) | 5·9 (2·5–13·9) |
| BV13.1 | 14·4 (4·4–44·0) | 5·3 (0·9–45·6) | 12·6 (5·9–42·1) |
| BV13.6 | 12·4*† (0·7–68·7) | 2·8 (0·3–10·7) | 4·4 (0·9–11·5) |
| BV14.1 | 11·4 (1·9–41·8) | 1·1 (0·4–10·9) | 9·9 (4·2–39·0) |
| BV17.1 | 10·9* (0·2–39·7) | 2·5 (0·2–11·7) | 6·5 (1·5–10·7) |
| BV21.3 | 23·4* (10·9–51·8) | 5·9 (2·4–8·2) | 11·6 (7·1–41·4) |
P<0·05 chronic hepatitis C versus self-limited hepatitis C.
P<0·05 chronic hepatitis C versus non-HCV disease controls.
Data are given as median and range of TCR BV positive cells in the CD45RO+ subsets.
Spontaneous and HCV antigen-induced cytokine expression in relation to TCR BV subsets
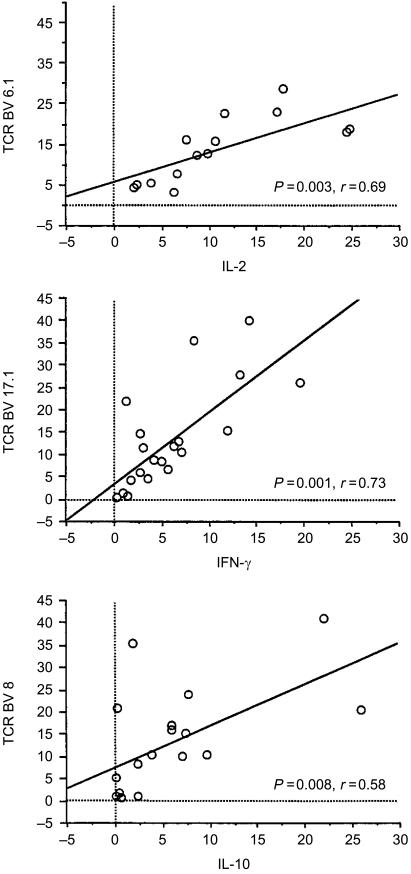
After a 60-hr culture a higher number of cells producing IFN-γ spontaneously was noted in patients with chronic hepatitis C (chronic hepatitis C: mean± SD 8·4±7·3%; self-limited disease: 1·6±1·0; disease controls: 3·7±3·6, P=0·042). Furthermore, we found correlations between the number of cytokine-producing T cells and the size of several T-cell receptor subsets (Fig. 1 and Table 3). The correlations between cytokines and TCR BV subsets were rechecked in randomly selected subjects for the TCR BV2.1-, BV3.1-, BV5.1-, BV6.1-, BV8- and BV13.6-positive T-cell subsets by direct triple-colour flow cytometric analysis of each combination (data not shown).
Figure 1.
Representative examples for the correlations between cytokine production and memory T-cell subsets. (a) The bivariate scatterplot shows the correlation of IL-2+ memory T cells with the fraction of the BV6.1+ T-cell subset Data are given as % positive CD45RO T cells. (b) The bivariate scatterplot shows the correlation of IFN-γ+ memory T cells with the fraction of the BV17.1+ T-cell subset. Data are given as % positive CD45RO T cells. (c) The bivariate scatterplot shows the correlation of IL-10+ memory T cells with the fraction of the BV8+ T-cell subset. Data are given as % positive CD45RO T cells. Data for other correlations are given in Table 3.
Table 3. Association between the size of TCR BV subsets and spontaneous cytokine production in patients with chronic hepatitis C.
| Chronic hepatitis C (n=27) | Self-limited disease (n=8) | Controls (n=16) | ||||
|---|---|---|---|---|---|---|
| r-value | P-value | r-value | P-value | r-value | P-value | |
| Correlation of IL-2+ cells with: | ||||||
| TCR BV2.1 | 0·66 | 0·006 | 0·115 | NS | 0·92 | 0·005 |
| TCR BV6.1 | 0·69 | 0·003 | 0·07 | NS | 0·96 | 0·001 |
| TCR BV14.1 | 0·63 | 0·011 | 0·38 | NS | 0·95 | 0·002 |
| TCR BV17.1 | 0·51 | 0·03 | −0·35 | NS | −0·25 | NS |
| TCR BV21.3 | 0·75 | 0·001 | 0·11 | NS | 0·83 | 0·04 |
| Correlation of IFN-γ+ cells with: | ||||||
| TCR BV2.1 | 0·73 | 0·001 | 0·38 | NS | 0·83 | 0·04 |
| TCR BV6.1 | 0·46 | NS | 0·42 | NS | 0·96 | 0·001 |
| TCR BV14.1 | 0·26 | NS | 0·83 | 0·008 | 0·86 | 0·03 |
| TCR BV17.1 | 0·73 | 0·001 | −0·04 | NS | 0·32 | NS |
| TCR BV21.3 | 0·71 | 0·001 | 0·21 | NS | 0·65 | NS |
| Correlation of IL-10+ cells with: | ||||||
| TCR BV5.1 | 0·37 | NS | 0·6 | NS | 0·2 | NS |
| TCR BV8 | 0·58 | 0·008 | 0·37 | NS | 0·62 | NS |
| TCR BV13.6 | 0·04 | NS | −0·22 | NS | 0·64 | 0·04 |
| TCR BV17.1 | 0·56 | 0·012 | 0·11 | NS | 0·68 | 0·03 |
| TCR BV21.3 | 0·75 | 0·001 | −0·08 | NS | −0·83 | 0·04 |
NS, not significant.
In the non-HCV-related controls and in patients with chronic hepatitis C we found significant correlations (P<0·05) between the production of IL-2 and the TCR subsets BV2.1, 6.1, 14.1 and BV21.3, indicating a general non HCV-specific phenomenon. Unexpectedly, in the patients with self-limited disease we did not find significant correlations for IL-2 and any investigated TCR subsets. Both control groups showed significant correlations between the TCR BV14.1+ and IFN-γ+ subsets which were not seen in patients with chronic hepatitis C. Furthermore, in the non-HCV-related controls the frequency of IL-10+ cells was correlated to the TCR BV13.6 and 17.1 subsets and inversely to the BV21.3 subset (P<0·05). By contrast, a significant correlation between IL-10+ T cells and BV8+ memory cells was exclusively seen in patients with chronic hepatitis C.
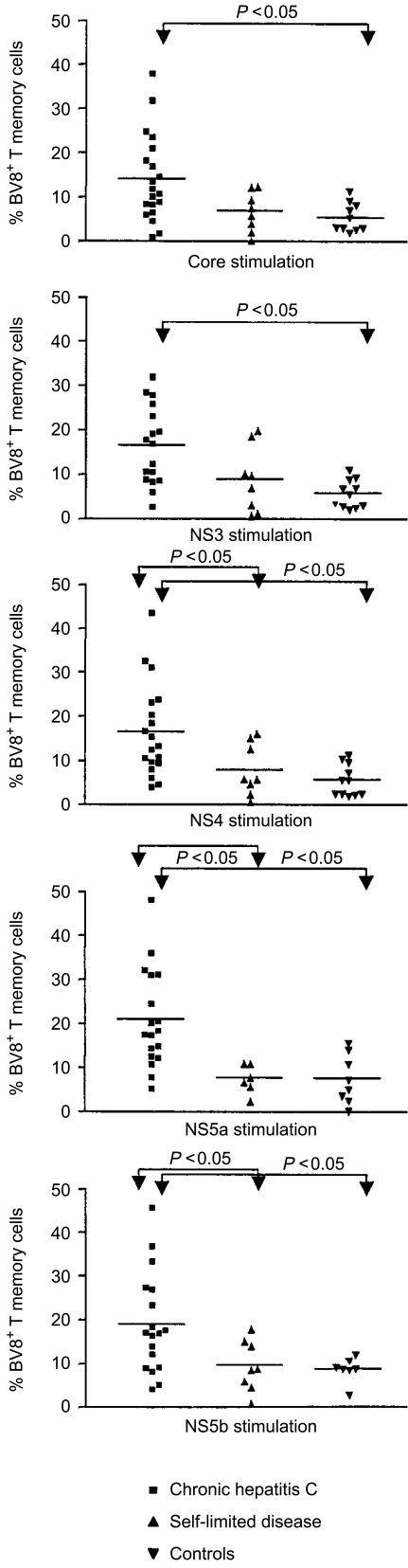
Antigen-specific stimulation led to stronger type 1 cytokine responses in the patients with self-limited disease, however, we did not find a specific TCR family associated with this cytokine response (data not shown). By contrast, when the PBMC of patients with chronic hepatitis C were stimulated in vitro with HCV antigens, there was a conspicuous response of TCR BV8+ T cells to all tested HCV antigens (P<0·05). This increment of BV8+ T cells after antigenic stimulation in patients with chronic hepatitis C was significant for core, NS4 and NS5b proteins (P=0·02, Wilcoxon signed-rank test).
A similar expansion of BV8+ T cells, however, was not observed in the patients with self-limited disease or in the non-HCV-related control group. Thus, we measured significantly higher proportions of BV8+ T cells in patients with chronic hepatitis C after HCV-specific stimulation than in the two control groups (Fig. 2).
Figure 2.
HCV-specific expansion of TCR BV8+ memory T cells. The scatterplots show the fraction of TCR BV8+ memory T cells after stimulation with recombinant HCV proteins (1 µg/ml) for 60 hr. Each dot represents the data from a single patient. The horizontal bar gives the mean of the group. Significant differences between the patient groups are indicated were appropriate.
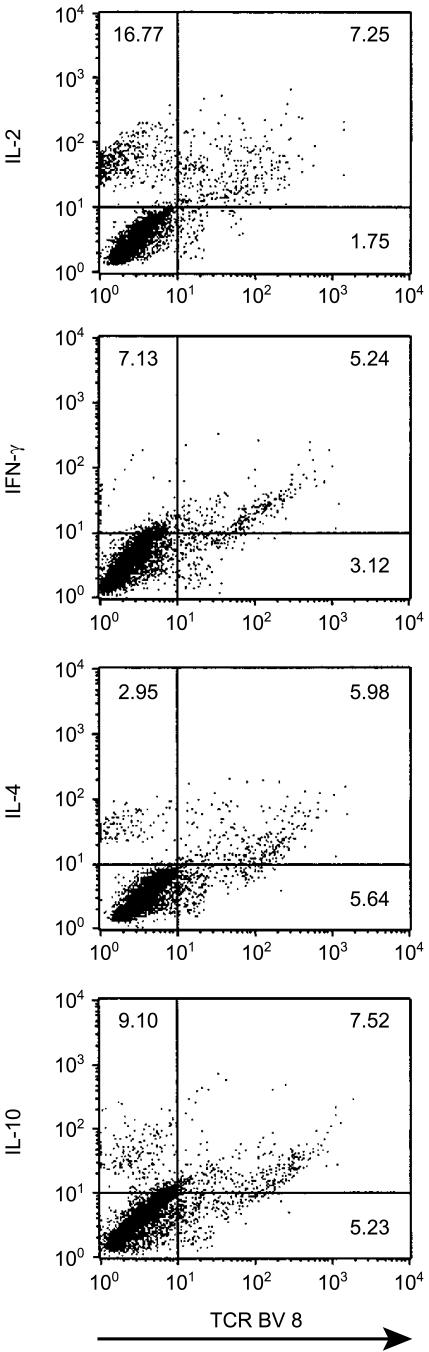
The marked expansion of BV8+ T cells in chronic hepatitis C was correlated to the number of IL-10-producing CD45RO+ T cells after stimulation with all HCV antigens except core (NS3 r=0·85, P<0·0001; NS4 r=0·58, P<0·009; NS5a r=0·98, P<0·0001; NS5b r=0·76, P<0·0002). Production of IL-10 in response to HCV antigens was confirmed in the BV8+ subset by direct triple-colour flow cytometric analysis. However, BV8+ T cells did not show a preferential cytokine type 2 profile but also produced IL-2, IL-4 and IFN-γ (Fig. 3).
Figure 3.
Cytokine induction in the TCR BV8+ subset. Representative dot plot analysis of TCR BV8+ memory T cells for antigen-specific cytokine production in patient B.R. suffering from chronic hepatitis C after stimulation with recombinant NS5b (1 µg/ml) for 60 hr. Quadrant statistics were set on the basis of the isotype controls. The plots demonstrate that in chronic hepatitis C BV8+ memory T cells can produce both type 1 (IL-2, IFN-γ) and type 2 (IL-4, IL-10) cytokines in response to HCV antigens.
After core-specific in vitro stimulation memory T cells of patients with chronic hepatitis C revealed significant correlations between the total numbers of IL-2- and IFN-γ-producing memory T cells and the TCRs BV2.1 (IL-2 r=0·70, P=0·009; IFN-γ r=0·92, P<0·0001) and BV6.1 (IL-2 r=0·57, P=0·054; IFN-γ r=0·84, P=0·0003), respectively. Non-HCV-related disease controls did not exhibit expansions of cognate T-cell subsets after stimulation with the HCV antigens. Control experiments showed that the cytokine-producing memory T cells expressed HLA-DR and were of CD4 phenotype (data not shown).
Discussion
Acute or chronic infection can affect the composition of the T cells in the peripheral blood.20–22 In our study the fractions of unstimulated TCR BV2.1+, BV6.1+, BV13.6+, BV17.1+ and BV21.3+ T cells tended to be higher in the patients with chronic hepatitis C than in the healthy subjects with previous self-limited HCV infection and the non-HCV-related controls. These differences were significant only between patients with chronic hepatitis C and the subjects with previous self-limited HCV infection for the TCR BV2.1+, BV6.1+, BV13.6+, BV17.1+ and BV21.3+ T-lymphocyte subsets. Interestingly, the size of these subsets was statistically correlated to the numbers of cells with ‘spontaneous’ type 1 cytokine production in the patients with chronic hepatitis C and the non-HCV-related controls but not in the patients with previous self-limited HCV infection.
With the exception of an apparently expanded TCR BV13.6+ T-cell subset, none of these findings appear to be specifically related to chronic hepatitis C infection, especially since none of the subsets increased after HCV-specific stimulation.
In contrast, stimulation by virtually all tested HCV antigens resulted in a marked antigen-specific expansion of BV8+ T cells in the patients with chronic hepatitis C, which was not seen in patients with self-limited disease or the disease controls. Moreover, after HCV-specific stimulation the numbers of IL-10-producing cells were apparently correlated to numbers of TCR BV8+ T memory cells. HCV-specific induction of IL-10 in TCR BV8+ T cells was confirmed at the single-cell level by direct flow cytometric analyis (Fig. 3). Thus, expansion of the BV8+ T-cell subset may reflect subtle changes in the TCR repertoire, which become apparent only after re-exposure to the antigen, analogous to previous findings in experimental lymphocytic choriomeningitis virus infections in mice.23 Moreover, altered HCV-specific immune responses of selected T-cell subsets of patients with chronic hepatitis C could contribute to down-regulation of pro-inflammatory cytokines via IL-10.24,25
Although restricted TCR involvement has been reported for intrahepatic T cells,26,27 previous studies on the peripheral blood TCR repertoire have reported a normal composition with respect to TCR BV families in chronic hepatitis C.28,29 In a recent study, Kashii reported possible hepatic compartmentalization of BV5.1+ T cells both by immunohistochemistry and polymerase chain reaction (PCR) techniques. Our stimulation experiments could not disclose BV5.1+ T cells as a major HCV-responsive T-cell subset in the peripheral blood.26 Differences in the immunogenetic background of the study population and sequence variations of the infecting HCV strains may account for this discrepancy. Furthermore, differences in methodology may have also contributed to such divergent results, as a recent comparison of flow cytometry and PCR-based TCR analysis indicated that PCR does not accurately estimate the percentage of peripheral blood lymphocytes expressing a given TCR BV chain.30
A key-finding of our study was the marked in vitro expansion of BV8+ T cells in the response to the HCV antigens. In preceding experiments the chosen conditions of tissue culture and stimulation with mitogens did not alter the TCR BV repertoire (ref. 31 and authors' unpublished observations). Moreover, expansion of BV8+ T cells was seen exclusively in patients with chronic hepatitis C. Thus, simple skewing of the TCR repertoire by tissue culture and the stimulation process itself can be excluded.
A bias due to HCV-associated autoimmunity can be excluded, since none of our patients had autoantibodies or cryoglobulinemia. It is also unlikely that the expansion of BV8+ T cells after stimulation with HCV antigens reflects the effects of a superantigen. Superantigens activate large fractions (5–20%) of the T-cell population via direct interaction with the BV domain of the TCR outside the antigen-binding groove.32–34 They either lead to in vivo expansion or disappearance of the associated TCR BV subset which becomes hyporesponsive to subsequent in vitro stimulation. This is in contrast to the in vitro expansion of the BV8+ subset that we observed after HCV-specific stimulation.
Antigen-specific stimulation may have unmasked circulating HCV-specific T cells trafficking to the site of inflammation. Failure to observe spontaneous expansion of BV8+ T cells in the peripheral blood can be reconciled with this hypothesis, because the majority of BV8+ T cells would not be HCV-specific and the in vivo expansion of some clones might be insufficient to detect for the family as a whole. Antigen-specific stimulation will expand such clones in vitro to a level that becomes detectable by flow cytometry. Interestingly, the BV8+ T-cell subset was the predominant intrahepatic T-cell population in a patient with chronic hepatitis C reported by Pham et al.35 Thus, antigen-specific stimulation may have unmasked HCV-specific T cells in our study, which would otherwise have been missed in the peripheral blood.28,29
We used synthetic proteins carrying several T-cell epitopes restricted by different HLA alleles. Thus, it was not unexpected that many of the HCV proteins induced expansion of BV8+ T cells apparently independently from MHC restriction. In addition, promiscuous binding to different MHC alleles, which has been described repeatedly for major histocompatibility complex class I and II restricted epitopes,36–39 may account for a broad recognition of T-cell epitopes specifically in hepatitis C.40–42 Finally, we cannot completely exclude the possibility that expansion of BV8+ T cells reflects stimulation of T cells with natural killer-like properties.
Our choice of TCR BV-specific antibodies allows only a selective view on few TCR BV subsets. Nonetheless, it provides evidence of differential TCR BV family involvement in the immune response to hepatitis C. Whether additional TCR BV families respond to HCV antigens in a similar fashion to BV8+ T cells awaits further studies.
Acknowledgments
This work was supported by grants from the Deutsche Forschungs Gemeinschaft (grant Spe483 1-1) and the Faculty of Medicine of the University of Bonn (BONFOR 107/27).
Abbreviations
- BV
variable beta
- HCV
hepatitis C virus
- TCR
T-cell receptor
References
- 1.Davis MM, Bjorkman PJ. T-cell antigen receptor genes and T-cell recognition. Nature. 1988;334:395–402. doi: 10.1038/334395a0. [DOI] [PubMed] [Google Scholar]
- 2.Gorochov G, Neumann AU, Kereveur A, et al. Perturbation of CD4+ and CD8+ T-cell repertoires during progression to AIDS and regulation of the CD4+ repertoire during antiviral therapy. Nat Med. 1998;4:215–21. doi: 10.1038/nm0298-215. [DOI] [PubMed] [Google Scholar]
- 3.Gray D. Immunological memory. Annu Rev Immunol. 1993;11:49–77. doi: 10.1146/annurev.iy.11.040193.000405. [DOI] [PubMed] [Google Scholar]
- 4.Zinkernagel RM. The role of antigen in maintaining T cell memory. Dev Biol Stand. 1994;82:189–91. [PubMed] [Google Scholar]
- 5.Beverly PC. Functional analysis of human T cell subsets defined by CD45 isoform expression. Semin Immunol. 1992;4:35–41. [PubMed] [Google Scholar]
- 6.Sprent J, Tough DF. Lymphocyte life-span and memory. Science. 1994;265:1395–400. doi: 10.1126/science.8073282. [DOI] [PubMed] [Google Scholar]
- 7.Akolkar PN, Gulwani-Akolkar B, Pergolizzi R, Bigler RD, Silver J. Influence of HLA genes on T cell receptor V segment frequencies and expression levels in peripheral blood lymphocytes. J Immunol. 1993;150:2761–73. [PubMed] [Google Scholar]
- 8.Malhotra U, Spielman R, Concannon P. Variability in T cell receptor V beta gene usage in human peripheral blood lymphocytes. Studies of identical twins, siblings, and insulin-dependent diabetes mellitus patients. J Immunol. 1992;149:1802–8. [PubMed] [Google Scholar]
- 9.Sad S, Mosmann TR. Single IL-2-secreting precursor CD4 T cell can develop into either Th1 or Th2 cytokine secretion phenotype. J Immunol. 1994;153:3514–22. [PubMed] [Google Scholar]
- 10.Imberti L, Cariani E, Bettinardi A, Zonaro A, Albertini A, Primi D. An immunoassay for specific amplified HCV sequences. J Virol Methods. 1991;34:233–43. doi: 10.1016/0166-0934(91)90103-7. [DOI] [PubMed] [Google Scholar]
- 11.Simmonds P, Rose KA, Graham S, et al. Mapping of serotype-specific, immunodominant epitopes in the NS-4 region of hepatitis C virus (HCV): use of type-specific peptides to serologically differentiate infections with HCV types 1, 2, and 3. J Clin Microbiol. 1993;31:1493–503. doi: 10.1128/jcm.31.6.1493-1503.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choo QL, Kuo G, Weiner AJ, Overby LR, Bradley DW, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–62. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 13.Hoffmann RM, Diepolder HM, Zachoval R, et al. Mapping of immunodominant CD4+ T lymphocyte epitopes of hepatitis C virus antigens and their relevance during the course of chronic infection. Hepatology. 1995;21:632–8. [PubMed] [Google Scholar]
- 14.Jung T, Schauer U, HeusSeries C, Neumann C, Rieger C. Detection of intracellular cytokines by flow cytometry. J Immunol Methods. 1993;159:197–207. doi: 10.1016/0022-1759(93)90158-4. [DOI] [PubMed] [Google Scholar]
- 15.North ME, Ivory K, Funauchi M, Webster AD, Lane AC, Farrant J. Intracellular cytokine production by human CD4+ and CD8+ T cells from normal and immunodeficient donors using directly conjugated anti-cytokine antibodies and three-colour flow cytometry. Clin Exp Immunol. 1996;105:517–22. doi: 10.1046/j.1365-2249.1996.d01-795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Picker LJ, Singh MK, Zdraveski Z, Treer JR, Waldrop SL, Bergstresser PR, Maino VC. Direct demonstration of cytokine synthesis heterogeneity among human memory/effector T cells by flow cytometry. Blood. 1995;86:1408–19. [PubMed] [Google Scholar]
- 17.Prussin C, Metcalfe DD. Detection of intracytoplasmic cytokine using flow cytometry and directly conjugated anti-cytokine antibodies. J Immunol Methods. 1995;188:117–28. doi: 10.1016/0022-1759(95)00209-x. [DOI] [PubMed] [Google Scholar]
- 18.Tartakoff AM. Perturbation of the structure and function of the Golgi complex by monovalent carboxylic ionophores. Meth Enzymol. 1983;98:47–59. doi: 10.1016/0076-6879(83)98138-7. [DOI] [PubMed] [Google Scholar]
- 19.Sander B, Andersson J, Andersson U. Assessment of cytokines by immunofluorescence and the paraformaldehyde-saponin procedure. Immunol Rev. 1991;119:65–93. doi: 10.1111/j.1600-065x.1991.tb00578.x. [DOI] [PubMed] [Google Scholar]
- 20.Abbott WG, Geursen A, Fraser JD, Marbrook J, Skinner MA, Tan PL. The influence of a maternal chronic hepatitis B virus infection on the repertoire of transcribed T-cell receptor beta chain variable region genes in human cord blood. Hepatology. 1995;22:1034–9. doi: 10.1016/0270-9139(95)90606-1. [DOI] [PubMed] [Google Scholar]
- 21.Dou HY, Wu JC, Peng WL, Chang C, Chi WK, Chu YD, Hu CP. Analysis of T cell receptor Vbeta gene usage during the course of disease in patients with chronic hepatitis B. J Biomed Sci. 1998;5:428–34. doi: 10.1007/BF02255931. [DOI] [PubMed] [Google Scholar]
- 22.Chen X, Cooksley G, Sing G. Distinct patterns of T cell receptor distribution of peripheral blood CD8+ cells during different stages of chronic infection with hepatitis B virus. Hum Immunol. 1998;59:199–211. doi: 10.1016/s0198-8859(98)00007-x. [DOI] [PubMed] [Google Scholar]
- 23.Selin LK, Lin MY, Kraemer KA, et al. Attrition of T cell memory: selective loss of LCMV epitope-specific memory CD8 T cells following infections with heterologous viruses. Immunity. 1999;11:733–42. doi: 10.1016/s1074-7613(00)80147-8. [DOI] [PubMed] [Google Scholar]
- 24.Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–93. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 25.Mosmann TR, Sad S. The expanding universe of T-cell subsets. Th1, Th2 and more. Immunol Today. 1996;17:138–46. doi: 10.1016/0167-5699(96)80606-2. [DOI] [PubMed] [Google Scholar]
- 26.Kashii Y, Shimizu Y, Nambu S, Minemura M, Okada K, Higuchi K, Watanabe A. Analysis of T-cell receptor Vbeta repertoire in liver-infiltrating lymphocytes in chronic hepatitis C. J Hepatol. 1997;26:462–70. doi: 10.1016/s0168-8278(97)80408-4. 10.1016/S0168-8278(97)80408-4. [DOI] [PubMed] [Google Scholar]
- 27.Nuti S, Rosa D, Valiante NM, et al. Dynamics of intra-hepatic lymphocytes in chronic hepatitis C. Enrichment for Valpha24 + T cells and rapid elimination of effector cells by apoptosis. Eur J Immunol. 1998;28:3448–55. doi: 10.1002/(SICI)1521-4141(199811)28:11<3448::AID-IMMU3448>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 28.Wang CC, Hwang LH, Yang PM, Chiang BL, Chen PJ, Chen DS. Unbiased usage of T-cell receptor beta variable region genes in peripheral blood cells of hepatitis C patients: no correlation with superantigen effect. J Med Virol. 1995;45:24–8. doi: 10.1002/jmv.1890450105. [DOI] [PubMed] [Google Scholar]
- 29.Cacoub P, Musset L, Hausfater P, et al. No evidence for abnormal immune activation in peripheral blood T cells in patients with hepatitis C virus (HCV) infection with or without cryoglobulinaemia. Multivirc Group. Clin Exp Immunol. 1998;113:48–54. doi: 10.1046/j.1365-2249.1998.00635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diu A, Romagne F, Genevee C, et al. Fine specificity of monoclonal antibodies directed at human T cell receptor variable regions: comparison with oligonucleotide-driven amplification for evaluation of V beta expression. Eur J Immunol. 1993;23:1422–9. doi: 10.1002/eji.1830230703. [DOI] [PubMed] [Google Scholar]
- 31.Arenz M, Herzog-Hauff S, Meyer zum Buschenfelde KH, Lohr HF. Antigen-independent in vitro expansion of T cells does not affect the T cell receptor V beta repertoire. J Mol Med. 1997;75:678–86. doi: 10.1007/s001090050152. [DOI] [PubMed] [Google Scholar]
- 32.Coffin JM. Superantigens and endogenous retroviruses: a confluence of puzzles. Science. 1992;255:411–13. doi: 10.1126/science.1310360. [DOI] [PubMed] [Google Scholar]
- 33.Herman A, Kappler JW, Marrack P, Pullen AM. Superantigens: mechanism of T-cell stimulation and role in immune responses. Annu Rev Immunol. 1991;9:745–72. doi: 10.1146/annurev.iy.09.040191.003525. [DOI] [PubMed] [Google Scholar]
- 34.Laurence J, Hodtsev AS, Posnett DN. Superantigen implicated in dependence of HIV-1 replication in T cells on TCR V beta expression. Nature. 1992;358:255–9. doi: 10.1038/358255a0. [DOI] [PubMed] [Google Scholar]
- 35.Pham BN, Degos F, Mosnier JF, Ollivier S, Sauvanet A, Erlinger S, Cohen JH. Restriction of V beta gene usage of liver-derived lymphocytes in chronic hepatitis B and C. Hum Immunol. 1996;49:56–63. doi: 10.1016/0198-8859(96)00053-5. [DOI] [PubMed] [Google Scholar]
- 36.Chen BP, Rothbard J, Parham P. Apparent lack of MHC restriction in binding of class I HLA molecules to solid-phase peptides. J Exp Med. 1990;172:931–6. doi: 10.1084/jem.172.3.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chicz RM, Urban RG, Gorga JC, Vignali DA, Lane WS, Strominger JL. Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J Exp Med. 1993;178:27–47. doi: 10.1084/jem.178.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Busch R, Strang G, Howland K, Rothbard JB. Degenerate binding of immunogenic peptides to HLA-DR proteins on B cell surfaces. Int Immunol. 1990;2:443–51. doi: 10.1093/intimm/2.5.443. [DOI] [PubMed] [Google Scholar]
- 39.Panina-Bordignon P, Tan A, Termijtelen A, Demotz S, Corradin G, Lanzavecchia A. Universally immunogenic T cell epitopes: promiscuous binding to human MHC class II and promiscuous recognition by T cells. Eur J Immunol. 1989;19:2237–42. doi: 10.1002/eji.1830191209. [DOI] [PubMed] [Google Scholar]
- 40.Erickson AL, Kimura Y, Igarashi S, et al. The outcome of hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity. 2001;15:883–95. doi: 10.1016/s1074-7613(01)00245-x. [DOI] [PubMed] [Google Scholar]
- 41.Langhans B, Lechmann M, Ihlenfeldt H, et al. A hepatitis C virus (HCV) core protein derived peptide inhibits HCV specific lymphocyte proliferation. Eur J Med Res. 2000;5:115–20. [PubMed] [Google Scholar]
- 42.Schweitzer S, Schneiders AM, Langhans B, et al. Flow cytometric analysis of peptide binding to major histocompatibility complex class I for hepatitis C virus core T-cell epitopes. Cytometry. 2000;41:271–8. doi: 10.1002/1097-0320(20001201)41:4<271::aid-cyto5>3.0.co;2-m. 10.1002/1097-0320(20001201)41:4<271::AID-CYTO53.0.CO;2-M. [DOI] [PubMed] [Google Scholar]