Abstract
We previously reported that inducible granulysin gene expression in a human monocytic cell line, THP-1 is dominantly dependent on transcription factor activator protein-1 (AP-1). Here, we further examined the precise regulatory mechanisms underlying granulysin gene expression using THP-1 cells treated with Acholeplasma laidlawii. Transfection of reporter gene constructs into THP-1 cells indicated that the presence of a positive regulatory element(s) is located from −329 to −85 base pairs, containing two distinct AP-1 binding sites and one nuclear factor-κB (NF-κB) binding site. Deletion or mutation of the NF-κB binding site failed to affect inducible promoter activity, whereas deletion or mutation of both the AP-1 binding sites abrogated the promoter activity. Interestingly, deletion of the putative CCAAT/enhancer binding protein β (C/EBPβ) binding site upstream of the positive regulatory element induced the augmentation of granulysin promoter activity. Electrophoretic mobility shift assays demonstrated that nuclear extract prepared from A. laidlawii-treated THP-1 cells generated a specific binding to oligonucleotides, including AP-1, C/EBPβ, and NF-κB element. Furthermore, over-expression of liver-enriched transcriptional activator protein, a subunit of C/EBPβ, augmented A. laidlawii-induced granulysin promoter activity, whereas over-expression of liver-enriched transcriptional inhibitory protein inhibited the promoter activity. NF-κB p50 homodimer had no transactivation property, although it bound to the NF-κB site. These results indicate that AP-1 and C/EBPβ, but not NF-κB participate in the regulation of inducible granulysin gene expression in THP-1 cells. Moreover, the Toll-like receptor 2-dependent signalling pathway may be involved in A. laidlawii-induced transactivation of the granulysin promoter. Thus, these results suggest that the gene expression of granulysin in macrophages would be exquisitely regulated by positive and negative transcription factors when microbial invasion occurs.
Introduction
An antimicrobial protein granulysin is constitutively expressed in cytotoxic T lymphocytes (CTL) and natural killer (NK) cells.1–3 CTL are reported to kill intracellular Mycobacterium tuberculosis by releasing granulysin from cytotoxic granules into the infected target cells following antigen recognition.4 On the other hand, the role of granulysin in NK cells in host defence remains to be defined. Granulysin lyses not only a broad range of microbes, including bacteria, fungi and parasites4–6 but also a variety of tumour cell lines by inducing apoptosis.7–9 Accordingly, granulysin appears to play a critical role in the defence mechanism against bacterial invasion.
Granulysin is constitutively expressed selectively in CTL and NK cells as described above, although a low level of granulysin mRNA was detected in a human T-cell line HuT-78.2 However, granulysin mRNA in NK cells was not augmented by stimulation with cytokines such as interleukin-2 (IL-2) and interferon-α (IFN-α).10 Similarly, granulysin mRNA in IL-2-dependent CTL lines is constitutively expressed.1 In contrast, unstimulated human peripheral blood lymphocytes (PBL) show a very low level of granulysin mRNA, but the level of granulysin mRNA of PBL cultured in a mixed lymphocyte culture or in the presence of phytohaemagglutinin is significantly increased.1 Little is known, however, about the regulatory mechanisms underlying granulysin gene expression at the level of transcription factors in both CTL and NK cells.
Recently, an antimicrobial peptide defensin, comparable to granulysin regarding killing mechanisms against microbes, has been investigated for the regulatory mechanisms of its expression. The expression of human α-defensin (human neutrophil peptide-1 and -3) gene in a human promyelocytic cell line, HL-60 is reported to be regulated by a transcription factor CCAAT/enhancer binding protein α (C/EBPα).11 Subsequently, the expression of human β-defensin-2 in human epithelial cells is demonstrated to be controlled by the pathways of mitogen-activated protein kinase, but not nuclear factor-κB (NF-κB).12 In contrast, the transcriptional regulation of the human β-defensin-2 gene using a mouse macrophage cell line, RAW264.7 is shown to be dependent on NF-κB.13 Thus, the expression of the defensin gene seems to be regulated by various transcription factors.
We previously reported that Acholeplasma laidlawii-induced transactivation of the granulysin promoter in a human monocytic cell line, THP-1, is dominantly dependent on transcription factor activator protein-1 (AP-1).14 In the present study, we further examined the precise regulatory mechanisms involved in A. laidalawii-induced granulysin promoter activity in THP-1 cells. Our results demonstrate that deletion or mutation of the AP-1 binding site abrogated the response to A. laidlawii stimulation. In contrast, the granulysin promoter activity was enhanced by deletion of the putative CCAAT/enhancer binding protein β (C/EBPβ) binding site. Electrophoretic mobility shift assays (EMSA) showed that nuclear extract prepared from A. laidlawii-treated THP-1 cells generated specific binding to DNA oligonucleotides, including AP-1, C/EBPβ, and the NF-κB element in the granulysin promoter. Furthermore, over-expression of liver-enriched transcriptional activator protein (LAP), a subunit of C/EBPβ, augmented A. laidlawii-induced granulysin promoter activity, whereas over-expression of liver-enriched transcriptional inhibitory protein (LIP) inhibited the promoter activity. The NF-κB p50 homodimer had no transactivation property, although it bound to the NF-κB binding site. Thus, these results indicate that AP-1 and C/EBPβ, but not NF-κB, participate in the regulation of inducible granulysin gene expression in THP-1 cells. Moreover, the signalling pathway of A. laidlawii-induced transactivation of the granulysin promoter seems to be Toll-like receptor (TLR) 2-dependent.
Materials and methods
Antibodies
Rabbit anti-C/EBPβ, anti-c-Jun/AP-1, and anti-NF-κB p65 polyclonal antibodies (pAb) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit anti-NF-κB p50 pAb was obtained from Rockland (Gilbertsville, PA). Mouse anti-human TLR2 monoclonal antibody (mAb) was obtained from Cascade Bioscience (Winchester, MA).15 Normal mouse immunoglobulin G2a (IgG2a) was purchased from PharMingen (San Diego, CA).
Cell line
The human monocytic cell line THP-l was maintained in RPMI-1640 medium (Nissui Pharmaceutical, Tokyo, Japan) supplemented with 10% heat-inactivated fetal calf serum, 2 mm l-glutamine, 0·15% sodium bicarbonate, 100 U/ml penicillin and 100 μg/ml streptomycin at 37° in 5% CO2.
Acholeplasma laidlawii
The A. laidlawii PG8 was cultured in Pleuropneumonia-like organism (PPLO) medium (Difco, Detroit, MI) and heat-inactivated for 30 min at 60° as described previously.16 It was then centrifuged for 30 min at 20 000 g, followed by lyophilization. For stimulation, the lyophilized A. laidlawii was suspended in the supplemented RPMI-1640 described above.
Construction of reporter plasmids
The granulysin gene upstream region from the start codon was cloned from THP-1 genomic DNA by polymerase chain reaction (PCR) using a common 3′ antisense primer, 5′-GGGCCCAGGTAGCCATGGTGGGGCAGCCGC-3′[from nucleotide +78 with a NcoI restriction site (underlined)] and various 5′ sense primers. The following 5′ sense primers were used: 5′-GGATCCGGATCCTGCCTATACGATTCCCACTCCC-3′[from nucleotide −1167 with a BamHI restriction site (underlined)], 5′-GGATCCGGATCCATTAGCAGGGCATGGTGGCG-3′[from nucleotide −967 with a BamHI restriction site (underlined)], 5′-GGATCCGGATCCTCCAGCAAAAGACTCCCTTC-3′[from nucleotide −757 with a BamHI restriction site (underlined)], 5′-GGATCCGGATCCCTACTGGGTGAGCCCTTGGAG-3′[from nucleotide −510 with a BamHI restriction site (underlined)], 5′-GGATCCGGATCCTATATTGTTAAATTAAAAAGC-3′[from nucleotide −329 with a BamHI restriction site (underlined)], 5′-GGATCCGGATCCAATATCTGGAACGTACTTGTA-3′[from nucleotide −239 with a BamHI restriction site (underlined)], 5′-GGATCCGGATCCACATCCCAGGCATCCTGGCAGC-3′[from nucleotide −193 with a BamHI restriction site (underlined)], 5′-GGATCCGGATCCTCTTCACACACTGGTATTTG-3′[from nucleotide −85 with a BamHI restriction site (underlined)], 5′-GGATCCGGATCCGGAGATGACATACAAAAAGGG-3′[from nucleotide −38 with a BamHI restriction site (underlined)], and 5′-GGATCCGGATCCAAAAGGGCAGGACCTGAG-3′[from nucleotide −24 with a BamHI restriction site (underlined)]. These primers were designed based on the granulysin genomic sequence.17,18 PCR was performed with KOD Plus DNA polymerase (Toyobo, Osaka, Japan) according to the manufacturer's protocol. The PCR profile included denaturation at 96° for 2 min, followed by 35 cycles of denaturation at 96° for 30 seconds, annealing at 55° for 30 seconds, extension at 68° for 1 min/kilobase, and a final extension at 68° for 5 min. The PCR products were digested with BamHI and NcoI and cloned into the pGL3-Basic luciferase reporter vector (Promega, Madison, WI). The recombinant constructs were designated pGL3−1167/+62, pGL3−967/+62, pGL3−757/+62, pGL3−510/+62, pGL3−329/+62, pGL3−239/+62, pGL3−193/+62, pGL3−85/+62, pGL3−38/+62, and pGL3−24/+62 (the positions are relative to the transcription start site). The pGL3−643/+62 construct was generated from the pGL3−1167/+62 construct by excision of the region upstream of the KpnI site (position −643). The potential AP-1 binding site [−277 to −271 base pairs (bp)] TGACTCA of the pGL3−329/+62 was mutated to TGAGCTC using PCR-based mutagenesis.19 The following primers were used: sense, 5′-GAGATGAGCTCCTTATACTTTTAGTATAAGTAT-3′ (bold indicates mutated sequences); and antisense, 5′-ACTTGACCCAAAAACTATTTTTAAAC-3′. The mutated construct was designated pGL3−329/+62 AP-1 mutant. The potential NF-κB binding site (−214 to −204 bp) GGGGTTTCTCC of the pGL3−329/+62 or pGL3−329/+62 AP-1 mutant was mutated to TACCTTTCTCC using PCR-based mutagenesis. The following primers were used: sense, 5′-TTGTACTGGTACCTTTCTCCCTCCATCGGCACATCCC-3′ (bold indicates mutated sequences); and antisense, 5′-GTACGTTCCAGATATTGCATGGG-3′. The mutated construct was designated pGL3−329/+62 NF-κB mutant and pGL3−329/+62 AP-1 + NF-κB mutant, respectively. The potential AP-1 binding site (−96 to −86 bp) CCTGACCTGCT of the pGL3−193/+62 was mutated to CCGAGCTCGCT using PCR-based mutagenesis. The following primers were used: sense, 5′-ACTGCCCGAGCTCGCTTCTTCACACACTGGTATTTG-3′ (bold indicates mutated sequences); and antisense, 5′-GGTAGCCCACAGGGTCCATGCCAC-3′. The mutated construct was designated pGL3−193/+62 AP-1 mutant. The mutated clone was verified by restriction pattern. The plasmids were purified with a Qiagen plasmid kit (Qiagen, Chatsworth, CA) and used for transient transfection.
Transient transfection and luciferase assay
Transient transfections were performed by the Effectene transfection reagent (Qiagen) according to the manufacturer's protocol. A total of 4 × 105 THP-1 cells were transfected transiently with 0·19 μg of reporter plasmid and 0·01 μg of pRL-TK or phRG-TK (Promega) as an internal control plasmid in 24-well plates (Costar, Cambridge, MA). After 24 hr, transfected cells were stimulated with 10 μg/ml A. laidlawii. After a further 24 hr of incubation, cells were lysed and assayed for luciferase activity using a Dual-Luciferase Reporter Assay System (Promega). Both firefly and Renilla luciferase activities were monitored with a Lumat LB9507 luminometer (Berthold, Wildbad, Germany). Normalized reporter activity is expressed as the firefly luciferase value divided by the Renilla luciferase value. Relative fold induction is calculated as the normalized reporter activity of the test sample divided by the untreated pGL3−1167/+62.
EMSA
Nuclear extracts were prepared from THP-1 cells as described previously.14 Synthetic oligonucleotides were used as probe for EMSA. The oligonucleotides were designed to generate a single 5′-G overhang to each end after annealing with their compliments. The following oligonucleotides were used: AP-1 (−279/−260), 5′-GATGACTCACTTATACTTTT-3′ (−279 to −260 bp); NF-κB (−221/−197), 5′-GTACTAAGGGGTTTCTCCCTCCATC-3′ (−221 to −197 bp); AP-1 (−109/−84), 5′-GGGCTACCACTGCCCTGACCTGCTTC-3′ (−109 to −84 bp); and C/EBPβ (−1008/−986), 5′-GGCCAACATGGTGAAACCCTGTC-3′ (−1008 to −986 bp). The double-stranded oligonucleotides were end-labelled with [α-32P]dCTP using the Klenow fragment of DNA polymerase I (Amersham-Pharmacia Biotech, Uppsala, Sweden). Labelled DNA probe was purified using Quick Spin Column G-25 (Roche, Basel, Switzerland). Purified DNA probe was adjusted to 10 000 counts per minute (c.p.m.)/μl and stored at 4° until use. For binding reactions, 5 μg of nuclear extract was incubated in a total volume of 23 μl of binding buffer [10 mm HEPES–KOH pH 7·9, 50 mm KCl, 5 mm MgCl2, 1 mm ethylenediaminetetraacetic acid (EDTA), 10% glycerol, 5 mm dithiothreitol, 0·5 μg of poly(dI-dC), 10 μg/ml leupeptin, 10 μg/ml aprotinin, 10 μg/ml pepstatin A, 1 mm phenylmethylsulphonyl fluoride, 1 mm Na3VO4] for 10 min at room temperature. Then, α-32P-end-labelled probe (20 000 c.p.m.) was added to the reaction mixture for an additional 30 min at room temperature. For competition assays, excess unlabelled oligonucleotides were preincubated with nuclear extract in the above binding buffer at room temperature for 10 min prior to the addition of the radiolabelled probe (20 000 c.p.m.). For antibody-mediated supershift assays, nuclear extracts were preincubated with 1 μg of appropriate antibodies or normal rabbit IgG (Inter-Cell Technologies, Hopewell, NJ) at 4° for 60 min before the addition of the radiolabelled probe. The reactions were loaded on a 5% polyacrylamide gel in 0·5 × Tris–borate–EDTA, and electrophoresed for 2 hr at 200 V before drying. The intensity of the DNA–protein complex bands was measured using a Phosphor-Imaging system (Fujifilm BAS-2000; Fujifilm, Tokyo, Japan).
Western blot
Fifty micrograms of THP-1 nuclear extract were separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) on a 12% gel and transferred onto a nitrocellulose membrane. Non-specific binding was inhibited by incubation of the membrane in 10% skimmed milk in Tris-buffered saline (TBS) [10 mm Tris (pH 7·6) and 0·15 m NaCl] containing 0·1% Tween-20 (TBS-T) for 1 hr at room temperature. The membrane was rinsed in TBS-T and incubated with the rabbit anti-C/EBPβ pAb (Santa Cruz) (1 : 4000 dilution) in 1% skimmed milk in TBS-T. After 1 hr incubation at room temperature, the membrane was incubated for 1 hr with peroxidase-conjugated goat-anti-rabbit immunoglobulin (Cappel Research, Durham, NC) in 1% skimmed milk in TBS-T at room temperature and developed with an ECL system (Amersham-Pharmacia Biotech).
Construction of LAP and LIP expression vector
THP-1 cells (2 × 106) were cultured in the presence of A. laidlawii at 10 μg/ml for 24 hr in six-well plates (Costar). The cells were collected and pelleted by centrifugation at 1500 g for 5 min at 4°. The pellets were used for total RNA extraction using the method of Gough.20 Synthesis of cDNA and PCR were performed using a RNA PCR kit (Takara, Ohtsu, Japan) and KOD Plus DNA polymerase (Toyobo) according to the manufacturer's instructions, respectively. The following LAP-specific 5′ sense primer was used: 5′-AAGCTTAAGCTTCCACCATGGAAGTGGCCAACTTC-3′ (underline indicates HindIII restriction site). The following LIP-specific 5′ sense primer was used: 5′-AAGCTTAAGCTTCCACCATGGCGGCGGGCTTCCCGTAC-3′ (underline indicates HindIII restriction site). The following common 3′ antisense primer was used: 5′-GGATCCGGATCCCTAGCAGTGGCCGGAGGC-3′ (underline indicates BamHI restriction site). The PCR profile included denaturation at 96° for 2 min, followed by 35 cycles of denaturation at 96° for 30 seconds, annealing at 60° for 30 seconds, extension at 68° for 1 min, and a final extension at 68° for 5 min. The PCR products were digested with HindIII and BamHI and cloned into a pcDNA3.1 (Invitrogen, Carlsbad, CA). The plasmids were purified with a Qiagen plasmid kit (Qiagen) and used for transient transfection.
Statistical evaluation
Data were analysed by using the Student's paired t-test. Data with a P-value of < 0·05 were considered significant.
Results
Functional analysis of the granulysin promoter in a human monocytic cell line, THP-1
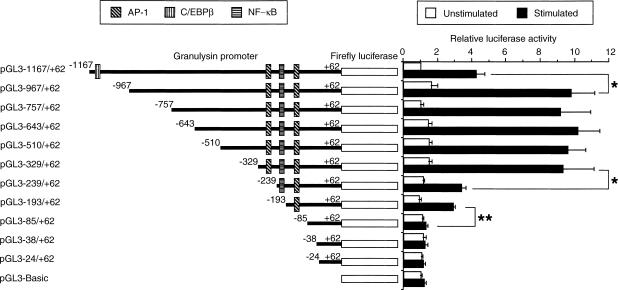
To examine elements necessary for inducible granulysin promoter activity, a 1·2-kb genomic DNA fragment immediately 5′ to the translation initiation site of the granulysin gene was cloned and fused to firefly luciferase gene. In addition, to localize the functional motif(s) necessary for inducible activity, serial 5′ deletions of the granulysin promoter were generated. THP-1 cells were transiently transfected with these luciferase reporter plasmids and then were left untreated or were stimulated with A. laidlawii at a concentration of 10 μg/ml. As shown in Fig. 1, the augmentation of inducible granulysin promoter activity was detected from pGL3–1167/+62 to pGL3–329/+62. The granulysin promoter activity then dropped sharply, which is consistent with the observation that the deleted region contained an AP-1 binding site (−277 to −271 bp, termed upstream AP-1 site) important for granulysin gene expression.14 In addition, the granulysin promoter activity continued to decrease from pGL3–239/+62 to pGL3–85/+62. These results indicate the presence of a positive regulatory element(s) located from −329 to −85 bp. Deletion of a putative NF-κB binding site (−214 to −204 bp) did not significantly affect inducible promoter activity, but deletion of another putative AP-1 binding site (−96 to −86 bp, termed downstream AP-1 site) abrogated the response to A. laidlawii stimulation. Further deletion (pGL3–38/+62 and pGL3–24/+62) had no effect on promoter activity. In contrast, the granulysin promoter activity was enhanced by deletion of the region between −1167 and −967 bp. Therefore, some negative regulatory element(s) may exist in this region. Indeed, the region located between −1003 and −990 bp contains a putative C/EBPβ binding site (Fig. 1), which is known to bind to a transcription repressor LIP.21 These results suggest that both AP-1 binding sites may be important for inducible granulysin promoter activity and a putative C/EBPβ binding site (−1003 to −990 bp) may participate in the negative regulation of granulysin promoter activity.
Figure 1.
Functional analysis of upstream region of granulysin gene. THP-1 cells (4 × 105 cells) were transfected with various truncated promoter constructs, as described in the Materials and Methods. After 24 hr, transfected cells were stimulated with or without 10 μg/ml A. laidlawii. After a further 24 hr of incubation, the cells were lysed and assayed for luciferase activity. Data are presented as relative luciferase activity. The results for each set of transfections were normalized for Renilla luciferase activity and to the full-length unstimulated promoter activity (pGL3–1167/+62). Values represent the mean ± SEM from three independent experiments. *P < 0·05; **P < 0·01.
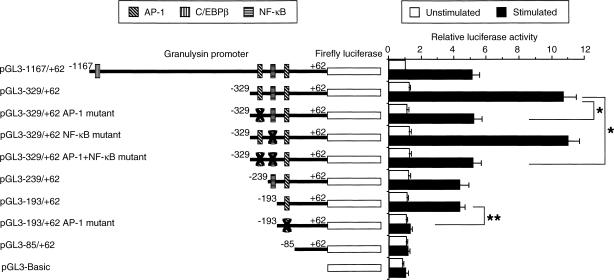
To confirm the functional significance of the region between −329 and −85 bp, the upstream AP-1 site and/or the NF-κB binding site (−214 to −204 bp) contained in the pGL3−329/+62 was mutated by PCR-based mutagenesis. In addition, the downstream AP-1 site of the pGL3−193/+62 was mutated by PCR-based mutagenesis. THP-1 cells were transfected with various truncated or mutated promoter constructs and luciferase activity was measured. As shown in Fig. 2, the nucleotide substitution in the upstream AP-1 site (pGL3−329/+62 AP-1 mutant) resulted in a reduction of promoter activity to the level of the pGL3−1167/+62, which is consistent with the previous observation.14 However, the nucleotide substitution in the NF-κB binding site (pGL3−329/+62 NF-κB mutant) did not significantly affect inducible promoter activity. In addition, the nucleotide substitution in the both the upstream AP-1 site and the NF-κB binding site (pGL3−329/+62 AP-1 + NF-κB mutant) resulted in a reduction of promoter activity to the level of the pGL3−329/+62 AP-1 mutant construct. On the other hand, the nucleotide substitution in the downstream AP-1 site (pGL3−193/+62 AP-1 mutant) abrogated the response to A. laidlawii stimulation. Therefore, the results in Fig. 2 support that both AP-1 binding sites may be involved in the up-regulation of inducible granulysin promoter activity.
Figure 2.
Suppression of granulysin promoter activity by mutation in the upstream and downstream AP-1 element, but not the NF-κB element. The upstream AP-1 binding site TGACTCA of the pGL3−329/+62 was mutated to TGAGCTC using PCR-based mutagenesis. Also, the potential NF-κB binding site GGGGTTTCTCC of the pGL3−329/+62 or pGL3−329/+62 AP-1 mutant was mutated to TACCTTTCTCC using PCR-based mutagenesis. In addition, the downstream AP-1 binding site CCTGACCTGCT of the pGL3−193/+62 was mutated to CCGAGCTCGCT using PCR-based mutagenesis. THP-1 cells (4 × 105 cells) were transfected with various truncated or mutated promoter constructs, as described in the Materials and Methods. After 24 hr, transfected cells were incubated with or without 10 μg/ml A. laidlawii for 24 hr. Then the cells were lysed and assayed for luciferase activity. Data are presented as in the legend to Fig. 1. *P < 0·05; **P < 0·01.
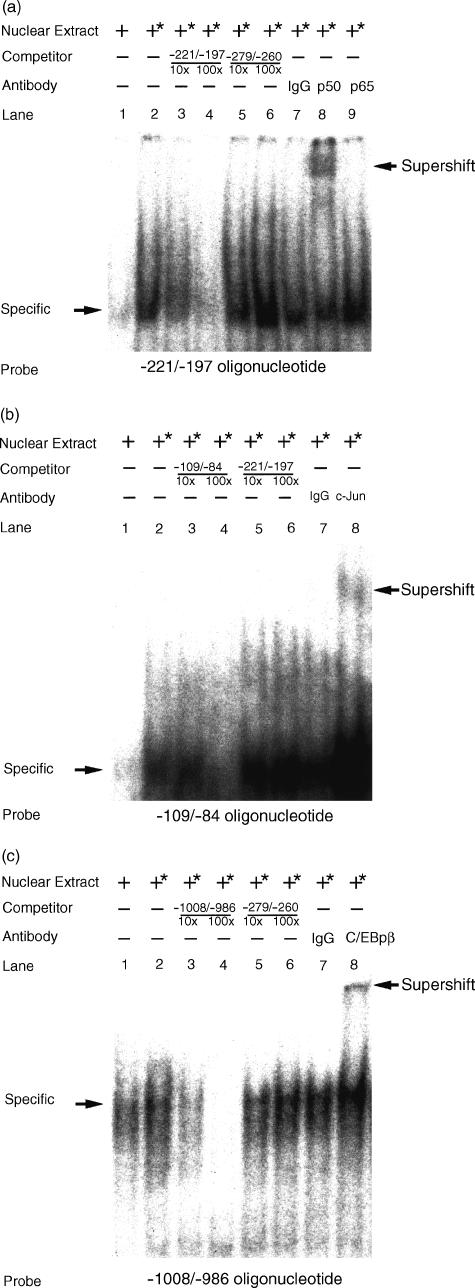
EMSA of the putative transcription factor binding sites in granulysin promoter
We previously reported that the AP-1 binds to the upstream AP-1 site.14 To demonstrate further specific protein binding to the putative NF-κB site, downstream AP-1 site, and C/EBPβ site, EMSA were performed. The double-strand oligonucleotides containing the putative NF-κB site, downstream AP-1 site, and C/EBPβ site of granulysin promoter were used as probes to detect a specific protein binding in the nuclear extract from THP-1 cells stimulated with A. laidlawii. As shown in Fig. 3(a), nuclear extract prepared from the THP-1 cells generated a specific binding to probe encompassing the NF-κB binding site (lane 2), whereas unstimulated nuclear extract from the cells failed to do so (lane 1). In competition assay, the presence of excess unlabelled wild-type probe efficiently out-competed the retarded band (lane 3–4), whereas a non-specific DNA probe containing an upstream AP-1 site failed to do so (lane 5–6). To identify the NF-κB protein, the nuclear extract from stimulated THP-1 cells was preincubated with NF-κB p50- or p65-specific antibody before the addition of radiolabelled probe. The specific retarded band was shifted in the presence of anti-NF-κB p50 antibody (lane 8). However, in the presence of anti-NF-κB p65 antibody (lane 9) and normal rabbit IgG (lane 7), the specific retarded band was not shifted. These results show that NF-κB p50 homodimer had no transactivation property, although it bound to the NF-κB site (−214 to −204 bp) in the granulysin promoter.
Figure 3.
EMSA of putative transcription factor binding sites in the upstream region of the granulysin gene. The following oligonucleotides were used: (a) NF-κB (−221/−197), 5′-GTACTAAGGGGTTTCTCCCTCCATC-3′ (−221 to −197 bp); (b) AP-1 (−109/−84), 5′-GGGCTACCACTGCCCTGACCTGCTTC-3′ (−109 to −84 bp); and (c) C/EBPβ (−1008/−986), 5′-GGCCAACATGGTGAAACCCTGTC-3′ (−1008 to −986 bp). A 32P-labelled, double-stranded oligonucleotide probe was incubated with nuclear extracts from THP-1 cells stimulated with or without 10 μg/ml A. laidlawii for 24 hr, as described in the Materials and Methods. The competitors were used in a 10- or 100-fold molar excess over labelled probes. −279/−260, −221/−197, −109/−84 and −1008/−986 indicate unlabelled probes. Supershift assays were perfomed with 1 μg of appropriate antibodies: rabbit anti-c-Jun/AP-1 antibody (c-Jun), rabbit anti-NF-κB p50 antibody (p50), rabbit anti-NF-κB p65 antibody (p65), and rabbit anti-C/EBPβ antibody (C/EBPβ) or normal rabbit IgG as a cotrol (IgG). The asterisk (*) indicates nuclear extract from A. laidlawii-stimulated THP-1 cells.
As shown in Fig. 3(b), nuclear extract prepared from THP-1 cells stimulated with A. laidlawii generated a specific binding to probe encompassing the downstream AP-1 site (lane 2), whereas unstimulated nuclear extract from the cells failed to do so (lane 1). In competition assay, the presence of excess unlabelled wild-type probe efficiently out-competed the retarded band (lane 4), whereas a non-specific DNA probe containing a NF-κB binding site failed to do so (lane 5–6). To identify the AP-1 protein, the nuclear extract from stimulated THP-1 cells was preincubated with c-Jun/AP-1-specific antibody before the addition of radiolabelled probe. The specific retarded band was shifted in the presence of anti-c-Jun/AP-1 antibody (lane 8). However, in the presence of normal rabbit IgG, the specific retarded band was not shifted (lane 7). These results show that the AP-1 binds to the downstream AP-1 site in the granulysin promoter. Therefore, these results support that two distinct AP-1 binding sites are important for the regulation of inducible granulysin promoter activity.
As shown in Fig. 3(c), nuclear extract prepared from THP-1 cells stimulated with A. laidlawii generated a specific binding to probe emcompassing the C/EBPβ binding site (lane 2). The level of the specific binding was enhanced as compared with that of unstimulated nuclear extract (lane 1). In competition assay, the presence of excess unlabelled wild-type probe efficiently out-competed the retarded band (lane 3–4), whereas a non-specific DNA probe containing an upstream AP-1 site failed to do so (lane 5–6). To identify the C/EBPβ protein, the nuclear extract from stimulated THP-1 cells was preincubated with C/EBPβ-specific antibody before the addition of radiolabelled probe. The specific retarded band was shifted in the presence of anti-C/EBPβ antibody (lane 8). However, the specific retarded band was not shifted in the presence of normal rabbit IgG (lane 7). These results show that the C/EBPβ binds to the putative C/EBPβ binding site (−1003 to −990 bp) in the granulysin promoter. Taken together, these results suggest that AP-1 and C/EBPβ, but not NF-κB play important roles in the regulation of inducible granulysin promoter activity.
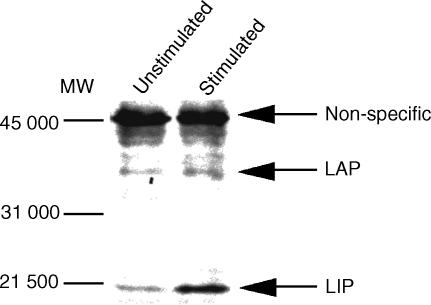
Induction of LIP in THP-1 cells stimulated with A. laidlawii
C/EBPβ consists of the positive regulator, LAP and the negative regulator, LIP.21 Honda et al. reported that the LIP produced in THP-1 cells treated with M. tuberculosis and lipopolysaccharide (LPS) showed repressor activity on human immunodeficiency virus-1 (HIV-1) long-terminal repeat (LTR) promoter.22 We next examined whether A. laidlawii induces LIP in THP-1 cells. To this aim, nuclear extract prepared from THP-1 cells stimulated with A. laidlawii were analysed for C/EBPβ proteins by Western blot. As shown in Fig. 4, THP-1 cells stimulated with A. laidlawii strongly induced LIP, compared to unstimulated cells. In contrast, the level of LAP was essentially constant. Therefore, these results indicate that A. laidlawii-induced LIP may participate in the negative regulation of inducible granulysin promoter activity.
Figure 4.
Western blot analysis of nuclear extracts prepared from THP-1 cells stimulated with A. laidlawii. Nuclear extracts from THP-1 cells stimulated with or without 10 μg/ml A. laidlawii for 24 hr were analysed for C/EBPβ protein by Western blot. LAP and LIP were detected with C/EBPβ antibody, as described in the Materials and Methods.
Over-expression of LIP led to a decrease in A. laidlawii-induced transactivation of the granulysin promoter
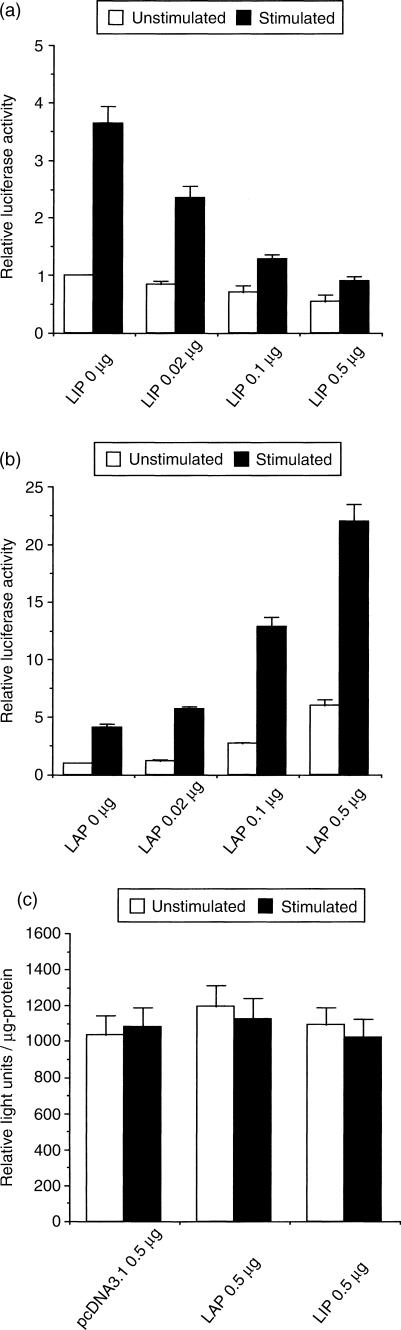
To examine further the roles of C/EBPβ proteins in the regulation of inducible granulysin promoter activity, we tested to see whether LIP suppresses A. laidlawii-induced transactivation of the granulysin promoter. To address this question, the cDNA for LIP was cloned, placed under the transcription control of the cytomegalovirus (CMV) promoter, and transiently co-transfected with pGL3−1167/+62 into THP-1 cells. As shown in Fig. 5(a), co-transfection of THP-1 cells with increasing amounts of a LIP expression vector inhibited basal and A. laidlawii-induced granulysin promoter activity in a dose-dependent manner. The observed inhibition was not due to non-specific inhibition of transcription by LIP because co-transfecting the CMV-LIP construct with phRG-TK driven by herpes simplex virus TK promoter, which has no C/EBPβ binding sites, did not significantly affect the luciferase activity (Fig. 5c). By contrast, over-expression of LAP increased basal and A. laidlawii-induced granulysin promoter activity in a dose-dependent manner (Fig. 5b). In these assays, we used only pGL3−1167/+62 construct containing C/EBPβ binding site (−1003 to −990 bp). Subsequently, we found another potential C/EBPβ binding site (−64 to −51 bp) in the granulysin promoter using the TRANSFAC database (http://transfac.gbf.de/). Indeed, over-expression of LIP decreased the promoter activity of pGL3−967/+62 and pGL3−329/+62 construct lacking in C/EBPβ binding site (−1003 to −990 bp) (data not shown). This site may function as a C/EBPβ binding site, however, the binding of C/EBPβ to this site remains to be elucidated. As shown in Fig. 3(c), transcription factor C/EBPβ binds to the putative C/EBPβ binding site (−1003 to −990 bp) in the granulysin promoter. In addition, LIP was strongly induced in nuclear extract prepared from THP-1 cells stimulated with A. laidlawii (Fig. 4). Furthermore, inducible granulysin promoter activity was enhanced by deletion of the region between −1167 and −967 bp, which contains the C/EBPβ binding site (Figs 1 and 2). Therefore, these results indicate that the LIP binding to C/EBPβ site in the granulysin promoter participates in the negative regulation of inducible granulysin promoter activity.
Figure 5.
Over-expression of LIP or LAP affects the granulysin promoter activity. (a) Basal and A. laidlawii-induced granulysin promoter activity is inhibited by over-expression of LIP. (b) Over-expression of LAP increases basal and A. laidlawii-induced granulysin promoter activity. (c) The herpes simplex virus TK promoter activity is not affected by over-expression of LAP or LIP. THP-1 cells (4 × 105 cells) were transfected with 0·19 μg of pGL3−1167/+62 and 0·01 μg of phRG-TK [in Fig. 5(c) except for only phRG-TK used], in addition to co-transfection with LAP or LIP expression vector. The pcDNA3.1 was used to ensure that all transfection mixtures had a total of 0·7 μg of DNA. After 24 hr, transfected cells were stimulated with or without 10 μg/ml A. laidlawii. After a further 24 hr of incubation, the cells were lysed and assayed for luciferase activity, as described in the Materials and Methods. Data are presented as in the legend to Fig. 1, except for the data in Fig. 5(c), which are presented as relative light units per μg protein.
Suppression of A. laidlawii-induced granulysin promoter activity by anti-TLR2 mAb
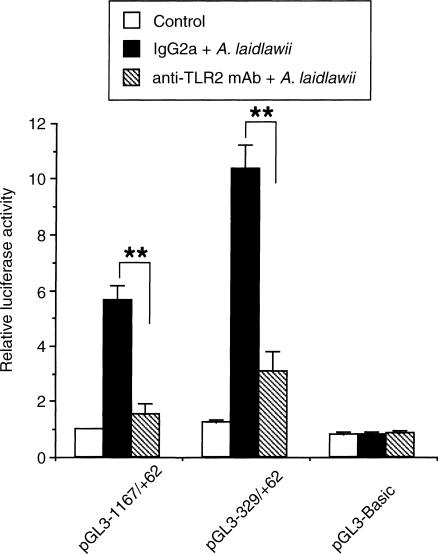
Mycoplasmas are wall-less bacteria that occur as commensals or pathogens in animals and humans.23 Mycoplasmas and their membranes are potent activators of macrophages, the active principal being lipoproteins24–26 and lipopeptides.27,28 Mycoplasmal lipoprotein and lipopeptide activates macrophages through a TLR2-dependent signalling pathway.29,30 In the next experiment, we examined the inhibitory effect of anti-TLR2 mAb on A. laidlawii-induced granulysin promoter activity. As shown in Fig. 6, A. laidlawii-induced transactivation of the granulysin promoter was significantly inhibited by pretreatment with anti-TLR2 mAb. These results suggest that the TLR2-dependent signalling pathway may be involved in A. laidlawii-induced transactivation of the granulysin promoter.
Figure 6.
Suppression of A. laidlawii-mediated transactivation of the granulysin promoter by anti-TLR2 mAb. THP-1 cells (4 × 105 cells) were transfected with pGL3−1167/+62 or pGL3−329/+62, as described in the Materials and Methods. After 24 hr, transfected cells were incubated with 10 μg/ml mouse anti-TLR2 mAb or normal mouse IgG2a as a control for 30 min prior to stimulation with or without 10 μg/ml A. laidlawii. After a further 24 hr of incubation, the cells were lysed and assayed for luciferase activity. Data are presented as in the legend to Fig. 1. **P < 0·01.
Discussion
In the present study we demonstrated that there are two distinct AP-1 binding sites (−277 to −271 bp and −96 to −86 bp) and a NF-κB binding site (−214 to −204 bp) in the granulysin promoter (Fig. 1). Deletion or mutation of both the AP-1 binding sites abrogated the response to A. laidlawii stimulation, whereas deletion or mutation of the NF-κB binding site failed to affect inducible promoter activity. Furthermore, deletion of the putative C/EBPβ binding site (−1003 to −990 bp) induced the augmentation of granulysin promoter activity (Figs 1 and 2). Thus, the transcription factors AP-1 and C/EBPβ function as critical regulators in the expression of the granulysin gene.
EMSA demonstrated that nuclear extract prepared from A. laidlawii-treated THP-1 cells generated specific binding to DNA oligonucleotides, including AP-1 and NF-κB element (Fig. 3a). However, NF-κB, binding NF-κB site (−214 to −204 bp) in the granulysin promoter, was the p50 homodimer without transactivation property (Fig. 3a). The members of NF-κB family include p50 (NF-κB1), p52 (NF-κB2), p65 (RelA), RelB, v-Rel, and c-Rel.31–33 In cells, NF-κB exists as homo- or heterodimers with distinct DNA-binding specificity. A heterodimer composed of p50 and p65 subunits is the most common dimer.31,32 The p65 protein has a transactivation domain, whereas the p50 protein is lacking in this domain. Therefore, the p50 homodimers are unable to transactivate, although they bind to DNA. The p50 homodimers have different binding specificities from p50–p65 heterodimers.34 They preferentially bind GGGGAT(T/C)CCC, as evidenced by screening with a pool of random oligonucleotides.34 The p65 homodimers and p50–p65 heterodimers cannot bind the p50 homodimers binding motif as well.34,35 The sequence of the NF-κB binding site of the granulysin promoter (GGGGTTTCTCC) shows high identity with the motif for the p50 homodimer. Therefore, these previous findings support our data that the p50 homodimer can bind to the NF-κB binding site (−214 to −204 bp) in the granulysin promoter.
In contrast to the augmentation of granulysin promoter activity via AP-1, but not NF-κB, we speculated that C/EBPβ may participate in the negative regulation of inducible granulysin promoter activity because deletion of the binding site for a C/EBPβ (located between −1003 and −990 bp) led to the enhancement of granulysin promoter activity (Figs 1 and 2). In addition, we showed that C/EBPβ binds to the putative C/EBPβ binding site (−1003 to −990 bp) in the granulysin promoter (Fig. 3c) and that LIP is significantly induced in THP-1 cells stimulated with A. laidlawii (Fig. 4). Furthermore, over-expression of LIP leads to a decrease in A. laidlawii-induced transactivation of the granulysin promoter (Fig. 5a). The C/EBPβ transcription factor is reported to be an important regulator of inflammation.36,37 The C/EBPβ gene has no introns, but a 37 000 MW stimulatory isoform (called LAP) and a 20 000 MW inhibitory isoform (called LIP) are produced from the same mRNA via a leaky ribosome scanning mechanism.21 Importantly, LIP functions as a repressor when the level of LIP expression is more than 20% of that of LAP expression.21 Honda et al. reported that the LIP produced in THP-1 cells treated with M. tuberculosis and LPS showed repressor activity on HIV-1 LTR promoter containing C/EBPβ binding sites.22 On the other hand, cytokine genes, such as tumour necrosis factor-α (TNF-α), also have C/EBPβ binding sites, and the transfection of LIP represses TNF-α promoter activity in myelomonocytic cells.38 Thus, these earlier studies support our data that the LIP binding to the C/EBPβ site in the granulysin promoter would suppress A. laidlawii-induced transactivation of granulysin promoter. Indeed, we have hardly detected the granulysin at the level of protein in this system (data not shown), and hence we tried to detect the granulysin protein in A. laidlawii-treated THP-1 cells, in which LAP is over-expressed.
The recognition of pathogens on the cell surface is mediated by receptors that are referred to as pattern-recognition receptors. These receptors recognize conserved molecular patterns (pathogen-associated molecular patterns or PAMPs) shared by large groups of micro-organisms.39,40 Recently, it has been well documented that the family of TLR plays important roles as pattern-recognition receptors. The TLR family is now comprised of at least 10 members in mammals.41–46 Components, including lipoprotein,47 peptidoglycan and lipoteichoic acid,48 induce intracellular signalling via TLR2, whereas LPS induces the signalling via TLR4.49 In the TLR signalling pathways, transcription factors such as AP-1 and NF-κB are activated and involved in cytokine gene expression.50 Feng and Lo reported that lipid extract of Mycoplasma penetrans induces TNF-α production and moreover activates both AP-1 and NF-κB in macrophages.51 We previously reported that A. laidlawii induces TNF-α production in THP-1 cells,52 and our recent preliminary results showed that components of A. laidlawii, capable of inducing TNF-α production, seem to be characteristic of lipoprotein (data not shown). In this study, we demonstrate that A. laidlawii-induced transactivation of the granulysin promoter was significantly inhibited by pretreatment with anti-TLR2 mAb (Fig. 6). Taken together, it is conceivable that the TLR2-dependent signalling pathway may be involved in A. laidlawii-induced transactivation of the granulysin promoter. On the other hand, Takeuchi et al. recently reported that TLR2 recognizes mycoplasmal lipopeptide co-operatively with TLR6.53 However, in our experimental systems the participation of TLR6 in A. laidlawii-induced transactivation of the granulysin promoter remains to be elucidated. Recently, Thoma-Uszynski et al. reported that bacterial lipoprotein activation of TLR2 leads to a nitric oxide-independent killing of intracellular M. tuberculosis in human monocytes and alveolar macrophages.54 However, the effector molecules responsible for this killing are unclear. Granulysin induced by bacterial lipoprotein may play important roles in this mechanism.
Our data demonstrate that treatment of THP-1 cells with A. laidlawii induces the activation of granulysin gene promoter via transcription factor AP-1, but not NF-κB, and that the TLR2-dependent signalling pathway may be involved in A. laidlawii-induced transactivation of the granulysin gene promoter. In contrast, transcription factor C/EBPβ participates in the negative regulation of inducible granulysin promoter activity. Thus, these results indicate that the gene expression of granulysin in macrophages would be acutely regulated by positive and negative transcription factors when microbial invasion occurs.
References
- 1.Jongstra J, Schall TJ, Dyer BJ, Clayberger C, Jorgensen J, Davis MM, Krensky AM. The isolation and sequence of a novel gene from a human functional T cell line. J Exp Med. 1987;165:601–14. doi: 10.1084/jem.165.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yabe T, McSherry C, Bach FH, Houchins JP. A cDNA clone expressed in natural killer and T cells that likely encodes a secreted protein. J Exp Med. 1990;172:1159–63. doi: 10.1084/jem.172.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pena SV, Hanson DA, Carr BA, Goralski TJ, Krensky AM. Processing, subcellular localization, and function of 519 (granulysin), a human late T cell activation molecule with homology to small, lytic, granule proteins. J Immunol. 1997;158:2680–8. [PubMed] [Google Scholar]
- 4.Stenger S, Hanson DA, Teitelbaum R, et al. An antimicrobial activity of cytolytic T cells mediated by granulysin. Science. 1998;282:121–5. doi: 10.1126/science.282.5386.121. [DOI] [PubMed] [Google Scholar]
- 5.Wang Z, Choice E, Kaspar A, Hanson D, Okada S, Lyu SC, Krensky AM, Clayberger C. Bactericidal and tumoricidal activities of synthetic peptides derived from granulysin. J Immunol. 2000;165:1486–90. doi: 10.4049/jimmunol.165.3.1486. [DOI] [PubMed] [Google Scholar]
- 6.Ernst WA, Thoma-Uszynski S, Teitelbaum R, et al. Granulysin, a T cell product, kills bacteria by altering membrane permeability. J Immunol. 2000;165:7102–8. doi: 10.4049/jimmunol.165.12.7102. [DOI] [PubMed] [Google Scholar]
- 7.Gamen S, Hanson DA, Kaspar A, Naval J, Krensky AM, Anel A. Granulysin-induced apoptosis. I. Involvement of at least two distinct pathways. J Immunol. 1998;161:1758–64. [PubMed] [Google Scholar]
- 8.Kaspar AA, Okada S, Kumar J, et al. A distinct pathway of cell-mediated apoptosis initiated by granulysin. J Immunol. 2001;167:350–6. doi: 10.4049/jimmunol.167.1.350. [DOI] [PubMed] [Google Scholar]
- 9.Pardo J, Perez-Galan P, Gamen S, et al. A role of the mitochondrial apoptosis-inducing factor in granulysin-induced apoptosis. J Immunol. 2001;167:1222–9. doi: 10.4049/jimmunol.167.3.1222. [DOI] [PubMed] [Google Scholar]
- 10.Mori S, Jewett A, Cavalcanti M, Murakami-Mori K, Nakamura S, Bonavida B. Differential regulation of human NK cell-associated gene expression following activation by IL-2, IFN-α and PMA/ionomycin. Int J Oncol. 1998;12:1165–70. doi: 10.3892/ijo.12.5.1165. [DOI] [PubMed] [Google Scholar]
- 11.Tsutsumi-Ishii Y, Hasebe T, Nagaoka I. Role of CCAAT/enhancer-binding protein site in transcription of human neutrophil peptide-1 and -3 defensin genes. J Immunol. 2000;164:3264–73. doi: 10.4049/jimmunol.164.6.3264. [DOI] [PubMed] [Google Scholar]
- 12.Krisanaprakornkit S, Kimball JR, Dale BA. Regulation of human β-defensin-2 in gingival epithelial cells: the involvement of mitogen-activated protein kinase pathways, but not the NF-kB transcription factor family. J Immunol. 2002;168:316–24. doi: 10.4049/jimmunol.168.1.316. [DOI] [PubMed] [Google Scholar]
- 13.Tsutsumi-Ishii Y, Nagaoka I. NF-kB-mediated transcriptional regulation of human β-defensin-2 gene following lipopolysaccharide stimulation. J Leukoc Biol. 2002;71:154–62. [PubMed] [Google Scholar]
- 14.Kida Y, Kuwano K, Zhang Y, Arai S. Acholeplasma laidlawii up-regulates granulysin gene expression via transcription factor activator protein-1 in a human monocytic cell line, THP-1. Immunology. 2001;104:324–32. doi: 10.1046/j.1365-2567.2001.01310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flo TH, Halaas O, Lien E, Ryan L, Teti G, Golenbock DT, Sundan A, Espevik T. Human toll-like receptor 2 mediates monocyte activation by Listeria monocytogenes, but not by group B streptococci or lipopolysaccharide. J Immunol. 2000;164:2064–9. doi: 10.4049/jimmunol.164.4.2064. [DOI] [PubMed] [Google Scholar]
- 16.Kuwano K, Ono S, Akashi A, Ohishi M, Shigematsu H, Arai S. Production of a T-cell clone which reacts with membrane proteins of Acholeplasma laidlawii. Microbiol Immunol. 1997;41:261–4. doi: 10.1111/j.1348-0421.1997.tb01198.x. [DOI] [PubMed] [Google Scholar]
- 17.Manning WC, O'Farrell S, Goralski TJ, Krensky AM. Genomic structure and alternative splicing of 519, a gene expressed late after T cell activation. J Immunol. 1992;148:4036–42. [PubMed] [Google Scholar]
- 18.Houchins JP, Kricek F, Chujor CS, Heise CP, Yabe T, McSherry C, Bach FH. Genomic structure of NKG5, a human NK and T cell-specific activation gene. Immunogenetics. 1993;37:102–7. doi: 10.1007/BF00216832. [DOI] [PubMed] [Google Scholar]
- 19.Imai Y, Matsushima Y, Sugimura T, Terada M. A simple and rapid method for generating a deletion by PCR. Nucl Acids Res. 1991;19:2785. doi: 10.1093/nar/19.10.2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gough NM. Rapid and quantitative preparation of cytoplasmic RNA from small numbers of cells. Anal Biochem. 1988;173:93–5. doi: 10.1016/0003-2697(88)90164-9. [DOI] [PubMed] [Google Scholar]
- 21.Descombes P, Schibler U. A liver-enriched transcriptional activator protein, LAP, and a transcriptional inhibitory protein, LIP, are translated from the same mRNA. Cell. 1991;67:569–79. doi: 10.1016/0092-8674(91)90531-3. [DOI] [PubMed] [Google Scholar]
- 22.Honda Y, Rogers L, Nakata K, et al. Type I interferon induces inhibitory 16-kD CCAAT/enhancer binding protein (C/EBP) β, repressing the HIV-1 long terminal repeat in macrophages: pulmonary tuberculosis alters C/EBP expression, enhancing HIV-1 replication. J Exp Med. 1998;188:1255–65. doi: 10.1084/jem.188.7.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baseman JB, Tully JG. Mycoplasmas: sophisticated, reemerging, and burdened by their notoriety. Emerg Infect Dis. 1997;3:21–32. doi: 10.3201/eid0301.970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feng SH, Lo SC. Induced mouse spleen B-cell proliferation and secretion of immunoglobulin by lipid-associated membrane proteins of Mycoplasma fermentans incognitus and Mycoplasma penetrans. Infect Immun. 1994;62:3916–21. doi: 10.1128/iai.62.9.3916-3921.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herbelin A, Ruuth E, Delorme D, Michel-Herbelin C, Praz F. Mycoplasma arginini TUH-14 membrane lipoproteins induce production of interleukin-1, interleukin-6, and tumor necrosis factor alpha by human monocytes. Infect Immun. 1994;62:4690–4. doi: 10.1128/iai.62.10.4690-4694.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kostyal DA, Butler GH, Beezhold DH. A 48-kilodalton Mycoplasma fermentans membrane protein induces cytokine secretion by human monocytes. Infect Immun. 1994;62:3793–800. doi: 10.1128/iai.62.9.3793-3800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muhlradt PF, Kiess M, Meyer H, Sussmuth R, Jung G. Isolation, structure elucidation, and synthesis of a macrophage stimulatory lipopeptide from Mycoplasma fermentans acting at picomolar concentration. J Exp Med. 1997;185:1951–8. doi: 10.1084/jem.185.11.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muhlradt PF, Kiess M, Meyer H, Sussmuth R, Jung G. Structure and specific activity of macrophage-stimulating lipopeptides from Mycoplasma hyorhinis. Infect Immun. 1998;66:4804–10. doi: 10.1128/iai.66.10.4804-4810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takeuchi O, Kaufmann A, Grote K, Kawai T, Hoshino K, Morr M, Muhlradt PF, Akira S. Cutting edge: preferentially the R-stereoisomer of the mycoplasmal lipopeptide macrophage-activating lipopeptide-2 activates immune cells through a toll-like receptor 2- and MyD88-dependent signaling pathway. J Immunol. 2000;164:554–7. doi: 10.4049/jimmunol.164.2.554. [DOI] [PubMed] [Google Scholar]
- 30.Nishiguchi M, Matsumoto M, Takao T, et al. Mycoplasma fermentans lipoprotein M161Ag-induced cell activation is mediated by Toll-like receptor 2: role of N-terminal hydrophobic portion in its multiple functions. J Immunol. 2001;166:2610–16. doi: 10.4049/jimmunol.166.4.2610. [DOI] [PubMed] [Google Scholar]
- 31.Siebenlist U, Franzoso G, Brown K. Structure, regulation and function of NF-κB. Annu Rev Cell Biol. 1994;10:405–55. doi: 10.1146/annurev.cb.10.110194.002201. [DOI] [PubMed] [Google Scholar]
- 32.Thanos D, Maniatis T. NF-κB: a lesson in family values. Cell. 1995;80:529–32. doi: 10.1016/0092-8674(95)90506-5. [DOI] [PubMed] [Google Scholar]
- 33.Baeuerle PA, Baltimore D. NF-κB: ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 34.Kunsch C, Ruben SM, Rosen CA. Selection of optimal κB/Rel DNA-binding motifs: interaction of both subunits of NF-κB with DNA is required for transcriptional activation. Mol Cell Biol. 1992;12:4412–21. doi: 10.1128/mcb.12.10.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Urban MB, Baeuerle PA. The 65-kD subunit of NF-κB is a receptor for IκB and a modulator of DNA-binding specificity. Genes Dev. 1990;4:1975–84. doi: 10.1101/gad.4.11.1975. [DOI] [PubMed] [Google Scholar]
- 36.Akira S, Isshiki H, Sugita T, et al. A nuclear factor for IL-6 expression (NF-IL6) is a member of a C/EBP family. EMBO J. 1990;9:1897–906. doi: 10.1002/j.1460-2075.1990.tb08316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Natsuka S, Akira S, Nishio Y, Hashimoto S, Sugita T, Isshiki H, Kishimoto T. Macrophage differentiation-specific expression of NF-IL6, a transcription factor for interleukin-6. Blood. 1992;79:460–6. [PubMed] [Google Scholar]
- 38.Pope RM, Leutz A, Ness SA. C/EBPβ regulation of the tumor necrosis factor α gene. J Clin Invest. 1994;94:1449–55. doi: 10.1172/JCI117482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Medzhitov R, Janeway CA. Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997;91:295–8. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 40.Hoffmann JA, Kafatos FC, Janeway CA, Ezekowitz RA. Phylogenetic perspectives in innate immunity. Science. 1999;284:1313–18. doi: 10.1126/science.284.5418.1313. [DOI] [PubMed] [Google Scholar]
- 41.Medzhitov R, Preston-Hurlburt P, Janeway CA. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–7. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 42.Rock FL, Hardiman G, Timans JC, Kastelein RA, Bazan JF. A family of human receptors structurally related to Drosophila Toll. Proc Natl Acad Sci USA. 1998;95:588–93. doi: 10.1073/pnas.95.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeuchi O, Kawai T, Sanjo H, Copeland NG, Gilbert DJ, Jenkins NA, Takeda K, Akira S. TLR6: a novel member of an expanding toll-like receptor family. Gene. 1999;231:59–65. doi: 10.1016/s0378-1119(99)00098-0. [DOI] [PubMed] [Google Scholar]
- 44.Chuang TH, Ulevitch RJ. Cloning and characterization of a sub-family of human toll-like receptors: hTLR7, hTLR8 and hTLR9. Eur Cytokine Netw. 2000;11:372–8. [PubMed] [Google Scholar]
- 45.Hemmi H, Takeuchi O, Kawai T, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–5. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 46.Chuang T, Ulevitch RJ. Identification of hTLR10: a novel human Toll-like receptor preferentially expressed in immune cells. Biochim Biophys Acta. 2001;1518:157–61. doi: 10.1016/s0167-4781(00)00289-x. [DOI] [PubMed] [Google Scholar]
- 47.Hirschfeld M, Kirschning CJ, Schwandner R, Wesche H, Weis JH, Wooten RM, Weis JJ. Cutting edge: inflammatory signaling by Borrelia burgdorferi lipoproteins is mediated by toll-like receptor 2. J Immunol. 1999;163:2382–6. [PubMed] [Google Scholar]
- 48.Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning CJ. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J Biol Chem. 1999;274:17406–9. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 49.Poltorak A, He X, Smirnova I, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–8. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 50.Aderem A, Ulevitch RJ. Toll-like receptors in the induction of the innate immune response. Nature. 2000;406:782–7. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 51.Feng SH, Lo SC. Lipid extract of Mycoplasma penetrans proteinase K-digested lipid-associated membrane proteins rapidly activates NF-κB and activator protein 1. Infect Immun. 1999;67:2951–6. doi: 10.1128/iai.67.6.2951-2956.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugama K, Kuwano K, Furukawa M, Himeno Y, Satoh T, Arai S. Mycoplasmas induce transcription and production of tumor necrosis factor in a monocytic cell line, THP-1, by a protein kinase C-independent pathway. Infect Immun. 1990;58:3564–7. doi: 10.1128/iai.58.11.3564-3567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takeuchi O, Kawai T, Muhlradt PF, Morr M, Radolf JD, Zychlinsky A, Takeda K, Akira S. Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int Immunol. 2001;13:933–40. doi: 10.1093/intimm/13.7.933. [DOI] [PubMed] [Google Scholar]
- 54.Thoma-Uszynski S, Stenger S, Takeuchi O, et al. Induction of direct antimicrobial activity through mammalian toll-like receptors. Science. 2001;291:1544–7. doi: 10.1126/science.291.5508.1544. [DOI] [PubMed] [Google Scholar]