Abstract
The human mast cell line (HMC)-1 cell line is growth-factor independent because of a constitutive activity of the receptor tyrosine kinase Kit. Such deregulated Kit activity has also been suggested causative in gastrointestinal stromal tumours (GISTs) and mastocytosis. HMC-1 is the only established continuously growing human mast cell line and has therefore been widely employed for in vitro studies of human mast cell biology. In this paper we describe two sublines of HMC-1, named HMC-1560 and HMC-1560,816, with different phenotypes and designated by the locations of specific mutations in the c-kit proto-oncogene. Activating mutations in the Kit receptor were characterized using the pyrosequencing™ method. Both sublines have a heterozygous T to G mutation at codon 560 in the juxtamembrane region of the c-kit gene causing an amino acid substitution of Gly-560 for Val. In contrast, only HMC-1560,816 cells have the c-kitV816 mutation found in mast cell neoplasms causing an Asp→Val substitution in the intracellular kinase domain. Kit was constitutively phosphorylated on tyrosine residues and associated with phosphatidylinositol 3′-kinase (PI 3-kinase) in both variants of HMC-1, but this did not lead to a constitutive phosphorylation of Akt or extracellular regulated protein kinase (ERK), which are signalling molecules normally activated by the interaction of stem cell factor (SCF) with Kit. The documentation and characterization of two sublines of HMC-1 cells provides both information on the biological consequences of mutations in Kit and recognition of the availability of what in reality are two distinct cultured human mast cell lines.
Introduction
Cell lines are frequently used in studies of mast cell biology. Only one human mast cell line has been described (HMC-1)1 thus contrasting with the availability of several growth-factor dependent, or independent, mast cell lines of murine or rat origin. The HMC-1 cell line was established from a patient with mast cell leukaemia, and these cells exhibit many characteristics of tissue mast cells, such as the expression of histamine, tryptase, heparin, and a similar cell surface antigen-profile.2,3 The major difference between HMC-1 cells and human tissue mast cells is the lack of FcεRI expression, although mast cells developed in vitro from human liver, bone marrow or cord blood have a reduced FcεRI expression compared with tissue mast cells.4–8 Because of the many advantages of using cell lines for studies of cell biology, HMC-1 cells have been widely used in studies of human mast cell function, for example refs 9–19.
One key characteristic of mast cells is the expression of the receptor for stem cell factor (SCF), i.e. Kit, a type III receptor tyrosine kinase encoded by c-kit, the cellular homologue of the viral oncogene v-kit.20 Signalling mediated by SCF and Kit is known to be of importance for the control of the proliferation, differentiation, migration, activation, and survival of mast cells.21,22 Mast cell lines often have a deregulated Kit receptor signalling due to mutations in the c-kit proto-oncogene. These alterations, that convert the receptor into a constitutive active state, have been reported for the RBL-2H3, P-815, C2, and HMC-1 mast cell lines.23–26 Activating mutations within the c-kit gene appear to be causative in most gastrointestinal stromal tumours (GISTs)27 and some types of mastocytosis,28–30 the latter of which is a heterogeneous group of disorders characterized by accumulation of mast cells in tissues. Several studies of cells obtained from patients with mastocytosis have described a point mutation in the catalytic domain of Kit (for reviews see Vliagoftis et al.31 and Nilsson et al.32). This missense mutation (c-kitV816) introduces an Asp → Val substitution into the Kit receptor, which makes the receptor constitutively phosphorylated and active.26,33–35 The V816 mutation is especially common in patients with adult onset sporadic mastocytosis and in patients in which mastocytosis is associated with hematologic disorders such as myelodysplastic or myeloproliferative syndromes.28–30 Mutations in other regions of Kit are rare in mastocytosis, although there is a report of two mastocytosis patients with a mutation (G560) in the juxtamembrane region between the transmembrane and tyrosine kinase part of Kit.36 Similarly, many GIST patients have activating mutations located between codon 550–560 in the juxtamembrane part of Kit.27,37
In this study we describe two variant sublines of HMC-1, which we designate HMC-1560 and HMC-1560,816. Both variants have a point mutation in the juxtamembrane domain (c-kitG560) causing an amino acid substitution of Gly-560 for Val but only HMC-1560,816 has the Asp-816→Val mutation, which can be found in mast cell neoplasms. The two HMC-1 sublines also exhibit different phenotypic and growth characteristics.
Materials and methods
Reagents
Human recombinant SCF was purchased from Pepro Tech (Rocky Hill, NJ), PIXY321 was provided by Immunex (Seattle, WA) and horseradish peroxidase conjugated donkey anti-rabbit immunoglobulin was purchased from Amersham Pharmacia Biotech (Little Chalfont, UK). The anti-Akt, Phospho-Akt (Ser473) and phospho-p44/42 mitogen-activated protein (MAP) kinase (Thr202/Thr204) antibodies were from New England Biolabs (Beverly, MA). The anti-extracellular protein kinase (ERK)-1 (K-23) antibody was from Santa Cruz Biotechnology (Santa Cruz, CA) and the 4G10 Ab was from Upstate Biotechnology (Lake Placid, NY). Ly 294002 was purchased from Calbiochem (Darmstadt, Germany).
Cell cultures
HMC-1 cells1,2 were cultured in Iscove's modified Dulbecco's medium (IMDM) supplemented with 10% fetal calf serum (FCS), 2 mm l-glutamine, 100 IU/ml penicillin, 50 µg/ml streptomycin, 1·2 mm α-thioglycerol and passaged every 3–4 days.
MO7e cells (a megakaryocytic cell line expressing normal Kit) were cultured as for HMC-1 cells with the addition of 10 ng/ml of PIXY321, a granulocyte–macrophage colony-stimulating factor (GM-CSF), interleukin (IL)-3 fusion protein. Human cord blood derived mast cells (CBMC) were developed in vitro as described.6
Cytochemistry
Cytocentrifuge preparations of HMC-1 cells were stained with May–Grünwald Giemsa for morphological analysis.
Cell cycle analysis
Cell cycle analysis was performed using Vindelöv's method for DNA measurements.38 Briefly, stained nuclei were analysed in a flow cytometer (FACScan, Becton Dickinson, Mountain View, CA) and the percentage of cells in each cycle phase was calculated by using the MacCycle software (Phoenix flow systems, San Diego, CA).
Karyotyping
Standard chromosome preparations were made of HMC-1 cells and conventional cytogenetic analysis was carried out on G-banded chromosomes.
Cell growth
Analysis of cell growth was performed by seeding triplicates of 5 × 104 cells/ml in 5 ml supplemented (see above) IMDM medium. Cells were counted every 24 hr using a Coulter Z1 cell counter (Coulter Electronics, Luton, UK).
The proliferation assay was performed by seeding triplicates of 100 µl cells (600 000 cells/ml) in cell culture medium. Ly 294002 was added at the beginning of the experiment. The cultures were incubated over night before addition of [3H]thymidine (0·5 µCi/well; Du Pont, Boston, MA). Incorporation of [3H]thymidine was measured after eight hours in a scintillation counter and expressed as mean counts per minute (c.p.m.).
Cell survival
Cell survival assay was performed by seeding 200 µl cells (600 000 cells/ml) in cell culture medium ± Ly 294002 at 10 or 50 µm. The cells were incubated for 48 h before the rate of survival was analysed using a flow cytometric assay and propidium iodide.
Flow cytometry
The cell surface antigen expression was analysed by indirect immunofluorescence using a FACScan (Becton Dickinson) as described.2 Monoclonal antibodies against the following cell surface markers were used: CD 13 (WM-15), CD 32 (IV.3), CD 44 (F-10-44-2), CD 54 (My13) from the 4th International Workshop of Human Leukocyte. The monoclonal antibody (mAb) against the α-chain of the FcεRI receptor (29C6)39 was kindly provided by Drs R. Chizzonite and F. Riske (Hoffmann-La Roche, Nutley, NJ); the antibody against Kit (YB5.B8)40 was a kind gift from Dr L. Ashman (Hanson Centre for Cancer Research, Adelaide, Australia), the mAbs against CD 49b (P1H5) and CD 49c (P1B5) were kindly provided by Prof K. Rubin (Uppsala University, Uppsala, Sweden), and the antibody against CD 18 (IB4) was a kind gift from Dr C. Lundberg (Pharmacia, Uppsala, Sweden). As negative controls, isotype-matched irrelevant antibodies were used (Dako Glostrup, Denmark).
Cell adhesion assay
For the adhesion assay, CytoMatrix cell adhesion strips (Chemicon International Inc. Temecula, CA) coated with human collagen type I, human collagen type IV, human fibronectin, human laminin, human vitronectin or human tenascin were used according to the manufacturer's recommendations. Strips coated with BSA were used for determination of background adhesion. HMC-1 cells were resuspended in serum-free IMDM medium and seeded at 50 × 103 cells/well ± 100 nm phorbol 12-myristate 13-acetate (PMA), incubated at 37° for 1 hr in a CO2 incubator and washed gently three times with PBS. One hundred µl of 0·2% crystal violet (Sigma) in 10% ethanol was then added to each well and incubated for 5 min at room temperature. The stain was removed and the wells were gently washed three times with phosphate-buffered saline (PBS). Cells were lysed by adding 100 µl of 1% sodium dodecyl sulphate (SDS) in PBS to each well, and the plate incubated on a shaker platform at room temperature until all cells were lysed and the stain dissolved. The absorbance was measured at 540 nm and an adhesion index was calculated by dividing the absorbance in wells coated with an extracellular matrix protein with the absorbance from the wells coated with bovine serum albumin (BSA).
Analysis of c-kit mutation
The analysis of the c-kit 560 and 816 mutation was performed using pyrosequencing™, a real-time pyrophosphate detection technology. Total RNA was prepared from HMC-1560, HMC-1560,816 and CBMC using TriPure isolation reagent (Roche, Basel, Switzerland). The RNA was then reverse-transcribed into complementary DNA (cDNA) using a first strand cDNA synthesis kit (Roche) and subjected to PCR amplification by a PCR core kit (Roche). Primers used in this study were obtained from Interactiva (Ulm, Germany). To be able to use the PCR product as a sequencing template, one of each PCR primer pair was covalently coupled to biotin. Primers (5′−3′ orientation) used were, biotin-CTT CCC GAA AGC TCC AG and CCC TGT TCA CTC CTT TGC, for the c-kitG560 mutation; and biotin-CAG GTG CCA TCC ACT TCA and CTT TCC TCG CCT CCA AGA for the c-kitV816 mutation. The amplification steps involved denaturation for 5 min at 94° followed by 30–35 cycles of denaturation for 30 s at 94°; annealing for 45 s at 63° for c-kitG560 at 65° for c-kitV816; extension for 30 s at 72°, and a final extension for 7 min at 72°. The PCR products were analysed on a 2% agarose gel, with the expected sizes for the c-kitG560 and c-kitV816 PCR products 248 bp and 174 bp, respectively. Biotinylated DNA fragments (20 µl) were attached to streptavidin-modified Dynabeads (Dynabeads™ M-280, Dynal A/S, Skøyen, Norway) according to a standard protocol by incubating at 65° for 10 min (shaking) in high salt (BW) buffer (0·05% Tween; 0·05 mm EDTA; 1 m NaCl; 5 mm Tris-HCl, pH 7·6). The Dynabeads were then captured using a PSQ 96 Sample Prep Tool with 96 magnetic ejectable microcylinders (Pyrosequencing AB, Uppsala, Sweden). This tool was also used for incubation (1 min) of the biotin–streptavidin complex in 0·5 m NaOH before washing in annealing buffer (200 mm Tris-HAc and 50 mm Mg-HAc). Subsequently, the samples were hybridized to 15 pmol of sequencing primers in annealing buffer at 80° for 2 min, followed by cooling to room temp. Sequencing primers (5′−3′ orientation) were AGT ACA GTG GAA GGT TG for the c-kitG560 mutation and TGT GAT TTT GGT CTA GCC for the c-kitV816 mutation. Pyrosequencing™ was performed using a SNP Reagent kit (Pyrosequencing AB) containing enzyme and substrate mixture, dATP-S, dCTP, dGTP and dTTP according to the manufacturer's instructions.
Mutation-specific restriction analysis
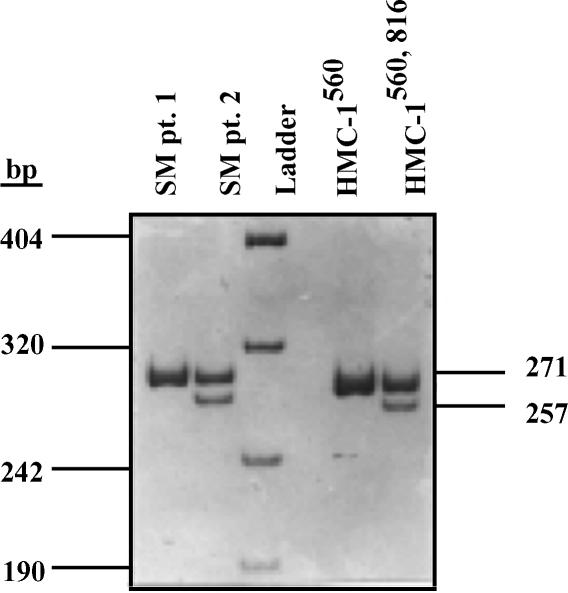
To analyse the A→T substitution at nt 8213 in c-kit, which produces a new HinfI restriction site (GANT-C), restriction endonuclease digestion of a PCR product was performed as described.29 Briefly, genomic DNA was extracted from peripheral blood mononuclear cells from two patients with mastocytosis, following informed consent, and from HMC-1 cells, and PCR-amplified. The amplified 332-bp single DNA fragment was digested with HinfI and separated on an 8% polyacrylamide gel. Since the V816 mutation creates a new HinfI site, predicted fragments were 257, 51 and 14 bp for the mutated and 271 and 51 for the wild type c-kit gene.
Immunoprecipitation and immunoblotting
The procedures of immunoprecipitation, gel electrophoresis, and immunoblotting were performed according to methods described.41 Briefly, lysates from untreated or SCF-treated cells were immunoprecipitated with a rabbit-polyclonal antibody against human Kit42 (kindly provided by Dr L. Rönnstrand, Ludwig Institute for Cancer Research, Uppsala, Sweden), collected with 50 µl of protein-A sepharose (Amersham Pharmacia Biotech) and washed in lysis buffer containing protease and phosphatase inhibitors. The immunoprecipitates were separated on 4–12% gradient polyacrylamide gels (Novex, San Diego, CA) and electrophoretically transferred from the gel to nitrocellulose membranes (Amersham Pharmacia Biotech). After blocking, immunoblotting was performed with 4G10 antiphosphotyrosine mAb and antibody recognizing the p85 subunit of phosphatidyl inositol (PI) 3-kinase and horseradish peroxidase-conjugated secondary antibodies (Amersham Pharmacia Biotech). p85 and phosphorylated Kit were visualized using enhanced chemiluminescence detection (LumiGLO, New England Biolabs). Phosphorylation analysis of Akt and ERK was carried out by separating total cell lysates on 4–12% gradient polyacrylamide gels (Novex); followed by transfer to nitrocellulose membranes (Amersham Pharmacia Biotech) and blocking. Blotting of membranes was performed using Abs specific for Akt, Phospho-Akt (Ser473), ERK-1 and phospho-p44/42 mitogen-activated protein (MAP) kinase. Proteins were visualized as described above.
Results
Cell morphology and proliferation rate
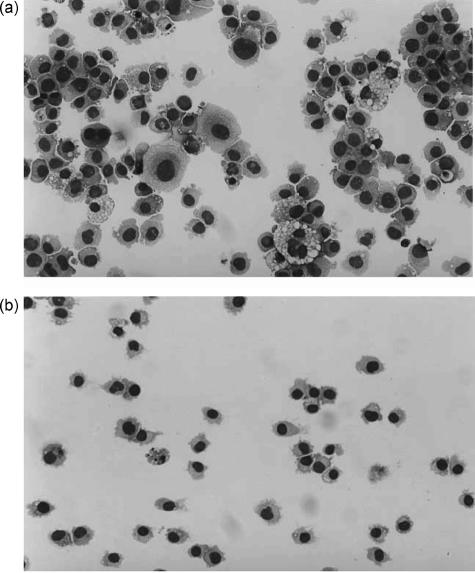
When culturing the HMC-1 sublines, we observed morphological and proliferative differences. HMC-1560 cells were heterogeneous in size and exhibited some homotypic aggregation (Fig. 1a). They were also more adherent than HMC-1560,816 cells, which grew in single cell suspension (Fig. 1b). HMC-1560,816 cells were smaller in size and tended to be more homogeneous (Fig. 1b).
Figure 1.
Morphologic analysis of HMC-1 cells. HMC-1560 (a) and HMC-1560,816 cells (b) were cytocentrifuged and stained with May–Grünwald Giemsa.
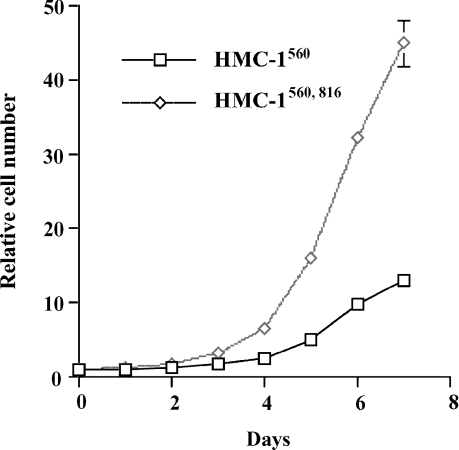
To measure cell growth in suspension, HMC-1560 and HMC-1560,816 cells were counted on days 1–7 after plating. Both HMC-1 variants grew without addition of growth factors. HMC-1560,816 cells exhibited a markedly increased proliferation rate compared with HMC-1560 cells. After 7 days there were approximately three times more cells in the HMC-1560,816 cultures compared with the number of cells in the HMC-1560 cultures (Fig. 2).
Figure 2.
Comparison of rate of proliferation for HMC-1560 and HMC-1560,816. Cells (5 × 104/ml) were seeded on day 0. Cell numbers on days 1–7 were determined using a Coulter cell counter. Relative cell number is the cell count divided by the number of seeded cells.
DNA profiles and karyotypes
We continued the characterization of the two HMC-1 sublines by cell cycle analysis. The DNA of HMC-1 cells was stained with propidium iodide and analysed by flow cytometry. HMC-1560,816 cells exhibited a normal diploid DNA profile, whereas the analysis of HMC-1560 cells indicated an increased DNA content (data not shown). This observation was then confirmed by karyotyping the cells that revealed that among 11 cells 4 were diploid (46 XX) and the remaining 7 were hyperdiploid with chromosome numbers ranging from 53 to 82. In contrast, the karyotype of analysed HMC-1560,816 cells were 46 XX (data not shown).
Phenotypic characterization of two HMC-1 sublines
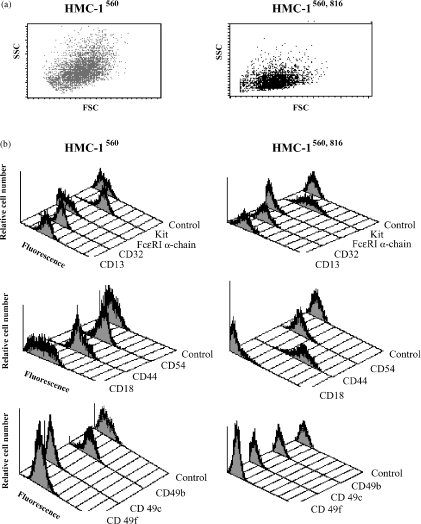
The observed morphological differences between the HMC-1 variants (Fig. 1) was verified by flow cytometer analysis (Fig. 3a) where HMC-1560 cells again were heterogeneous in size. Then, in order to characterise the differences in surface antigen expression of HMC-1560 and HMC-1560,816 cultures, cells were analysed using a panel of mAbs and flow cytometry (Fig. 3b). Both HMC-1560 and HMC-1560,816 cells displayed surface expression of CD13, CD32, CD44, CD54, and CD117 (Kit). HMC-1560 cells had a higher level of expression of the integrins CD18, CD49b, and CD49f. Both variants were negative for expression of the α-chain of the high affinity immunoglobulin E (IgE)-receptor complex.
Figure 3.
Flow cytometry analysis of expression of cell surface antigens on HMC-1560 and HMC-1560,816 cells. (a) The forward and light scatter of HMC-1560 cells (left) and HMC-1560,816 cells (right). Cells were stained as shown in (b) for expression of CD13, CD32, the α-chain of FcεRI, and Kit (first grouping), CD18, CD44 and CD54 (second grouping) and CD49b, CD49c and CD49f (bottom panel). Isotype-matched antibodies were used as controls.
Adhesion of HMC-1 cells to extracellular matrix proteins
In an adhesion assay, HMC-1560 cells, but not HMC-1560,816 cells, adhered to collagen I and collagen IV coated surfaces, while both adhered to fibronectin and laminin coated surfaces. After activation with a phorbol ester (PMA) HMC-1560, but not HMC-1560,816 cells, adhered to vitronectin and tenascin (data not shown).
c-kit mutations in HMC-1 cells
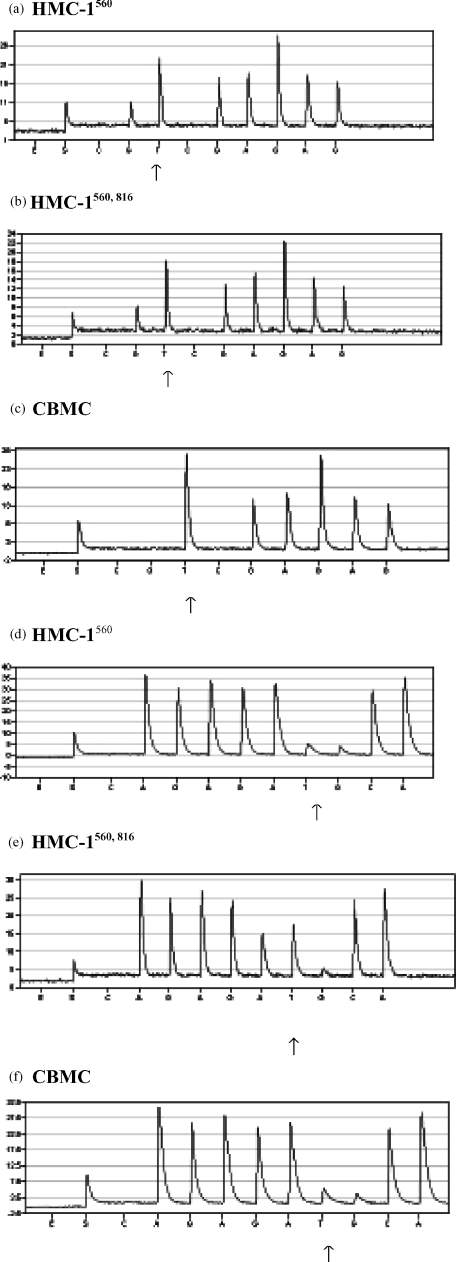
Kit has been reported to be constitutively activated in HMC-1 cells by two point mutations resulting in amino acid substitutions of Gly-560 for Val and Val-816 for Asp in the intracellular domain.26 These mutations are thought to cause malignant transformation of cells. However, the mechanism by which Kit is constitutively activated does not appear to be consistent and different mutations are found in distinct tumours/cancers. We thus evaluated the two HMC-1 variants for c-kitG560 and c-kitV816 mutations in cDNA. Evaluation of sequencing data was achieved by comparing peak heights in the pyrogram, where samples heterozygous for a point mutation produce half-height peaks for both allelic positions, whereas homozygous samples have a single base peak of full height.43 Target sequences were derived from the nucleotide sequence and organization of the human c-kit gene.44 Both variants of the HMC-1 cell line exhibited a heterozygous T→G mutation at codon 560 (Fig. 4a,b). This can be seen as a peak of half height for the G nucleotide, in comparison the CBMC wt sequence lack incorporation of G at this position (Fig. 4c). The c-kitV816 mutation was also found to be heterozygous in HMC-1. However, this A to T substitution was only found in HMC-1560,816 cells (Fig. 4d–f).
Figure 4.
Analysis of mutations at codon 560 (a–c) and at codon 816 (d–f). RNA from HMC-1560 cells (a,d), HMC-1560,816 cells (b,e) and CBMC (c,f) was extracted and used for RT–PCR. The PCR products were analysed for the 560 and 816 point mutations by means of pyrosequencing™. The arrows in a–c indicates the T→G substitution at position 560, and the arrows in d–f indicate the A→T substitution at position 816. E = enzyme, S = substrate.
The 816 mutation results in a new HinfI cleavage site in the c-kit gene.29 This enabled us to confirm the existence of this mutation in HMC-1560,816. A 322-bp PCR product of genomic c-kit DNA was cleaved with HinfI and digested PCR fragments were separated on a polyacrylamide gel. As predicted from the sequencing data, only HMC-1560,816 cells contained the restriction site and hence the point mutation at codon 816. The point mutation in HMC-1560,816 cells and a mutation in one of the mastocytosis patients were both found to be heterozygous (Fig. 5).
Figure 5.
c-kit mutations in DNA from HMC-1 variants and patients with systemic mastocytosis (SM). In c-kit, the A-to-T substitution at nucleotide 8213 of exon 17 creates a new HinfI restriction site. Genomic DNA from patients with systemic mastocytosis and from HMC-1560 and HMC-1560,816 cells was extracted and a segment of the DNA including this site was amplified by primers complimentary to the sequence of introns 16 and 17 of the c-kit gene. The PCR product was digested with HinfI and separated on polyacrylamide gel. Predicted sizes of digested fragments are 271 and 51 bp for wild type, and 257, 51 and 14 bp for mutated c-kit. Samples are: lane 1, patient 1 with indolent mastocytosis; lane 2, patient 2 with mastocytosis with an associated hematologic disorder; lane 3, ladder; lane 4, HMC-1560; and lane 5, HMC-1560 816.
Constitutive tyrosine phosphorylation of Kit
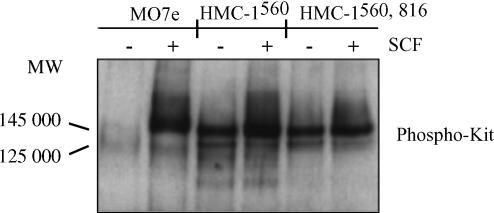
Ligand binding is known to induce tyrosine phosphorylation of Kit. In order to determine the effect of SCF on mutated and normal Kit, the two HMC-1 variants and MO7e cells were investigated for tyrosine phosphorylation of Kit. Both the mature, glycosylated (145 000 MW) and the immature (125 000 MW) forms of c-kit gene products were identified and found to be phosphorylated on tyrosine in HMC-1560 and HMC-1560,816 cells (Fig. 6). This phosphorylation on tyrosine was constitutive. However, stimulation with SCF increased the baseline phosphorylation. Unstimulated MO7e cells were found to have unphosphorylated receptors but in the presence of SCF the mature 145 000 MW form of Kit was phosphorylated.
Figure 6.
Phosphorylation of Kit in HMC-1 and MO7e cells. Cells were treated with SCF (100 ng/ml) for 15 min at 37° and lysed. Kit was immunoprecipitated with a rabbit anti-human-Kit antibody and protein A-sepharose beads from the cell lysates. Immunoblot was performed with the antiphosphotyrosine mAb 4G10.
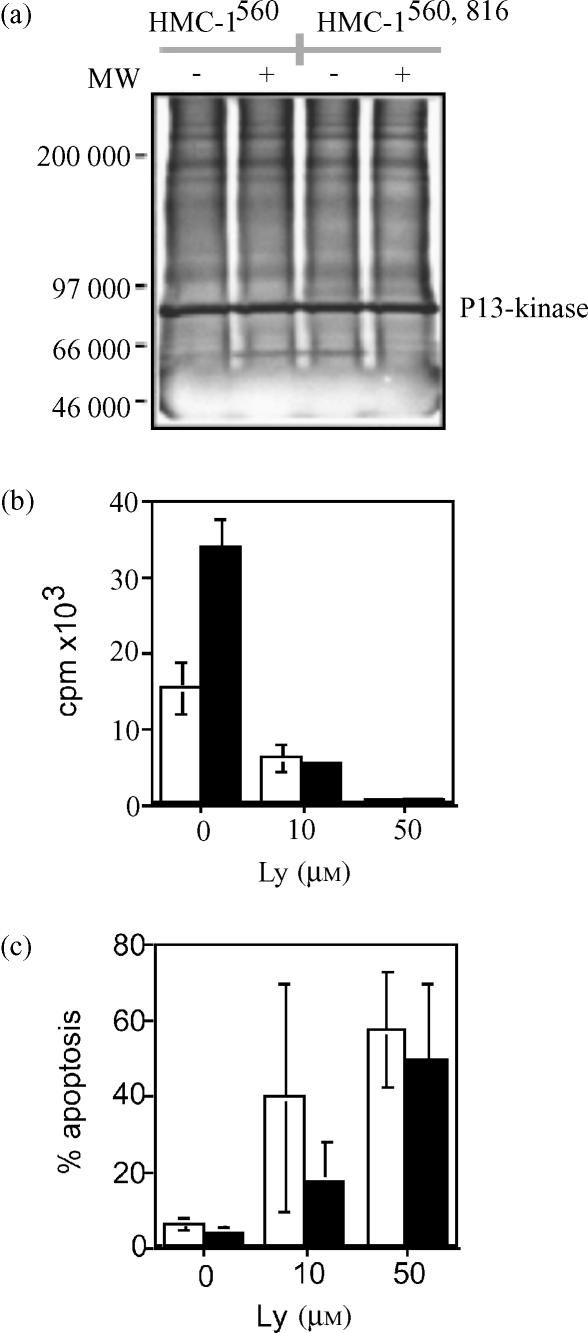
Kit phosphorylation is known to activate PI 3-kinase- and MAP kinase pathways, considered to be involved in the regulation of cell survival and proliferation of mast cells. We therefore analysed the association of the p85 subunit of PI 3-kinase to Kit, as well as the phosphorylation status of Akt and ERK in the two HMC-1 variants. Kit association with PI 3-kinase was investigated by immunoprecipitation of Kit followed by SDS–polyacrylamide gel electrophoresis (PAGE) and immunoblotting with anti-p85 antibodies. An identical pattern was found for the two variants of HMC-1. Kit was associated with the p85 subunit in a SCF independent manner (Fig. 7a).
Figure 7.
Intracellular signalling downstream of Kit in HMC-1 cells. (a) Kit immunoprecipitates were separated on a gel and analysis of association of c-Kit with PI 3-kinase was performed by immunoblotting with an antibody recognising the p85 regulatory subunit of PI 3-kinase. The effect of the PI3-kinase inhibitor Ly 29400 on HMC-1 proliferation (b) and apoptosis (c) was investigated. Open bars represent HMC-1560; and filled bars HMC-1560, 816.
Subsequently, the effect of a PI 3-kinase inhibitor, Ly 294002, was investigated on the proliferation and survival. As shown in Figs 7(b,c), treatment with Ly inhibited proliferation and induced apoptosis in both HMC-1560 and HMC-1560,816 cells.
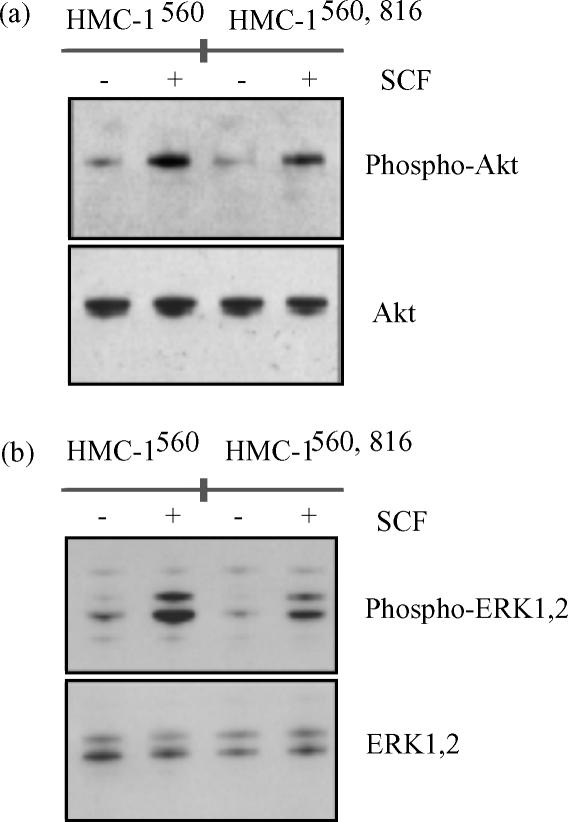
We next investigated the phosphorylation of Akt in total cell lysates. Akt is a serine-threonine kinase involved in an anti-apoptotic signalling pathway downstream of PI 3-kinase.45 Although PI 3-kinase was constitutively associated with Kit, Akt exhibited only minor phosphorylation in the absence of SCF. Addition of SCF induced Akt phosphorylation in both HMC-1560 and HMC-1560,816 cells (Fig. 8a). Phosphorylation of ERK, in the Ras-Raf-MAP kinase pathway, also showed a similar pattern in HMC-1560 and HMC-1560,816 cells. Unstimulated cells exhibited only a modest or little phosphorylation mainly on ERK2. However, stimulation with 100 ng/ml SCF for 10 min induced substantial phosphorylation of both ERK1 and ERK2 (Fig. 8b).
Figure 8.
Phosphorylation of Akt and ERK. Cells were treated with SCF (100 ng/ml) for 15 min and lysed. Total cell lysates were analysed for phosphorylation of Akt (a) or ERK (b). Antibodies against total AKT and ERK, respectively, were used for control of loading.
Discussion
The HMC-1 cell line is the only established human mast cell line. The purpose of this study was to characterise the phenotype, the set of disease associated gain-of function mutations in the c-kit gene, and the signalling outcome of these mutations in two sublines of the HMC-1 cell line. Based on their c-kit mutations, we have decided to designate these two variants as HMC-1560 and HMC-1560,816. When culturing these cells we observed differences in both morphology (Fig. 1) and cellular growth rate (Fig. 2). The two variants also exhibited different karyotypes, as many HMC-1560 cells were found to be hyperdiploid. Such aneuploidy has been observed in virtually all analysed solid cancers (reviewed by Li et al.46). However, aneuploidy among HMC-1 cells was not observed during the initial establishment and characterization made by Butterfield et al. in 19881 and therefore may represent an outcome of long-term continuous in vitro culture. The pattern of cell surface antigen expression also differed between the two variants. Interestingly, by point mutation analysis using the pyrosequencing™ method it was found that HMC-1560 and HMC-1560,816 exhibited a divergent set of mutations in the c-kit gene. Corresponding mutations in mouse, c-kitG559 and c-kitV814 have been reported to activate Kit in a SCF independent manner in transfected cells and may alter Kit substrate specificity.26,33–35,47 HMC-1560 cells were found to be heterozygous for c-kitG560 but negative for c-kitV816. In contrast, HMC-1560,816 cells were heterozygous for both mutations (Figs 4 and 5).
These mutations are associated with the development of different diseases. Mutation at codon 560 or alterations in the juxtamembrane region near this codon is mainly associated with GISTs.27 Similarly, malignant canine mastocytomas often have mutations in the juxtamembrane region of Kit.25 In contrast, human mast cell disorders are generally negative for mutations in the juxtamembrane region, with the exception of one report of indolent mastocytosis patients with an activating c-kit mutation at codon 560.36 However, the other activating point mutation, c-kitV816 is common in mastocytosis. It is hypothesized that this mutation is necessary for the persistence or progression of adult onset mastocytosis and a subset of paediatric cases.28
Point mutations in the c-kit proto-oncogene and the subsequent activation of Kit are thought to change the cell into a more tumorigenic type with a higher rate of proliferation. As the two variants of HMC-1 cells express different forms of mutated Kit, we investigated the rate of cell growth for the two HMC-1 sublines. We found that HMC-1560,816 cells had a higher rate of proliferation than that of HMC-1560 cells. This may be due to the presence of the activating V816 mutation in HMC-1560,816, as IC-2 and FDCP-1 cells transfected with murine c-kitV814 also exhibit a higher proliferation rate than cells transfected with murine c-kitG559.33,34 Furthermore, mast cells cultured from patients with indolent or aggressive variants of mastocytosis had a higher rate of proliferation if compared with mast cells from normal subjects.48 Interestingly, the V816 mutation also appears to be involved in SCF-induced migration of mast cell progenitor cells. We have previously reported that cells from mastocytosis patients having this mutation migrate to a higher extent towards SCF if compared with cells having wild type Kit.49 In that study we also used the HMC-1 cell lines but referred to them as HMC-1.1 and HMC-1.2, respectively.49
Subsequently, we analysed the rate of tyrosine phosphorylation of Kit. Both HMC-1 variants were found to have the same magnitude of constitutive phosphorylation. To determine how this Kit phosphorylation might affect downstream kinase signalling pathways, Kit immunoprecipitates were analysed for association with PI 3-kinase. These experiments revealed that in both HMC-1 variants, the regulatory p85 subunit of PI 3-kinase was constitutively associated with Kit. Thus, there appears to be a loss of normal regulated activity of Kit in HMC-1 cells. Our results are in agreement with Furitsu et al. who described that c-kit560,816 in HMC-1 cells is autophosphorylated and associated with PI3-kinase.26
Similar findings have been reported for cells transfected with c-kit816. D816V c-kit mutant have been described to be constitutively associated with PI3-kinase, leading to higher levels of PI3-kinase activity than unstimulated wild-type c-kit.50 In another study it was shown that transfected MO7e cells had a constitutive activation of Akt, which could be blocked with a PI 3-kinase inhibitor.51 In the same study they showed that the Ras pathway is not activated in transfected cells. Surprisingly, although Kit is constitutively phosphorylated and associated with p85, neither of the two variants of HMC-1 had a constitutive phosphorylation of Akt or ERK (Fig. 8). Therefore, we speculate that Akt and ERK signalling pathways do not play a major role in the growth factor independent proliferation and survival observed in HMC-1 cells (Fig. 2). This is in agreement with previous observations that c-kitV816 does not lead to ERK phosphorylation and that ERK activity is not sufficient for induction of mast cell mitogenesis.,51,52 SCF-induced proliferation has instead been reported to be dependent of PI 3-kinase and Src, which converged to activate Rac1 and JNK.52
In summary, these characterizations help us to better understand the mechanism behind the transforming capacity of the juxtamembrane and c-kitV816 mutations found in GIST and mastocytosis, respectively. We are currently using HMC-1560 and HMC-1560,816 cells in a study of the impact of Kit inhibitors on deregulated cellular responses due to activating point mutations in different regions of Kit. The availability of two distinct cultured human mast cell lines will also be useful in future studies of mast cell biology.
Acknowledgments
We wish to thank Tekli Semere for technical help, Anna-Lotta Schiller (Pyrosequencing AB) for assistance in the pyrosequencing analysis and Marie Fischer for critical comments on the manuscript. This work was supported by grants from the Swedish Cancer Society, Hans von Kantzow's foundation, Ollie and Elof Ericsson's foundation, King Gustaf V's 80 years foundation, Selanders foundation and Göran Gustafsson's Foundation.
Abbreviations
- HMC-1
human mast cell line 1
- PI 3-kinase
phosphatidylinositol 3′-kinase
- ERK
extracellular regulated protein kinase
- SCF
stem cell factor
- FcεRI
high affinity IgE-receptor I
- GIST
gastrointestinal stromal tumour
- GM-CSF
granulocyte–macrophage colony-stimulating factor
- CBMC
cord blood derived mast cells
- PMA
phorbol-12- myristate-13-acetate
References
- 1.Butterfield JH, Weiler D, Dewald G, Gleich GJ. Establishment of an immature mast cell line from a patient with mast cell leukemia. Leuk Res. 1988;12:345–55. doi: 10.1016/0145-2126(88)90050-1. [DOI] [PubMed] [Google Scholar]
- 2.Nilsson G, Blom T, Kusche-Gullberg M, Kjellen L, Butterfield JH, Sundstrom C, Nilsson K, Hellman L. Phenotypic characterization of the human mast-cell line HMC-1. Scand J Immunol. 1994;39:489–98. doi: 10.1111/j.1365-3083.1994.tb03404.x. [DOI] [PubMed] [Google Scholar]
- 3.Valent P, Bettelheim P. Cell surface structures on human basophils and mast cells: biochemical and functional characterization. Adv Immunol. 1992;52:333–423. doi: 10.1016/s0065-2776(08)60879-2. [DOI] [PubMed] [Google Scholar]
- 4.Irani AM, Nilsson G, Miettinen U, Craig SS, Ashman LK, Ishizaka T, Zsebo KM, Schwartz LB. Recombinant human stem cell factor stimulates differentiation of mast cells from dispersed human fetal liver cells. Blood. 1992;80:3009–21. [PubMed] [Google Scholar]
- 5.Li L, Macpherson JJ, Adelstein S, Bunn CL, Atkinson K, Phadke K, Krilis SA. Conditioned media from a cell strain derived from a patient with mastocytosis induces preferential development of cells that possess high affinity IgE receptors and the granule protease phenotype of mature cutaneous mast cells. J Biol Chem. 1995;270:2258–63. doi: 10.1074/jbc.270.5.2258. [DOI] [PubMed] [Google Scholar]
- 6.Nilsson G, Blom T, Harvima I, Kusche-Gullberg M, Nilsson K, Hellman L. Stem cell factor-dependent human cord blood derived mast cells express alpha- and beta-tryptase, heparin and chondroitin sulphate. Immunology. 1996;88:308–14. doi: 10.1111/j.1365-2567.1996.tb00020.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nilsson G, Forsberg K, Bodger MP, Ashman LK, Zsebo KM, Ishizaka T, Irani AM, Schwartz LB. Phenotypic characterization of stem cell factor-dependent human foetal liver-derived mast cells. Immunology. 1993;79:325–30. [PMC free article] [PubMed] [Google Scholar]
- 8.Toru H, Ra C, Nonoyama S, Suzuki K, Yata J, Nakahata T. Induction of the high-affinity IgE receptor (Fc epsilon RI) on human mast cells by IL-4. Int Immunol. 1996;8:1367–73. doi: 10.1093/intimm/8.9.1367. [DOI] [PubMed] [Google Scholar]
- 9.Burd PR, Thompson WC, Max EE, Mills FC. Activated mast cells produce interleukin 13. J Exp Med. 1995;181:1373–80. doi: 10.1084/jem.181.4.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feoktistov I, Biaggioni I. Adenosine A2b receptors evoke interleukin-8 secretion in human mast cells. An enprofylline-sensitive mechanism with implications for asthma. J Clin Invest. 1995;96:1979–86. doi: 10.1172/JCI118245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gauchat JF, Henchoz S, Mazzei G, et al. Induction of human IgE synthesis in B cells by mast cells and basophils. Nature. 1993;365:340–3. doi: 10.1038/365340a0. [DOI] [PubMed] [Google Scholar]
- 12.Grabbe J, Welker P, Moller A, Dippel E, Ashman LK, Czarnetzki BM. Comparative cytokine release from human monocytes, monocyte-derived immature mast cells, and a human mast cell line (HMC-1) J Invest Dermatol. 1994;103:504–8. doi: 10.1111/1523-1747.ep12395649. [DOI] [PubMed] [Google Scholar]
- 13.Legler DF, Loetscher M, Jones SA, Dahinden CA, Arock M, Moser B. Expression of high- and low-affinity receptors for C3a on the human mast cell line, HMC-1. Eur J Immunol. 1996;26:753–8. doi: 10.1002/eji.1830260405. [DOI] [PubMed] [Google Scholar]
- 14.Moller A, Lippert U, Lessmann D, et al. Human mast cells produce IL-8. J Immunol. 1993;151:3261–6. [PubMed] [Google Scholar]
- 15.Nilsson G, Butterfield JH, Nilsson K, Siegbahn A. Stem cell factor is a chemotactic factor for human mast cells. J Immunol. 1994;153:3717–23. [PubMed] [Google Scholar]
- 16.Selvan RS, Butterfield JH, Krangel MS. Expression of multiple chemokine genes by a human mast cell leukemia. J Biol Chem. 1994;269:13893–8. [PubMed] [Google Scholar]
- 17.Sillaber C, Strobl H, Bevec D, et al. IL-4 regulates c-kit proto-oncogene product expression in human mast and myeloid progenitor cells. J Immunol. 1991;147:4224–8. [PubMed] [Google Scholar]
- 18.Valent P, Bevec D, Maurer D, et al. Interleukin 4 promotes expression of mast cell ICAM-1 antigen. Proc Natl Acad Sci USA. 1991;88:3339–42. doi: 10.1073/pnas.88.8.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Virgolini I, Li SR, Yang Q, et al. Characterization of LDL and VLDL binding sites on human basophils and mast cells. Arterioscler Thromb Vasc Biol. 1995;15:17–26. doi: 10.1161/01.atv.15.1.17. [DOI] [PubMed] [Google Scholar]
- 20.Besmer P, Murphy JE, George PC, et al. A new acute transforming feline retrovirus and relationship of its oncogene v-kit with the protein kinase gene family. Nature. 1986;320:415–21. doi: 10.1038/320415a0. [DOI] [PubMed] [Google Scholar]
- 21.Galli SJ, Zsebo KM, Geissler EN. The kit ligand, stem cell factor. Adv Immunol. 1994;55:1–96. doi: 10.1016/s0065-2776(08)60508-8. [DOI] [PubMed] [Google Scholar]
- 22.Nilsson G, Metcalfe DD. Contemporary issues in mast cell biology. Allergy Asthma Proc. 1996;17:59–63. doi: 10.2500/108854196778645074. [DOI] [PubMed] [Google Scholar]
- 23.Tsujimura T, Furitsu T, Morimoto M, Kanayama Y, Nomura S, Matsuzawa Y, Kitamura Y, Kanakura Y. Substitution of an aspartic acid results in constitutive activation of c-kit receptor tyrosine kinase in a rat tumor mast cell line RBL-2H3. Int Arch Allergy Immunol. 1995;106:377–85. doi: 10.1159/000236870. [DOI] [PubMed] [Google Scholar]
- 24.Tsujimura T, Furitsu T, Morimoto M, Isozaki K, Nomura S, Matsuzawa Y, Kitamura Y, Kanakura Y. Ligand-independent activation of c-kit receptor tyrosine kinase in a murine mastocytoma cell line P-815 generated by a point mutation. Blood. 1994;83:2619–26. [PubMed] [Google Scholar]
- 25.Ma Y, Longley BJ, Wang X, Blount JL, Langley K, Caughey GH. Clustering of activating mutations in c-KIT's juxtamembrane coding region in canine mast cell neoplasms. J Invest Dermatol. 1999;112:165–70. doi: 10.1046/j.1523-1747.1999.00488.x. [DOI] [PubMed] [Google Scholar]
- 26.Furitsu T, Tsujimura T, Tono T, et al. Identification of mutations in the coding sequence of the proto- oncogene c-kit in a human mast cell leukemia cell line causing ligand- independent activation of c-kit product. J Clin Invest. 1993;92:1736–44. doi: 10.1172/JCI116761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirota S, Isozaki K, Moriyama Y, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–80. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 28.Longley BJ, Metcalfe DD, Tharp M, Wang X, Tyrrell L, Lu SZ, Heitjan D, Ma Y. Activating and dominant inactivating c-KIT catalytic domain mutations in distinct clinical forms of human mastocytosis. Proc Natl Acad Sci USA. 1999;96:1609–14. doi: 10.1073/pnas.96.4.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagata H, Worobec AS, Oh CK, Chowdhury BA, Tannenbaum S, Suzuki Y, Metcalfe DD. Identification of a point mutation in the catalytic domain of the protooncogene c-kit in peripheral blood mononuclear cells of patients who have mastocytosis with an associated hematologic disorder. Proc Natl Acad Sci USA. 1995;92:10560–4. doi: 10.1073/pnas.92.23.10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Worobec AS, Semere T, Nagata H, Metcalfe DD. Clinical correlates of the presence of the Asp816Val c-kit mutation in the peripheral blood mononuclear cells of patients with mastocytosis. Cancer. 1998;83:2120–9. doi: 10.1002/(sici)1097-0142(19981115)83:10<2120::aid-cncr10>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 31.Vliagoftis H, Worobec AS, Metcalfe DD. The protooncogene c-kit and c-kit ligand in human disease. J Allergy Clin Immunol. 1997;100:435–40. doi: 10.1016/s0091-6749(97)70131-3. [DOI] [PubMed] [Google Scholar]
- 32.Nilsson G, Costa JJ, Metcalfe DD. Mast cells and basophils. In: Gallin JI, Snyderman R, editors. Inflammation: Basic Principles and Clinical Correlates. 3. Philadelphia: Lippincott-Raven; 1999. pp. 97–117. [Google Scholar]
- 33.Hashimoto K, Tsujimura T, Moriyama Y, et al. Transforming and differentiation-inducing potential of constitutively activated c-kit mutant genes in the IC-2 murine interleukin-3-dependent mast cell line. Am J Pathol. 1996;148:189–200. [PMC free article] [PubMed] [Google Scholar]
- 34.Kitayama H, Kanakura Y, Furitsu T, et al. Constitutively activating mutations of c-kit receptor tyrosine kinase confer factor-independent growth and tumorigenicity of factor-dependent hematopoietic cell lines. Blood. 1995;85:790–1004. [PubMed] [Google Scholar]
- 35.Kitayama H, Tsujimura T, Matsumura I, et al. Neoplastic transformation of normal hematopoietic cells by constitutively activating mutations of c-kit receptor tyrosine kinase. Blood. 1996;88:995–1004. [PubMed] [Google Scholar]
- 36.Buttner C, Henz BM, Welker P, Sepp NT, Grabbe J. Identification of activating c-kit mutations in adult-, but not in childhood-onset indolent mastocytosis: a possible explanation for divergent clinical behavior. J Invest Dermatol. 1998;111:1227–31. doi: 10.1046/j.1523-1747.1998.00414.x. [DOI] [PubMed] [Google Scholar]
- 37.Taniguchi M, Nishida T, Hirota S, Isozaki K, Ito T, Nomura T, Matsuda H, Kitamura Y. Effect of c-kit mutation on prognosis of gastrointestinal stromal tumors. Cancer Res. 1999;59:4297–300. [PubMed] [Google Scholar]
- 38.Vindelov LL, Christensen IJ, Nissen NI. A detergent-trypsin method for the preparation of nuclei for flow cytometric DNA analysis. Cytometry. 1983;3:323–7. doi: 10.1002/cyto.990030503. [DOI] [PubMed] [Google Scholar]
- 39.Riske F, Hakimi J, Mallamaci M, et al. High affinity human IgE receptor (Fc epsilon RI). Analysis of functional domains of the alpha-subunit with monoclonal antibodies. J Biol Chem. 1991;266:11245–51. [PubMed] [Google Scholar]
- 40.Lerner NB, Nocka KH, Cole SR, Qiu FH, Strife A, Ashman LK, Besmer P. Monoclonal antibody YB5.B8 identifies the human c-kit protein product. Blood. 1991;77:1876–83. [PubMed] [Google Scholar]
- 41.Sundström M, Alfredsson J, Olsson N, Nilsson G. Stem Cell Factor-Induced Migration of Mast Cells Requires p38 Mitogen- Activated Protein Kinase Activity. Exp Cell Res. 2001;267:144–51. doi: 10.1006/excr.2001.5239. [DOI] [PubMed] [Google Scholar]
- 42.Blume-Jensen P, Claesson-Welsh L, Siegbahn A, Zsebo KM, Westermark B, Heldin CH. Activation of the human c-kit product by ligand-induced dimerization mediates circular actin reorganization and chemotaxis. EMBO J. 1991;10:4121–8. doi: 10.1002/j.1460-2075.1991.tb04989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alderborn A, Kristofferson A, Hammerling U. Determination of single-nucleotide polymorphisms by real-time pyrophosphate DNA sequencing. Genome Res. 2000;10:1249–58. doi: 10.1101/gr.10.8.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giebel LB, Strunk KM, Holmes SA, Spritz RA. Organization and nucleotide sequence of the human KIT (mast/stem cell growth factor receptor) proto-oncogene. Oncogene. 1992;7:2207–17. [PubMed] [Google Scholar]
- 45.Blume-Jensen P, Janknecht R, Hunter T. The kit receptor promotes cell survival via activation of PI 3-kinase and subsequent Akt-mediated phosphorylation of Bad on Ser136. Curr Biol. 1998;8:779–82. doi: 10.1016/s0960-9822(98)70302-1. [DOI] [PubMed] [Google Scholar]
- 46.Li R, Sonik A, Stindl R, Rasnick D, Duesberg P. Aneuploidy vs. gene mutation hypothesis of cancer: recent study claims mutation but is found to support aneuploidy. Proc Natl Acad Sci USA. 2000;97:3236–41. doi: 10.1073/pnas.040529797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piao X, Bernstein A. A point mutation in the catalytic domain of c-kit induces growth factor independence, tumorigenicity, and differentiation of mast cells. Blood. 1996;87:3117–23. [PubMed] [Google Scholar]
- 48.Rottem M, Okada T, Goff JP, Metcalfe DD. Mast cells cultured from the peripheral blood of normal donors and patients with mastocytosis originate from a CD34+/Fc epsilon RI− cell population. Blood. 1994;84:2489–96. [PubMed] [Google Scholar]
- 49.Taylor ML, Dastych J, Sehgal D, Sundstrom M, Nilsson G, Akin C, Mage RG, Metcalfe DD. The Kit-activating mutation D816V enhances stem cell factor-dependent chemotaxis. Blood. 2001;98:1195–9. doi: 10.1182/blood.v98.4.1195. [DOI] [PubMed] [Google Scholar]
- 50.Chian RJ, Young S, Danlkovitch-Miagkova A, Rönnstrand L, Leonard E, Ferrao P, Ashman L, Linnekin D. Phosphatidylinositol 3 kinase contributes to the transformation of hematopoietic cell by the D816V c-Kit mutant. Blood. 2001;98:1365–73. doi: 10.1182/blood.v98.5.1365. [DOI] [PubMed] [Google Scholar]
- 51.Ning ZQ, Li J, Arceci RJ. Signal transducer and activator of transcription 3 activation is required for Asp (816) mutant c-Kit-mediated cytokine-independent survival and proliferation in human leukemia cells. Blood. 2001;97:3559–67. doi: 10.1182/blood.v97.11.3559. [DOI] [PubMed] [Google Scholar]
- 52.Timokhina I, Kissel H, Stella G, Besmer P. Kit signaling through PI 3-kinase and Src kinase pathways: an essential role for Rac1 and JNK activation in mast cell proliferation. EMBO J. 1998;17:6250–62. doi: 10.1093/emboj/17.21.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]