Abstract
DNA vaccines induce immune responses against encoded proteins, and have clear potential for cancer vaccines. For B-cell tumours, idiotypic (Id) immunoglobulin encoded by the variable region genes provides a target antigen. When assembled as single chain Fv (scFv), and fused to an immunoenhancing sequence from tetanus toxin (TT), DNA fusion vaccines induce anti-Id antibodies. In lymphoma models, these antibodies have a critical role in mediating protection. For application to patients with lymphoma, two questions arise: first, whether pre-existing antibody against TT affects induction of anti-scFv antibodies; second, whether individual human scFv fusion sequences are able to fold consistently to generate antibodies able to recognize private conformational Id determinants expressed by tumour cells. Using xenogeneic vaccination with scFv sequences from four patients, we have shown that pre-existing anti-TT immunity slows, but does not prevent, anti-Id antibody responses. To determine folding, we have monitored the ability of nine DNAscFv–FrC patients' vaccines to induce xenogeneic anti-Id antibodies. Antibodies were induced in all cases, and were strikingly specific for each patient's immunoglobulin with little cross-reactivity between patients, even when similar VH or VL genes were involved. Blocking experiments with human serum confirmed reactivity against private determinants in 26–97% of total antibody. Both immunoglobulin G1 (IgG1) and IgG2a subclasses were present at 1·3 : 1–15 : 1 consistent with a T helper 2-dominated response. Xenogeneic vaccination provides a simple route for testing individual patients' DNAscFv–FrC fusion vaccines, and offers a strategy for production of anti-Id antibodies. The findings underpin the approach of DNA idiotypic fusion vaccination for patients with B-cell tumours.
Introduction
Low grade follicular lymphoma (FL) is susceptible to chemotherapy, but tends to relapse, leading to the poor impact of treatment on overall survival.1 The attraction of intervening with immunotherapeutic strategies during remission in order to attack residual tumour cells is obvious. Initial attempts focused on antibody therapy, using the idiotype (Id) determinants of the surface immunoglobulin of the tumour cell as a target.2 Both polyclonal and monoclonal anti-Id showed significant effects,3,4 and these were confirmed in a small clinical trial.5 However, the logistical difficulties of preparing customized anti-Id antibodies have hampered the wide application of this promising approach.
An alternative approach is to use active vaccination with Id antigen. While this relies on the patient's ability to respond, it should lead to a continuing polyclonal attack on all the available target epitopes. Anti-Id antibody appears to directly induce apoptosis via the B-cell receptor (BCR),6 and cellular responses may mediate additional effector pathways.7 Vaccination with Id protein showed remarkable protective effects in preclinical models,8 and is being used with some success to vaccinate patients in remission from FL.9,10 However, individual Id proteins are still difficult to prepare, and logistical arguments remain.
DNA vaccines appear to offer a solution, since the variable region genes, VH and VL, which encode the tumour Id determinants, can be readily isolated.11 The operation of DNA vaccines depends on expression of the encoded protein in the injected cells, and presentation of the protein to the immune system.12 This is assisted by the presence of CpG dinucleotide repeats in the backbone of bacterial DNA, which are recognized by the innate immune system.13 Currently it is unclear as to whether antigen transfer from the transfected muscle or skin cells, or antigen expressed by directly transfected antigen-presenting cells is critical for induction of immunity, and possibly both are involved.14 However, the outcome is activation of all arms of the immune response,15 and DNA vaccines have clear potential both for protection against infectious diseases and for cancer vaccines.12
For Id determinants, genes encoding a whole immunoglobulin molecule, or the variable regions alone, can be used.16,17 Assembly as single chain Fv avoids the complexity of inserting two chains into the vector, but folding of the molecule to display the Id determinants present in the natural tumour immunoglobulin is less predictable. A further complexity is that Id determinants alone are weakly immunogenic, and that promotion of immunogenicity requires expression of scFv as a fusion protein with an immunoenhancing molecule.16,18,19 We chose to attach a gene encoding fragment C of TT for this purpose.19 This design induces protective anti-Id antibodies against mouse lymphomas, indicating that folding of scFv is not hampered in the fusion protein.17
However, two questions relevant to application to a large cohort of patients remained: first the effect of pre-existing immunity against TT on anti-Id responses against a range of human scFv fusion vaccines; second, whether the conformational integrity of different human scFv fusion proteins was maintained. We have investigated both these points in the present report.
Materials and methods
Patients' tumour and serum samples
To analyse the quality of folding of scFv, it was necessary to have available the natural immunoglobulin protein. We therefore chose nine cases of myeloma where tumour-derived immunoglobulin protein was present in serum. Anti-coagulated bone marrow aspirates and serum were obtained, after consent, from nine patients with diagnosis of multiple myeloma referred to the Division of Haematology, Nottingham, UK. In eight cases, serum monoclonal immunoglobulin G (IgG) was obtained by affinity chromatography using HiTrap Protein G HP 1 ml columns (Amersham Pharmacia Biotech, Uppsala, Sweden), following the manufacturers' instructions. Serum monoclonal IgA was obtained from one case by ammonium sulphate precipitation followed by ion exchange chromatography.
Identification of tumour VH and VL genes
Preparation of cDNA from bone marrow mononuclear cells and identification of tumour VH transcripts was as described.20 Similarly to VH, tumour Vκ and Vλ light chain transcripts were identified by polymerase chain reaction (PCR) using a mixture of 5′ primers specific for each of the Vκ or Vλ leader sequences (Vκ- or Vλ-leader mix) together with a 3′ primer specific for either Cκ or Cλ region, according to the tumour isotype. PCR conditions, separation of PCR products for cloning, and identification and analysis of tumour V genes were as previously reported.20,21 The tumour VH and VL sequences were confirmed from at least a second independent PCR with identical conditions.
Assembly and characterization of DNA scFv–FrC vaccines
Tumour-derived V genes were assembled as scFv by a two-step PCR procedure as described,11 using specific primers incorporating SfiI and NotI sites, and Pfu DNA polymerase for amplification (Promega, Madison, WI). The DNA vaccine was prepared by cloning scFv into pVAC2 plasmid. pVAC2 is a mammalian expression vector derived from pcDNA3 (Invitrogen, Paisley, UK), designed to allow direct cloning of scFv into the 5′-SfiI and 3′-NotI sites. The SfiI site is located in the VH1 family leader sequence 3′ to the pCMV promoter. The 3′-NotI site is located in a seven amino acid linker sequence (Ala-Ala-Ala-Gly-Pro-Gly-Pro) fused directly to the downstream sequence for fragment C (FrC) of TT. This design enables expression of an in-frame VH1 leader–scFv–FrC fusion protein. Cloned scFv ligated in pVAC2 was used to transform XL1-blue bacterial competent cells (Stratagene, Amsterdam, The Netherlands). Clones positive for inserts were sequenced to verify the scFv. For large scale purification of vaccine plasmids the QIAGEN plasmid Giga kit (Qiagen, Crawley, UK) was used. Prior to use, vaccines were precipitated, washed and diluted in sterile saline solution at 0·5 mg/ml. DNA scFv-FrC vaccines were checked for expression and size by the in vitro TNT Quick Coupled Transcription/Translation System kit (Promega). The secretion of human scFv–FrC fusion protein was assessed by transfection of COS-7 cells with DNA using FuGENE 6 Transfection reagent (Roche, Lewes, UK), following the manufacturer's instructions. A vaccine containing the gene encoding the full length sequence of FrC with a leader sequence (p.FrC) was used as a positive control.19 Supernatants were collected 72 hr after transfection and the concentration of scFv–FrC fusion protein in the supernatant was estimated by enzyme-linked immunosorbent assay (ELISA), as described.17
Vaccination protocol and measurement of antibody responses
Groups of six to nine C57BL/6 mice, bred in house, were vaccinated at 6–10 weeks of age with 50 µg DNA vaccine in normal saline injected into two sites in the quadriceps muscles. Injections were at days 0, 21 and 42 and tail bleeds were taken at day 21 prior to the second immunization, with a final bleed at day 63 to measure antibody responses. IgG antibody levels against fragment C were measured by ELISA, as described.19 Anti-Id IgG antibodies against patients' autologous or heterologous tumour-derived Igs were measured by ELISA, as reported,17 using patients' idiotypic IgG or IgA (1 µg/ml) for coating and a sheep anti-mouse IgG for detection (The Binding Site, Birmingham, UK). In all cases, antibody levels in individual mice were measured and the mean values±SEM calculated. Prevaccination of mice with TT was performed by intramuscular injection of 8 IU of TT adsorbed onto aluminium hydroxide (BP Pasteur Merieux, Lyon, France) in 100 µl saline, at day −42. Aluminium hydroxide (Sigma, Poole, UK) only was administered in the control groups of mice. At day 0, mice were bled to measure anti-FrC antibody titres and the DNA vaccination procedure was started.
Assessment of specificity of anti-Id antibodies
Cross-reactivity of anti-Id antibodies with unrelated Id immunoglobulins was measured using serum pooled at day 63, assigned in each case to an arbitrary value of 10 000 units/ml to act as a comparative standard. To assess the proportion of antibodies directed against private Id determinants, pooled antisera from mice immunized with DNAscFv–FrC vaccines, collected at day 63, were incubated in 50% normal human serum for 1 hr. This procedure blocks reactivity with public Id determinants.22 Blocked or control antisera were then tested for reactivity with patients' Id immunoglobulin by ELISA. The percentage of anti-Id antibodies against the private determinants was calculated as the ratio (reactivity of blocked antiserum/reactivity of unblocked antiserum)×100. Reproducibility of the assays was tested on four selected sera, and values for inhibition varied by ≤5%.
Results
Identification and assembly of tumour-derived V genes
The VH and VL genes used by the tumour cells were identified in each case as repeated identical VH–D–JH and VL–JL sequences obtained after cloning of PCR products (Table 1). All sequences showed extensive somatic mutation, typical of follicular and post-follicular malignancies. To ensure relevance for follicular lymphoma, we included four cases (GH, JC, PC and FA) which contained potential glycosylation motifs in VH. These sites are a feature of FL but are found in only 8% of cases of myeloma.23 Following assembly of the scFv–FrC fusion vaccines, an in vitro transcription/translation system was used to show that all constructs expressed protein of the expected size (∼80 000 MW) (data not shown). Transfection of COS-7 cells and assay of FrC in the supernatants confirmed secretion of the fusion protein for each construct.17
Table 1. Analysis of tumour derived V genes*.
| VH | VL | ||||||
|---|---|---|---|---|---|---|---|
| Patient | Immunoglobulin | V-gene | % homology* | JH | V-gene | % homology* | JL |
| I GH | IgGλ | V4–59 | 90·9 | JH4b | VλIII (IGLV3S6) | 89·6 | Jλ2 |
| II JC | IgAκ | V3–23 | 93·8 | JH4b | VκIII (A27) | 95·1 | Jκ2 |
| III GS | IgGκ | V3–7 | 88·8 | JH4b | VκI (012/02) | 88·9 | Jκ5 |
| IV JW | IgGκ | V2–70 | 96·6 | JH4b | VκIII (38 k/L6) | 97 | Jκ2 |
| V MW | IgGκ | V5–51 | 95·5 | JH4b | VκI (018/08) | 95·8 | Jκ2 |
| VI PR | IgGλ | V2(S12–4) | 93·3 | JH3b | VλI (1c/DPL2) | 94·7 | Jλ7 |
| VII WD | IgGκ | V2(S12–4) | 92·5 | JH5b | VκI (02/DPK9) | 90·9 | Jκ1 |
| VIII FA | IgGκ | V2(S12–7) | 86·6 | JH4b | VκIII (38 k/L6) | 95·4 | Jκ4 |
| IX PC | IgGκ | V1–3 | 88·4 | JH6b | VκI (02/DPK9) | 95·8 | Jκ3 |
Nucleotide sequences have been deposited in the GenBank/EMBL database: accession numbers are AF442750–67.
Effect of pre-existing immunity against TT on anti-Id antibody responses
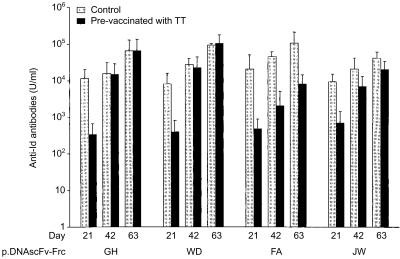
Vaccination of mice with TT induced 2000–4000 IU/ml of anti-FrC antibody as measured 6 weeks later. At this point (day 0), vaccination with the DNAscFv–FrC constructs from four patients was begun, and levels of antibody reacting with the patients' Id immunoglobulin were measured at days 21, 42 and 63. Anti-Id antibodies were detectable against four of four individual vaccines. However, in all cases, anti-Id titres were reduced at day 21 in mice prevaccinated with TT, as compared to those vaccinated with the alum control (Fig. 1). Following a further injection of DNA vaccine on day 21, there was a rapid rise in anti-Id levels in the TT-immune mice, so that by day 42 differences between the two groups were diminishing. This trend continued after the second boost (Fig. 1), and indicates that pre-existing anti-TT antibodies consistently slow the response to the Id antigen in the fusion gene, but that this can be largely overcome with further injections.
Figure 1.
Effect of prevaccination with TT on induction of anti-Id antibodies by patients' DNAscFv–FrC vaccines. Groups of six mice were prevaccinated with TT/alum (black columns) or alum (grey columns) at day −42. Mice were injected patient's DNAscFv–FrC vaccine at day 0, 21 and 42 and bled at days 21, 42 and 63 to measure anti-Id antibody levels. Bars indicate SEM.
Analysis of the anti-Id responses induced by DNA scFv–FrC fusion vaccines
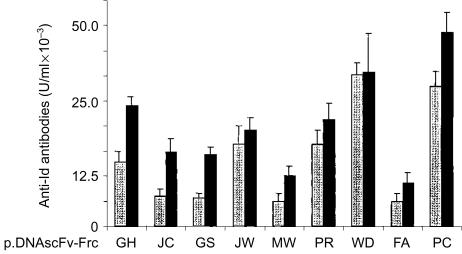
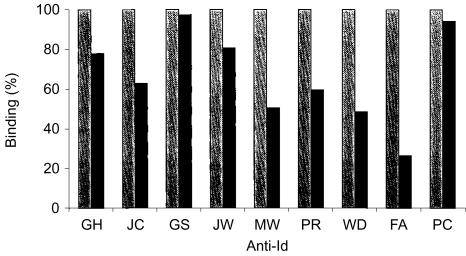
Induction of xenogeneic anti-Id antibodies able to recognize the tumour immunoglobulin derived from the corresponding patient's serum was achieved for 9/9 DNA scFv–FrC fusion vaccines (Fig. 2). Reactivity was clearly detectable after a single injection (day 21), but generally increased following further injections. Specificity of the antibodies was assessed by testing reactivity with Id immunoglobulin from other patients. Cross-reactivity between the patients' immunoglobulins was remarkably low (Table 2), even when the patient's tumour immunoglobulin was encoded by the same VH gene (PR/WD), or the same VL gene (WD/PC and JW/FA). A more stringent test of cross-reactivity was provided by adding an equal volume of normal human serum to the pre-plateau serum dilution used in the assay. This will block antibodies against determinants present in the repertoire of normal immunoglobulin, especially those commonly expressed such as V3–23 (JC), V5–51 (MW), V4–59 (GH) and V3–7 (GS).24 The results (Fig. 3) show that all cases had a residual activity considered to be directed against private Id determinants expressed on the Id immunoglobulin. It appears therefore that the anti-Id antibodies recognize determinants arising from VH–VL combination, CDR3 sequences or somatic mutations, with little tendency to focus on the framework regions of the V genes. The finding that the anti-Id response induced by DNA scFv–FrC is generally focused on private Id determinants expressed by the tumour-derived immunoglobulin indicates that folding of the scFv mimics that of the natural Id immunoglobulin.
Figure 2.
Anti-Id antibodies induced by DNA scFv-FrC vaccines. Each pair of columns represents mean antibody levels induced by individual DNA human scFv-FrC vaccines. For each pair, the grey column indicates levels at day 21, black column at day 63. Anti-Id levels were measured in individual mice. Mean values are represented in arbitrary units to compare antibody levels at day 21 and 63. Bars indicate SEM.
Table 2. Evaluation of specificity of anti-Id antibodies for patients' Id immunoglobulins.
| Immune serum | Paraprotein | GH | JC | GS | JW | MW | PR | WD | FA | PC | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| V4–59 | JH4b | ||||||||||
| GH | 10000 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| Vλ3 (IGLV3S6) | Jλ2 | ||||||||||
| V3–23 | JH4b | ||||||||||
| JC | 0 | 10000 | 0 | 0 | 0 | 0 | 310 | 310 | 250 | ||
| VK3 (A27) | JK2 | ||||||||||
| V3–7 | JH4b | ||||||||||
| GS | 0 | 0 | 10000 | 0 | 0 | 0 | 0 | 0 | 0 | ||
| V K1 (012/2) | JK5 | ||||||||||
| V2–70 | JH4b | ||||||||||
| JW | 0 | 0 | 0 | 10000 | 0 | 0 | 0 | 0 | 0 | ||
| VK3 (38k/L6) | JK2 | ||||||||||
| V5–51 | JH4b | ||||||||||
| MW | 0 | 0 | 0 | 0 | 10000 | 0 | 0 | 0 | 0 | ||
| VK1 (018/08) | JK2 | ||||||||||
| V2 (S12–4) | JH3b | ||||||||||
| PR | NT | NT | NT | NT | NT | 10000 | 0 | 467 | 0 | ||
| Vλ1 (1c/DPL2) | Jλ7 | ||||||||||
| V2 (S12–4) | JH5b | ||||||||||
| WD | NT | NT | NT | NT | NT | 0 | 10000 | 113 | 0 | ||
| VK1 (02/DPK9) | JK1 | ||||||||||
| V2 (S12–7) | JH4b | ||||||||||
| FA | NT | NT | NT | NT | NT | 0 | 0 | 10000 | 0 | ||
| VK3 (38k/L6) | JK4 | ||||||||||
| V1–3 | JH6b | ||||||||||
| PC | NT | NT | NT | NT | NT | 0 | 272 | 108 | 10000 | ||
| VK1 (02/DPK9) | JK3 |
NT, serum not tested against tumour immunoglobulin.
Figure 3.
Assessment of the specificity of anti-Id responses for private Id determinants. Binding to each patient's Id immunoglobulin (left column) represents reactivity with determinants on the whole variable region (arbitrarily 100%). The presence of 50% normal human serum blocks reactivity with public determinants, leaving only antibodies against private Id determinants (right column). In each case, there was evidence for reactivity against private Id expressed by natural Id immunoglobulin, demonstrating optimal folding of the scFv expressed in vivo.
IgG subclasses of antibodies induced by DNA scFv–FrC vaccines
In order to assess the nature of the antibody response induced by the DNAscFv–FrC vaccines, the IgG1 and IgG2a components were compared. The ratios of IgG1 : IgG2a for anti-Id varied from 1·3 : 1 to 15 : 1 (median 5·6 : 1). In all cases, both IgG subclasses were detected, with a consistent dominance of IgG1. These findings point to a mixed T helper 1 (Th1)/Th2 response with a possible excess of Th2. In mice prevaccinated with TT, the suppression of anti-Id antibody was particularly evident in the IgG2a subclass, with three out of four of the patients' vaccines (GH, FA and JW) failing to induce detectable IgG2a antibody at day 21. However, recovery of IgG2a production was evident after further injections (data not shown).
Discussion
The Id determinants of B-cell tumours present ideal targets for immune attack. Private determinants are unique to the neoplastic B cell, and induction of specific immunity against them carries no autoimmune potential. Because surface immunoglobulin-negative lymphoma is rare, the BCR may also be associated with survival of tumour cells, as it is for normal B cells.25 Our recent finding that the majority of the variable regions of tumour cells of FL have acquired potential glycosylation sites suggests that there may be an interaction with stromal cells of the germinal centre.23 If this is required for maintenance of the tumour cell, attacking the surface immunoglobulin would target an essential molecule. Vaccination has the advantage over passive monoclonal anti-Id antibody therapy in that a polyclonal response against all available Id determinants should be induced,26 so that the only escape route of the target cell is to delete surface immunoglobulin.27
Preclinical models of Id vaccines have revealed the importance of anti-Id antibodies in attacking tumour cells, and it is clear that they can signal via the BCR and induce apoptosis.6 Induction of anti-Id antibodies will require delivery of Id protein with conformational integrity. The role of T cells in anti-Id immunity against lymphoma is less defined, but folded protein may also be an advantage in induction of T cell responses via DNA delivery.28,29 For clinical application, our goal therefore is to use DNA vaccines to activate anti-Id immunity by delivery of folded individual patients' scFv proteins. The requirement to include an immunoenhancing sequence has been met by fusing a gene from FrC to the scFv.19
The phenomenon of epitope suppression, where mice primed with a carrier protein showed reduced anti-hapten antibody responses to a subsequent injection of hapten–carrier conjugate, is well known.30 It also appears to occur when the carrier protein is TT.31 There is therefore the potential problem for DNAscFv–FrC fusion vaccination of encountering pre-existing antibody against FrC in patients immune to TT. We initially investigated this in a mouse myeloma model.17 In preimmune mice, only a minor diminution in anti-Id antibody level was noted at day 63, but the effect on the kinetics of response was not assessed. The current analysis of responses to four different DNA human scFv vaccines reveals a consistent suppression of anti-Id antibody following the primary injection. However, in all cases, a second injection led to an accelerated response, so that by day 63 the prevaccinated group and the controls were approaching equivalence, confirming the findings in the mouse myeloma model.
The next question concerned the conformational integrity of the scFv–FrC fusion protein. It is clearly possible for scFv to fold to recreate the specificity of the parental antibody,32 although optimization of folding and stability in different expression systems has often been necessary.33 We were concerned that the variety of scFv sequences derived from B-cell malignancies would introduce uncertainty about the quality of the individual vaccines. The leader sequence would allow secretion of the scFv fusion protein, but the relative contribution of secreted protein or protein transferred directly from muscle cell to antigen-presenting cells is unknown.29 Remarkably, this design appears capable of inducing anti-Id antibodies focused largely on the private Id determinants of the corresponding natural human tumour-derived immunoglobulin. This mirrors observations in a syngeneic model using synthetic mouse scFvs with different VH and VL combinations.34
The profile of IgG subclasses differs from that induced by vaccination with exogenous Id immunoglobulin/complete Freund's adjuvant where the response is almost entirely IgG1.17 Involvement of Th1 CD4+ T cells following DNA delivery is expected, and is at least partly a result of the presence of CpG dinucleotide repeats in the backbone, which lead to production of interleukin-2 and interferon-γ.35
Clearly, DNAscFv–FrC vaccines are capable of activating antibody responses influenced by both Th1 and Th2 cells, with a predominance of Th2. In mice prevaccinated against TT, the apparent selective suppression of IgG2a at day 21 mirrors previous findings in a hapten–carrier system.36 It supports the suggestion that interferon-γ-producing Th1 cells are particularly affected in this setting.
In summary, DNAscFv–FrC vaccines can operate in the context of pre-existing immunity against TT, and the fusion format apparently allows folding of the scFv component to display Id determinants comparable with those in the natural immunoglobulin. For lymphoma, this opens the way to routine preparation and testing of DNA fusion vaccines for clinical trial. It provides the confidence that this design can induce anti-Id antibodies known to be powerful mediators of attack on surface immunoglobulin of neoplastic B cells.
Acknowledgments
F. F. and C. A. K. contributed equally to this study. The authors would like to thank the Multiple Myeloma Research Foundation (USA), Associazione Italiana contro le Leucemie (Italy), Tenovus (UK), the Leukaemia Research Fund (UK) and the Cancer Research Campaign (UK) for support.
References
- 1.Horning SJ, Negrin RS, Hoppe RT, Rosenberg SA, Chao NJ, Long GD, Brown BW, Blume KG. High-dose therapy and autologous bone marrow transplantation for follicular lymphoma in first complete or partial remission: results of a phase II clinical trial. Blood. 2001;97:404–9. doi: 10.1182/blood.v97.2.404. [DOI] [PubMed] [Google Scholar]
- 2.Stevenson GT, Stevenson FK. Antibody to a molecularly-defined antigen confined to a tumour cell surface. Nature. 1975;254:714–6. doi: 10.1038/254714a0. [DOI] [PubMed] [Google Scholar]
- 3.Hamblin TJ, Abdul-Ahad AK, Gordon J, Stevenson FK, Stevenson GT. Preliminary experience in treating lymphocytic leukaemia with antibody to immunoglobulin idiotypes on the cell surfaces. Br J Cancer. 1980;42:495–502. doi: 10.1038/bjc.1980.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller RA, Maloney DG, Warnke R, Levy R. Treatment of B-cell lymphoma with monoclonal anti-idiotype antibody. N Engl J Med. 1982;306:517–22. doi: 10.1056/NEJM198203043060906. [DOI] [PubMed] [Google Scholar]
- 5.Meeker TC, Lowder J, Maloney DG, Miller RA, Thielemans K, Warnke R, Levy R. A clinical trial of anti-idiotype therapy for B cell malignancy. Blood. 1985;65:1349–63. [PubMed] [Google Scholar]
- 6.Yefenof E, Picker LJ, Scheuermann RH, Vitetta ES, Street NE, Tucker TF, Uhr JW. Induction of B cell tumor dormancy by anti-idiotypic antibodies. Curr Opin Immunol. 1993;5:740–4. doi: 10.1016/0952-7915(93)90130-k. [DOI] [PubMed] [Google Scholar]
- 7.Wilson A, George AJ, King CA, Stevenson FK. Recognition of a B cell lymphoma by anti-idiotypic T cells. J Immunol. 1990;145:3937–43. [PubMed] [Google Scholar]
- 8.George AJ, Stevenson FK. Prospects for the treatment of B cell tumors using idiotypic vaccination. Int Rev Immunol. 1989;4:271–310. doi: 10.3109/08830188909044783. [DOI] [PubMed] [Google Scholar]
- 9.Hsu FJ, Caspar CB, Czerwinski D, Kwak LW, Liles TM, Syrengelas A, Taidi-Laskowski B, Levy R. Tumor-specific idiotype vaccines in the treatment of patients with B-cell lymphoma – long-term results of a clinical trial. Blood. 1997;89:3129–35. [PubMed] [Google Scholar]
- 10.Bendandi M, Gocke CD, Kobrin CB, et al. Complete molecular remissions induced by patient-specific vaccination plus granulocyte–monocyte colony-stimulating factor against lymphoma. Nat Med. 1999;5:1171–7. doi: 10.1038/13928. [DOI] [PubMed] [Google Scholar]
- 11.Hawkins RE, Zhu D, Ovecka M, Winter G, Hamblin TJ, Long A, Stevenson FK. Idiotypic vaccination against human B-cell lymphoma. Rescue of variable region gene sequences from biopsy material for assembly as single-chain Fv personal vaccines. Blood. 1994;83:3279–88. [PubMed] [Google Scholar]
- 12.Gurunathan S, Klinman DM, Seder RA DNA. vaccines: immunology, application and optimization. Annu Rev Immunol. 2000;18:927–74. doi: 10.1146/annurev.immunol.18.1.927. [DOI] [PubMed] [Google Scholar]
- 13.Wagner H. Bacterial CpG DNA activates immune cells to signal infectious danger. Adv Immunol. 1999;73:329–68. doi: 10.1016/s0065-2776(08)60790-7. [DOI] [PubMed] [Google Scholar]
- 14.Corr M, von Damm A, Lee DJ, Tighe H. In vivo priming by DNA injection occurs predominantly by antigen transfer. J Immunol. 1999;163:4721–7. [PubMed] [Google Scholar]
- 15.Ulmer JB, Donnelly JJ, Parker SE, et al. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science. 1993;259:1745–9. doi: 10.1126/science.8456302. [DOI] [PubMed] [Google Scholar]
- 16.Syrengelas AD, Chen TT, Levy R. DNA immunization induces protective immunity against B-cell lymphoma. Nat Med. 1996;2:1038–41. doi: 10.1038/nm0996-1038. [DOI] [PubMed] [Google Scholar]
- 17.King CA, Spellerberg MB, Zhu D, et al. DNA vaccines with single-chain Fv fused to fragment C of tetanus toxin induce protective immunity against lymphoma and myeloma. Nat Med. 1998;4:1281–6. doi: 10.1038/3266. [DOI] [PubMed] [Google Scholar]
- 18.Biragyn A, Tani K, Grimm MC, Weeks S, Kwak LW. Genetic fusion of chemokines to a self tumor antigen induces protective, T-cell dependent antitumor immunity. Nat Biotechnol. 1999;17:253–8. doi: 10.1038/6995. [DOI] [PubMed] [Google Scholar]
- 19.Spellerberg MB, Zhu D, Thompsett A, King CA, Hamblin TJ, Stevenson FK. DNA vaccines against lymphoma: promotion of anti-idiotypic antibody responses induced by single chain Fv genes by fusion to tetanus toxin fragment C. J Immunol. 1997;159:1885–92. [PubMed] [Google Scholar]
- 20.Forconi F, Sahota SS, Raspadori D, Mockridge CI, Lauria F, Stevenson FK. Tumor cells of hairy cell leukemia express multiple clonally related immunoglobulin isotypes via RNA splicing. Blood. 2001;98:1174–81. doi: 10.1182/blood.v98.4.1174. [DOI] [PubMed] [Google Scholar]
- 21.Thompsett AR, Ellison DW, Stevenson FK, Zhu D. V (H) gene sequences from primary central nervous system lymphomas indicate derivation from highly mutated germinal center B cells with ongoing mutational activity. Blood. 1999;94:1738–46. [PubMed] [Google Scholar]
- 22.Stevenson FK, Stevenson GT. Therapeutic strategies for B cell malignancies involving idiotype–anti-idiotype interactions. Int Rev Immunol. 1986;1:303–33. doi: 10.3109/08830188609056611. [DOI] [PubMed] [Google Scholar]
- 23.Zhu D, McCarthy H, Ottensmeier CH, Johnson PW, Hamblin TJ, Stevenson FK. Acquisition of potential N-glycosylation sites in the immunoglobulin variable region by somatic mutation is a distinctive feature of follicular lymphoma. Blood. 2002. pp. 2562–8. [DOI] [PubMed]
- 24.Brezinschek HP, Brezinschek RI, Lipsky PE. Analysis of the heavy chain repertoire of human peripheral B cells using single-cell polymerase chain reaction. J Immunol. 1995;155:190–202. [PubMed] [Google Scholar]
- 25.Lam KP, Kuhn R, Rajewsky K. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell. 1997;90:1073–83. doi: 10.1016/s0092-8674(00)80373-6. [DOI] [PubMed] [Google Scholar]
- 26.Caspar CB, Levy S, Levy R. Idiotype vaccines for non-Hodgkin's lymphoma induce polyclonal immune responses that cover mutated tumor idiotypes: comparison of different vaccine formulations. Blood. 1997;90:3699–706. [PubMed] [Google Scholar]
- 27.George AJ, Spellerberg MB, Stevenson FK. Idiotype vaccination leads to the emergence of a stable surface Ig-negative variant of the mouse lymphoma BCL1, with different growth characteristics. J Immunol. 1988;140:1695–701. [PubMed] [Google Scholar]
- 28.Doms RW, Lamb RA, Rose JK, Helenius A. Folding and assembly of viral membrane proteins. Virology. 1993;193:545–62. doi: 10.1006/viro.1993.1164. [DOI] [PubMed] [Google Scholar]
- 29.Rice J, King CA, Spellerberg MB, Fairweather N, Stevenson FK. Manipulation of pathogen-derived genes to influence antigen presentation via DNA vaccines. Vaccine. 1999;17:3030–8. doi: 10.1016/s0264-410x(99)00171-1. [DOI] [PubMed] [Google Scholar]
- 30.Herzenberg LA, Tokuhisa T, Herzenberg LA. Carrier-priming leads to hapten-specific suppression. Nature. 1980;285:664–7. doi: 10.1038/285664a0. [DOI] [PubMed] [Google Scholar]
- 31.Di John D, Wasserman SS, Torres JR, et al. Effect of priming with carrier on response to conjugate vaccine. Lancet. 1989;2:1415–8. doi: 10.1016/s0140-6736(89)92033-3. [DOI] [PubMed] [Google Scholar]
- 32.Huston JS, Levinson D, Mudgett-Hunter M, et al. Protein engineering of antibody binding sites: recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc Natl Acad Sci USA. 1988;85:5879–83. doi: 10.1073/pnas.85.16.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Worn A, Pluckthun A. Stability engineering of antibody single-chain Fv fragments. J Mol Biol. 2001;305:989–1010. doi: 10.1006/jmbi.2000.4265. [DOI] [PubMed] [Google Scholar]
- 34.Benvenuti F, Burrone OR. Anti-idiotypic antibodies induced by genetic immunisation are directed exclusively against combined V (L)/V (H) determinants. Gene Ther. 2001;8:1555–61. doi: 10.1038/sj.gt.3301546. [DOI] [PubMed] [Google Scholar]
- 35.Klinman DM, Yi AK, Beaucage SL, Conover J, Krieg AM. CpG motifs present in bacteria DNA rapidly induce lymphocytes to secrete interleukin 6, interleukin 12, and interferon gamma. Proc Natl Acad Sci USA. 1996;93:2879–83. doi: 10.1073/pnas.93.7.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renjifo X, Wolf S, Pastoret PP, et al. Carrier-induced, hapten-specific suppression: a problem of antigen presentation? J Immunol. 1998;161:702–6. [PubMed] [Google Scholar]