Abstract
CD8+ T lymphocytes play a major role in the clearance of bovine respiratory syncytial virus (BRSV), an important respiratory pathogen of young calves that shares many of the epidemiological and pathological features of human respiratory syncytial virus (HRSV) in infants. Recombinant vaccinia virus (rVV) and recombinant fowlpox virus (rFPV), expressing individual BRSV proteins, were used to demonstrate that the F, N and M2 proteins were the major antigens recognized by bovine CD8+ T cells in major histocompatibility complex (MHC)-defined cattle. BRSV protein recognition by CD8+ T cells was analysed using cytotoxic T lymphocyte (CTL) assays or by the production of interferon-γ (IFN-γ) following restimulation with BRSV proteins. Strong recognition of the G protein by CD8+ T cells was observed in cattle that had been vaccinated with rVV expressing this protein and subsequently challenged with BRSV. Although there is variation in the number of expressed MHC genes in cattle with different class I haplotypes, this did not appear to influence BRSV protein recognition by CD8+ T cells. Knowledge of the antigenic specificity of BRSV-specific CD8+ T cells will facilitate the qualitative and quantitative analysis of BRSV-specific CD8+ T-cell memory in cattle and help to ensure that potential vaccines induce a qualitatively appropriate CD8+ T-cell response.
Introduction
Bovine respiratory syncytial virus (BRSV) is a major cause of lower respiratory tract disease in young calves and has also been associated with outbreaks of respiratory disease in dairy cows.1,2 BRSV is closely related to human respiratory syncytial virus (HRSV), which is the predominant cause of hospitalization of infants with pneumonia and bronchiolitis, and the epidemiology and pathogenesis of infection with these viruses are similar.1 There are two major problems that have hampered the development of effective bovine and human RSV vaccines. First, prior vaccination can enhance the severity of disease following infection, indicating that the immune response can contribute to the pathogenesis of disease;3,4 and second, natural infection does not provide complete protection against reinfection.1,5 The role of the immune response in protection and pathogenesis of RSV infections has been studied extensively in a murine model of HRSV. Such studies have demonstrated that both CD8+ and CD4+ T cells not only play a role in virus clearance, but also contribute to lung pathology.6–9 However, the utility of this model is limited because the disease produced in mice infected with HRSV differs in a number of important aspects from that in the natural host. BRSV is therefore not only an important pathogen in its own right, but is also a valuable model for HRSV.
Studies in calves have demonstrated that the F and G surface glycoproteins, and the N protein, are major protective antigens of BRSV;10 that neutralizing, fusion-inhibiting antibodies to the F protein can mediate protection;11 and that CD8+ T cells play a crucial role in virus clearance.12 Therefore, calves in which CD8+ T cells have been depleted showed delayed virus clearance and increased pulmonary pathology, similar to that seen in HRSV-infected immunocompromised individuals.13 In contrast, the kinetics of BRSV infection was unaffected in calves depleted of CD4+ or γδ T cells.12 BRSV-specific, major histocompatibility complex (MHC) class I-restricted cytotoxic T lymphocytes (CTLs) can be detected in the blood and lungs of calves 7–10 days after infection with BRSV,14 and CD8+ T-cell numbers are increased in the nasal passages, trachea and lungs at this time.15 Taken together, these studies indicate that an effective vaccine should prime both virus-specific CD8+ T cells and antibody.
The lack of complete protection against reinfection with RS viruses may be related to antigenic variation between virus strains, short-lived immunological memory and/or a defect in T-cell effector function at the site of virus replication. Given the crucial role of CD8+ T cells in clearance of BRSV, the short duration of protection following BRSV infection may be caused by poor CD8+ T-cell memory. Knowledge of the antigenic specificity of BRSV-specific CD8+ T cells will allow the development of methodologies to investigate qualitative and quantitative aspects of BRSV-specific CD8+ T-cell responses and investigation of factors that regulate the generation and long-term survival of memory CTLs. CD8+ T-cell responses tend to be focused on a limited number of dominant epitopes in individual animals. Consequently, the response of different animals may be directed towards different proteins. Therefore, it is also important to determine the extent to which the MHC influences the antigenic specificity of BRSV-specific CD8+ T-cell responses.
Following HRSV infection of BALB/c mice, the major target for HRSV-specific CTLs is the M2 protein,16 and CD8+ T cells mediate protection against HRSV in mice vaccinated with recombinant vaccinia virus (rVV) expressing the M2 protein.17,18 However, resistance induced by rVV-M2 is short-lived.19 CTLs from HRSV-infected BALB/c mice also recognize the HRSV F and N proteins, but there is little or no recognition of the SH, M, P or G proteins.16 Identification of Kd-restricted epitopes in the HRSV M220 and F proteins21 has provided the opportunity to quantify antigen-specific CTLs in the lungs of HRSV-infected mice and to analyse the kinetics of pulmonary HRSV-specific CTLs following infection.21 These studies have demonstrated that HRSV infection in mice inhibits pulmonary CD8+ T-cell effector activity by interfering with T-cell receptor (TCR)-mediated signalling.21,22 Such an effect may help to explain the limited duration of protective immunity in RSV infections.
The major target protein of HRSV recognized by human CTLs appears to be the N protein.23,24 Human CTLs also recognize the SH, F, M and NS1 proteins, but few of the individuals studied recognized the M2, G, P or NS2 proteins. Recognition of the F and M2 proteins appears to be associated with recent infection and there does not appear to be any correlation between MHC phenotype and HRSV protein recognition.24 There is little information on the epitopes recognized by HRSV-specific human CD8+ T cells. Two human leucocyte antigen (HLA)-restricted CTL epitopes in the HRSV F protein, and one in the N protein, have been identified.25,26 Identification of the HLA B7-restricted epitope in the HRSV N protein allowed the construction of HLA-B7–RSV–NP tetrameric complexes that were used to stain human CD8+ T cells.25 These studies demonstrated that only a small number of CD8+ T cells are specific for this HRSV N-protein epitope in the peripheral blood of healthy adults.
The BRSV proteins recognized by bovine CD8+ T cells have not been identified. In this study, rVV and/or recombinant fowlpox virus (rFPV) expressing the F, G, N and M2 proteins of BRSV were used in CTL assays or in assays to detect interferon-γ (IFN-γ) production by CD8+ T cells to identify BRSV proteins recognized by bovine CD8+ T cells in MHC-defined cattle. MHC class I expression in cattle is unusually complex. International workshops have identified approximately 50 class I specificities,27 and it was assumed that the majority of these represented products at a single locus. More recent molecular studies have shown that haplotypes may express one, two or three class I genes from a putative total of five, in any combination.28 At present, only about 40 full-length cDNA sequences are available and thus DNA-based typing methods are of limited use. However, a polymerase chain reaction using sequence-specific primers (PCR–SSP) has been developed29 for typing (at the level of expressed genes) the inbred cattle herd produced at the IAH (Compton, Newbury, UK). Many of the animals used in this study were those in which MHC class I haplotypes have been completely characterized at the level of expressed genes. Knowledge of the BRSV proteins and epitopes recognized by bovine CD8+ T cells will increase our understanding of the role of CD8+ T cells in RSV infections and will help to ensure that vaccination induces a qualitatively appropriate CTL response.
Materials and methods
Cattle
Calves that were used to investigate the MHC restriction of the primary BRSV-specific CTL response were BRSV seronegative, gnotobiotic or specific pathogen-free (SPF) calves, housed in a barrier-maintained building. The haplotypes of these calves were characterized at the level of the expressed class I genes. Calves were infected intranasally (i.n.) and intratracheally (i.t.) with 2 × 105 plaque-forming units (PFU) of the Snook strain of BRSV.30
The BRSV proteins recognized by memory CD8+ T cells were analysed in five different groups of cattle (Table 1). Group I (animals 1– 4), comprised 18–24-month-old-cattle (housed in a barrier-maintained building) that had been infected i.n. and i.t. with 2 × 105 PFU of the Snook strain of BRSV on three or four occasions. Groups II and III were cattle that had been vaccinated as gnotobiotic calves, on one or two occasions, 8 weeks apart, by intradermal scarification with 1–2 × 108 PFU of rVV expressing either the F (Group II) or the G (Group III) protein of the 391–2 strain of BRSV.31,32 These cattle were then housed conventionally and either infected experimentally, as described above (animals 5–10), or infected naturally with BRSV (animal 11). Group IV cattle were 4–6-year-old conventionally reared animals that had been immunized subcutaneously (s.c.) with 106 glutaraldehyde-fixed NM7 cells plus Quil A at 6–12 months of age (animals 12–14). The NM7 cell line is a bovine nasal mucosa cell line persistently infected with the 127 strain of BRSV.33 Cattle 12 and 13 were further immunized with NM7 cells, as described previously, at 7 and 9 months following the first immunization, and all animals may have also undergone natural BRSV infections. Group V was a control group of uninfected gnotobiotic calves, 4 weeks of age (calves 15–17). All cattle used were seronegative for BRSV at the start of the experiment and tested negative for bovine viral diarrhoea virus (BVDV).
Table 1.
Calves used for studying memory CD8+ T-cell responses to bovine respiratory syncytial virus (BRSV)
| Calf no. | Haplotype | Immunogen |
|---|---|---|
| Group I | ||
| 1 | A18*/A17 | BRSV × 3 |
| 2 | A18*/A17 | BRSV × 4 |
| 3 | A18*/A31* | BRSV × 3 |
| 4 | A20/A20 | BRSV × 2 |
| Group II | ||
| 5 | A18/A20 | rVV-F, BRSV |
| 6 | A18/A10 | rVV-F, BRSV |
| 7 | A18/A18 | rVV-F, BRSV |
| 8 | A31*/A31* | rVV-F × 2, BRSV |
| Group III | ||
| 9 | A14/W9 | rVV-G, BRSV |
| 10 | A31*/A31* | rVV-G × 2†, BRSV |
| 11 | A14/A14 | rVV-G, BRSV |
| Group IV | ||
| 12 | A18*/A31* | NM7, BRSV |
| 13 | A18*/A31* | NM7, BRSV |
| 14 | A18*/A18* | NM7, BRSV |
| Group V | ||
| 15 | ND | None |
| 16 | ND | None |
| 17 | ND | None |
Haplotype characterized at the level of the expressed genes. All other haplotypes were only determined serologically.
The calf was also inoculated subcutaneously with 40 µg of purified G protein prior to the BRSV challenge.
ND, not determined; rVV, recombinant vaccinia virus.
The majority of animals used in this study were from an inbred herd, generated by back-crossing, in which several MHC class I haplotypes have been completely characterized at the level of expressed genes: A18, A31, A11, A10, A14.28 The haplotypes are named according to serological specificity, but it should be noted that although a single specificity is designated in each case, the number of genes expressed is variable. A18 and A10 each express a single gene (HD6, JSP.1), A31 and A11 each express two genes (HD1, HD7 and D18.2, D18.3) and A14 expresses three genes (D18.1, D18.4, D18.5) (Table 2). Animals were typed either by PCR–SSP at the IAH or by complement-dependent microcytotoxicity testing, using alloantisera, by Dr Mike Stear (Glasgow University, Glasgow, UK). In the case of MHC types found outside the inbred herd (i.e. A17, w9, A20), there is no information available regarding the number or nature of class I genes expressed.
Table 2.
Class I genes expressed on different haplotypes within Institute for Animal Health (IAH; Compton, Newbury, UK) cattle
| Haplotype | Class I genes expressed |
|---|---|
| A18 | HD6 |
| A31 | HD1, HD7 |
| A11 | D18.2, D18.3 |
| A10 | JSP.1 |
| A14 | D18.1, D18.4, D18.5 |
| A20 | Unknown |
| A17 | Unknown |
| W9 | Unknown |
All experiments were performed under the regulations of the Home Office Scientific Procedures Act (1986).
Viruses
The Snook strain of BRSV30 was isolated from a calf with pneumonia and passaged once in calf testis cells and six times in calf kidney (CK) cells. Stocks of virus for inoculation of cattle were prepared in primary CK cells and stored in liquid nitrogen. Titres of BRSV were determined by plaque assay on secondary CK cells, as described previously.33 Recombinant VV expressing the F, G, N and M2 proteins of the 391–2 strain of BRSV were obtained from Dr G. Wertz (University of Alabama, Birmingham, AL)10,31,32 and grown and assayed on HuTK– 143B cells. For inoculation of calves, rVV was first purified over a 35% sucrose gradient.
Recombinant FPV, expressing F, N and M2 of the Snook strain, was constructed using the recombination vector, pEFL29, obtained from Dr M. Skinner (IAH).34 Briefly, cDNAs containing full-length N, M2 or F genes were generated by reverse transcription on mRNA extracted from BRSV Snook-infected fetal CK cells followed by PCR amplification. Primers were designed to introduce BamHI, EcoRI and SmaI restriction sites at the 5′ and 3′ ends of the full-length genes. The cDNAs were then cloned, using standard techniques, into the BamHI/EcoRI site of pGEM.TA (Promega, Madison, WI). Restriction enzyme digestion products of the expected size were obtained using BamHI and EcoRI. The genes were excised from pGEM.TA using SmaI, and blunt-end cloned into the SmaI site (downstream of the p7.5 promoter) of pEFL29, which contains the lacZ gene. Cloned plasmids with the F, N or M2 genes in the correct orientation with respect to the p7.5 promoter were selected and recombinants made using FP9.35 Recombinant FPV was identified as blue plaques in monolayers of chicken embryo fibroblasts (CEFs) overlaid with agarose containing 5-bromo-4-chloro-3-indolyl-b-d-galactosidase. Recombinant FPV was plaque-purified four times, and expression of the N, M2 or F proteins was confirmed by indirect immunofluorescence on rFPV-infected CEFs and/or by enzyme-linked immunosorbent assay (ELISA) analysis using lysates of rFPV-infected CEFs and monoclonal antibodies (mAbs) 6 (anti-N), 8 (anti-M2) or 19 (anti-F).36,37
Effector cells
Peripheral blood mononuclear cells (PBMC) were isolated from heparinized blood by centrifugation at 1200 g for 40 min at 20° over Histopaque 1083 (Sigma, St. Louis, MO). Lung mononuclear cells were obtained by enzyme digestion, as described previously12 before centrifugation over Histopaque. Cells from the interface were washed three times with phosphate-buffered saline (PBS) and resuspended in complete medium: RPMI-1640 with Glutamax-1 (Gibco BRL, Paisley, UK), containing 10% heat-inactivated fetal calf serum (FCS), 5 × 10−5 m 2-mercaptoethanol, 100 µg/ml streptomycin and 100 U/ml penicillin.
Cytotoxicity assays
Cytotoxicity assays were carried out as described previously.14 Briefly, 50 µl of 51Cr-labelled target cells was mixed with 100 µl of effector cells to give effector : target (E : T) cell ratios ranging from 6 : 1 to 100 : 1, in triplicate wells of a V-shaped microtitre plate. The cells were gently pelleted and incubated for 4 hr at 37° in 5% CO2 in air before 25 µl of supernatant was removed from each well and 51Cr release calculated, in counts per minute (c.p.m.), in a gamma counter. The percentage specific lysis was calculated as follows:
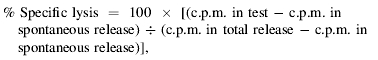 |
using the mean value of triplicate wells, provided the individual values did not exceed 20% of the mean value. Spontaneous and total release were determined from wells containing target cells and 100 µl of medium or 1% sodium dodecyl sulphate (SDS), respectively.
Target cells
Target cells were autologous skin or testicular fibroblasts infected at 0·5 PFU/cell with the 391–2 strain of BRSV37 by rolling at 37° in a 1-ml volume for 18 hr. Target cells were cultured as an adherent monolayer for a further 24 hr, trypsinized to provide a single-cell suspension, labelled for 1 hr with sodium chromate (51Cr) (40 µCi/106 cells) and washed by centrifugation. Stable mouse L-cell transfectants expressing the cattle class I gene, HD6, infected with BRSV, were used as targets in some assays. HD6 is the only class I gene expressed on the A18 haplotype. BRSV infection of target cells was confirmed by immunostaining of cells (fixed with 80% acetone in water) using a mAb, 19, specific for the BRSV F protein, followed by peroxidase-labelled goat anti-mouse antibody (1 : 20 dilution: Kirkegaard and Perry Laboratories Inc., Guild Ford, UK) and 3,3′-diaminobenzidine tetrahydrochloride (DAB; Sigma) as substrate. Cells infected with rVV at a multiplicity of infection (MOI) of 5 for 1 hr at 37° and labelled with 80 µCi of 51Cr for a further 1 hr, were also used as targets in some assays. After labelling with 51Cr, cells were washed twice in medium and incubated at 37° for a further 2 hr prior to the cytotoxicity assay.
In vitro stimulation of lymphocytes
Stimulator cells were either NM7 cells or, more usually, autologous fibroblasts infected with BRSV (as described above), or autologous concanavalin A (Con A) blasts (PBMC incubated for 48 hr in medium containing 10 µg/ml Con A) infected with rVV at a MOI of 5 overnight in 1-ml volumes. All stimulator cells were glutaraldehyde-fixed, as described previously,39 and incubated with effector cells at an E : T ratio of 5 : 1 for 7 days in complete medium. On day 4, 50% of the medium was removed and replaced with complete medium containing 12% T-cell growth factor (supernatant from 48-hr Con A-stimulated PBMC treated with methyl α-d-mannpyranoside; Sigma) and 20 U/ml of recombinant human IL-2 (Sigma). Target cells and virus stocks were free from BVDV and mycoplasmas.
Flow cytometric analysis of intracellular IFN-γ
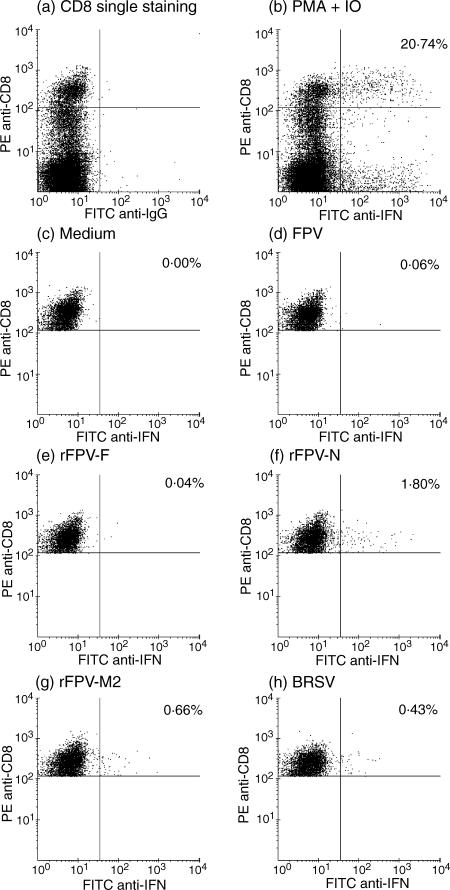
PBMC (4–6 × 106) were infected for 2 hr by rolling at 37° in 1 ml of complete RPMI with BRSV (as for target cells), with rVV at a MOI of 5, or with rFPV at a MOI of 2. As controls, PBMC were incubated with medium alone or with uninfected cell lysates. Cells were washed and aliquoted (2 × 106/well) in duplicate and incubated at 37° for 8 hr for assays in which PBMC were stimulated with rVV, or for 22 hr in assays where stimulation with BRSV or rFPV was carried out. The shorter incubation time-period for rVV was because rVV reduced the viability of the PBMC if left overnight. IFN-γ was retained within the cell by the addition of Brefeldin A (1 µg/ml; Calbiochem, Nottingham, UK) for the final 4 hr of the assays. As controls, PBMC were incubated for 8 or 22 hr, as appropriate, with phorbol 12-myristate 13-acetate (PMA, 0·02 µg/µl; Sigma). Ionomycin (0·4 µg/ml; Calbiochem) was added for the final 5 hr and Brefeldin A for the final 4 hr. Following incubation, all cells were washed in buffer [1% bovine serum albumin (BSA), 0·1% sodium azide in PBS], fixed in 1% paraformaldehyde in PBS for 10 min at 20°, washed and frozen in 10% dimethyl sulphoxide (DMSO)/90% FCS at −70°. To detect CD8+ IFNγ+ lymphocytes, cells were stained with mAb CC63 (anti-bovine CD8α)40 or an isotype-matched control mAb, for 10 min at 20°, washed and stained with R-phycoerythrin (PE)-labelled goat anti-mouse IgG2a for 10 min at 20°. After washing, cells were permeabilized with Permeabilization solution™ (Becton-Dickinson, Oxford, UK) for 10 min at 20°. Cells were then washed and stained for IFN-γ using mAb CC30241 at 1 µg/ml, or an isotype control. After 1 hr of incubation at 4°, cells were stained with fluorescein isothiocanate (FITC)-labelled goat anti-mouse immunoglobulin G1 (IgG1) (1 : 400 dilution) for 15 min at 20°, washed, resuspended in buffer and analysed using a FACSCalibur flow cytometer (Becton-Dickinson). Data analysis was performed using FCSexpress software (DeNovo Software, Ontario, Canada); the gate was set to include live mononuclear cells based on light-scattering properties and excluded cell doublet and triplets. Quadrants were set to include only those cells expressing a high level of CD8, as shown in Fig. 4(a). The means of duplicate samples were taken to give results based on a minimum of 6 × 104 high CD8 events. The variation between the duplicate samples was usually less than 12% of the mean value.
Figure 4.
Two-colour flow cytometric analysis of intracellular interferon-γ (IFN-γ) production by peripheral blood CD8+ T cells taken from an A20 homozygous calf (animal 4) which had been experimentally infected with bovine respiratory syncytial virus (BRSV) ≈ 12 months previously. Quadrants were set to include only CD8 high T cells by (a) single staining for CD8 and (b) by double staining for CD8 and IFN-γ, following stimulation in the presence of phorbol 12-myristate 13-acetate (PMA) and ionomycin (IO). PBMC were stimulated in vitro for 22 hr in the presence of: (c) medium alone; (d) control wild-type fowlpox virus (FPV); (e) recombinant (r)FPV-F; (f)rFPV-N; (g) rFPV-M2; or (h) BRSV (Snook). The values shown in the upper right quadrants represent the percentage of CD8+ high cells that are IFN-γ+.
Results
Presentation to BRSV-specific CTLs by class I alleles expressed on the haplotypes A18, A31 and A14
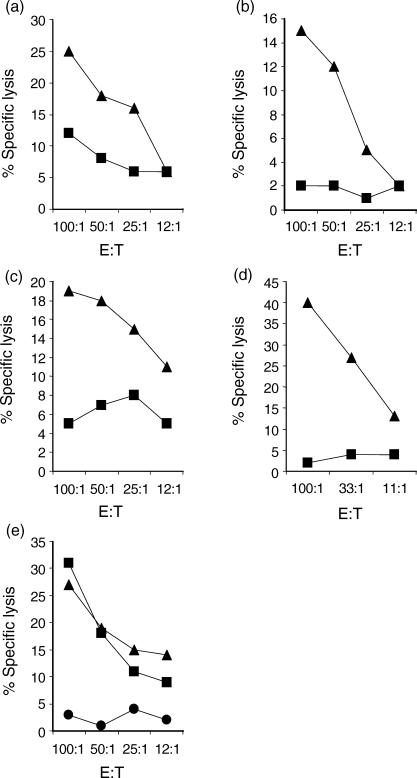
It was generally difficult to elicit CTL activity following in vitro restimulation of memory T cells with BRSV. Therefore, primary CTLs were used in the majority of studies to investigate the ability of MHC class I alleles expressed by A18, A31 and A14 haplotypes to present BRSV to CD8+ T cells. PBMC from A18/A31 calves, infected 10 days previously with BRSV, lysed BRSV-infected A18/A18 targets (Fig. 1a, 1b), indicating that A18, and therefore the HD6 gene product, could present BRSV peptides to CD8+ T cells. This was further confirmed by the ability of lung lymphocytes, from an A18/A10 calf, restimulated in vitro with BRSV-infected, autologous fibroblasts, to lyse BRSV-infected mouse L-cells expressing the HD6 gene (Fig. 2).
Figure 1.
Presentation of bovine respiratory syncytial virus (BRSV) by class I alleles expressed on haplotypes A18, A31 and A14. (a) Lysis of A18/A18 homozygous (▴) or A20/A11 (▪) BRSV-infected cells by peripheral blood mononuclear cells (PBMC) from calf no. 3397 (A18/A31) infected 10 days previously with BRSV. (b) Lysis of A18/A18 homozygous (▴) or A10/w8 (▪) BRSV-infected cells by PBMC from calf no. 7292 (A18/A31) infected 10 days previously with BRSV. (c) Lysis of A31/A31 homozygous (▴) or untyped (▪) BRSV-infected cells by PBMC from calf no. 3796 (A19/A31) infected 9 days previously with BRSV. (d) Lysis of A31/A31 homozygous (▴) or A14/A11 (▪) BRSV-infected cells by lung lymphocytes from calf no. 3395 (A31/A31), infected 10 days previously and restimulated in vitro with BRSV-infected autologous fibroblasts. (e) Lysis of A14/A31 (▴), A14/A11 (▪) or A20/A11 (•) BRSV-infected cells by lung lymphocytes from calf no. 6130 (A14/A31) infected 10 days previously with BRSV. E : T, effector : target ratio.
Figure 2.
Lysis of bovine respiratory syncytial virus (BRSV)-infected HD6 transfected mouse L-cells by BRSV-specific cytotoxic T lymphocytes (CTL). The percentage specific lysis of BRSV-infected HD6 transfected mouse L-cells (▪), uninfected HD6 transfected cells (□), BRSV-infected A18/A18 fibroblasts (▴) and uninfected A18/A18 fibroblasts (▵) by lung lymphocytes from calf no. 4795 (A18/A10), infected 11 days previously with BRSV and restimulated in vitro with BRSV-infected autologous fibroblasts. E : T, effector : target ratio.
BRSV-infected target cells, homozygous for A31, were lysed by PBMC from an A31/A19 calf, infected 9 days previously with BRSV, and by lung lymphocytes, from an A31/A31 calf, that had been restimulated in vitro with BRSV-infected autologous fibroblasts (Fig. 1c, 1d). These findings indicate that either the HD1 and/or HD7 MHC class I gene products (expressed on the A31 haplotype) can present BRSV peptides. Lung lymphocytes from an A14/A31 calf infected 10 days previously with BRSV, were able to lyse both MHC-matched (A14/A31) and partially matched (A14/A11), BRSV-infected cells, but not mismatched (A20/A11) cells (Fig. 1e). Therefore, one or more of the three MHC class I gene products expressed by A14 can present BRSV peptides. These studies demonstrated that at least some of the MHC class I genes expressed by each of the A18, A31 and A14 haplotypes can present BRSV peptides to BRSV-specific, MHC-restricted CTL.
BRSV-specific CTL recognize F, N and M2 proteins
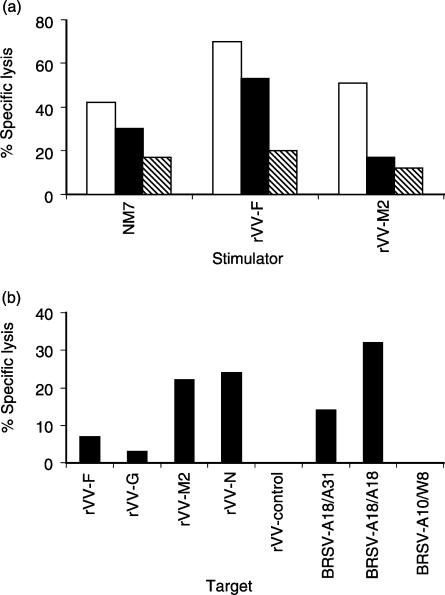
In order to investigate the BRSV proteins recognized by BRSV-specific CTLs, A18/A14 lung lymphocytes, from a calf infected 10 days previously with BRSV, were restimulated in vitro with either BRSV-infected cells (NM7 cells) or with autologous Con A blasts infected with either rVV-F or rVV-M2. Following restimulation, lymphocytes were analysed for their ability to lyse BRSV-infected, MHC-matched (A18/A14), partially MHC-matched (A14/A11) or MHC-mismatched (A11/A20) targets (Fig. 3a). Virus-stimulated lymphocytes lysed BRSV-infected A18/A14 and A14/A11 targets to a greater extent than infected-mismatched (A11/A20) cells, indicating that BRSV proteins were presented by A18 and/or A14 and by A14. Similarly, the F protein of BRSV was able to stimulate CTLs that lysed both BRSV-infected A18/A14 and A14/A11 targets, demonstrating that gene products expressed by both A18 and A14 haplotypes can present the F protein. Lung lymphocytes restimulated with the rVV-M2 lysed BRSV-infected A18/A14 cells, but not A14/A11 targets, suggesting that in this calf, only A18 MHC class I, i.e. the HD6 gene product, presented the M2 protein. Therefore, the HD6 gene product is able to present both the F protein and the M2 protein to CTL.
Figure 3.
Recognition of the F, N and M2 proteins by major histocompatibility complex (MHC) class I A14- and A18-restricted bovine respiratory syncytial virus (BRSV)-specific cytotoxic T lymphocytes (CTL). (a) Lysis of BRSV-infected, MHC-matched A18/A14 (white bars), partially MHC-matched A14/A11 (black bars) or MHC-mismatched A11/A20 (hatched bars) target cells by lung lymphocytes from an A18/A14 calf, infected 10 days previously with BRSV and restimulated in vitro with glutaraldehdye-fixed BRSV-infected NM7 fibroblasts, recombinant vaccinia virus (rVV)-F-infected concanavalin A (Con A) blasts or rVV-M2-infected Con A blasts [effector : target (E : T) ratio of 100 : 1] (b) Lysis of A18/A18 targets infected with rVV expressing individual BRSV proteins, or with BRSV-infected, MHC-matched (A18/A31) partially matched (A18/A18) and mismatched (A10/W8) targets, by peripheral blood mononuclear cells (PBMC) from an A18/A31 calf, infected 7 days previously with BRSV (E : T ratio of 100 : 1).
PBMC from an A18/A31 calf, infected with BRSV 7 days previously, were analysed for their ability to lyse A18/A18 targets infected with rVV expressing different BRSV proteins. BRSV-specific CTL from this calf recognized the M2 and N proteins to a greater extent than the F or G proteins of BRSV (Fig. 3b). Similar results were also obtained with PBMC from this calf on day 8 of BRSV infection. Therefore, MHC class I HD6-restricted CTLs recognized the M2 and N proteins.
BRSV-specific induction of IFN-γ by CD8+ T cells
As restimulation of memory T cells with BRSV in vitro did not reproducibly generate BRSV-specific CTLs, the ability of BRSV to activate memory CD8+ T cells for IFN-γ production was analysed by flow cytometry to detect intracellular IFN-γ. Only cells expressing high levels of CD8 were analysed (Fig. 4a). Following restimulation with BRSV, the proportion of CD8+ low cells producing IFN-γ was negligible (results not shown). An example of the profiles obtained by flow cytometry for PBMC from a BoLA A20 homozygous animal (no. 4, Table 1), which had been previously infected with BRSV, is shown in Fig. 4. Stimulation of PBMC with PMA and ionomycin, induced IFN-γ production in both CD8+ and CD8– cells, with 20·74% of CD8+ T cells producing IFN-γ (Fig. 4b). Following restimulation of PBMC with medium (Fig. 4c) or uninfected CK lysates (results not shown), the proportion of CD8+ IFN-γ+ T cells was ≤ 0·04% of all CD8+ T cells. In contrast, following restimulation with BRSV, 0·43% of CD8+ T cells were IFN-γ+ (Fig. 4h).
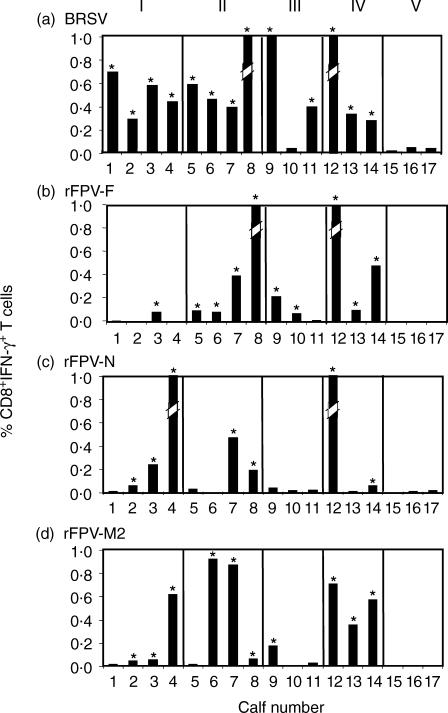
The frequency of BRSV-specific CD8+ T cells was analysed in a total of 14 cattle that had been primed either by BRSV infection and/or by vaccination with rVV-F, rVV-G or a glutaraldehyde-inactivated BRSV vaccine (Table 1). The responses of three naïve, gnotobiotic calves were also analysed. Virus-specific responses were determined following subtraction of values obtained after restimulation of PBMC with uninfected cell lysates. A minimum of two assays were carried out over a 4-month period for all of the cattle, except in the naïve group where only one assay was performed, and representative data is shown in Fig. 5a. The reproducibility of the assay following restimulation with BRSV was good as the majority of duplicate assays over the 4-month period gave similar results. However, there were two notable exceptions – animals 7 and 12, where the percentage of CD8+ T cells, which were IFN-γ+ following restimulation with BRSV, increased from 0·38% to 1·04% and from 1·51% to 4·85%, respectively, between the two assays. As these cattle had been housed conventionally, it is possible that they were reinfected with BRSV between the two assays, but serum samples were not taken at the time to confirm this.
Figure 5.
Recognition of bovine respiratory syncytial virus (BRSV) proteins by CD8+ memory T cells. The percentages of CD8+ T cells positive for intracellular interferon-γ (IFN-γ) production are shown. Peripheral blood mononuclear cells (PBMC) from each of the calves within the five groups (Table 1) were stimulated with: (a) BRSV (b) recombinant fowlpox virus (rFPV)-F; (c) rFPV-N; and (d) rFPV-M2. The broken bars represent values that exceed the maximum value shown. The actual values for these are as follows: calf no. 4 response to N = 1·74; calf no. 8 response to F = 2·55 and to BRSV = 1·86; calf no. 12 response to F = 1·09, to N = 2·09 and to BRSV = 4·85. *Values that are considered significant.
CD8+ T cells from 13 of 14 cattle primed with BRSV showed a detectable IFN-γ response following restimulation in vitro with BRSV. The frequency of BRSV-specific CD8+ IFN-γ+ T cells in these 13 animals ranged from 0·26% to 4·85% of CD8+ T cells. CD8+ T cells from animal 10, which had been vaccinated with rVV-G and infected with BRSV on two occasions, did not produce IFN-γ in response to BRSV. This animal did not respond to BRSV in a lymphocyte-proliferation assay, although it had developed BRSV-specific serum antibodies (results not shown). CD8+ T cells from naïve calves did not produce IFN-γ after restimulation, in vitro, with BRSV. The frequency of CD8+ IFN-γ+ T cells was ≤ 0·04% in all naïve, seronegative calves.
BRSV protein-specific induction of IFN-γ by CD8+ T cells
Recognition of BRSV proteins by CD8+ T cells was analysed in the 14 BRSV-primed cattle by investigating the ability of rFPV or rVV expressing individual BRSV proteins to stimulate production of IFN-γ by CD8+ T cells. Activation of CD8+ T cells by rFPV expressing BRSV proteins was greater than that induced by rVV. The frequencies of CD8+ IFN-γ+ T cells induced 8 hr after restimulation of PBMC from BRSV-infected or NM7-vaccinated cattle with rVV were low, and responses were not significantly greater than those induced by the control rVV. In contrast, the responses of cattle vaccinated with rVV were high. However, the frequencies of CD8+ IFN-γ+ T cells induced after stimulation with rVV expressing BRSV proteins were not significantly greater than those induced by the control rVV. The exception to this was that seen in two animals (10 and 11) where, following subtraction of the value induced by the control rVV, 0·08% and 0·25% of CD8+ T cells produced IFN-γ in response to rVV-G.
An example of the profiles obtained by flow cytometry for PBMC from a BoLA A20 homozygous individual (animal 4, Table 1) induced 22 hr after restimulation, in vitro, with rFPV is shown in Fig. 4. In this animal, the proportion of CD8+ T cells producing IFN-γ following restimulation with wild-type FPV was 0·06% (Fig. 4d). In contrast, the frequencies of CD8+ IFN-γ+ T cells induced by stimulation with rFPV-N or rFPV-M2 were 1·80% and 0·66%, respectively (Fig. 4f, 4g). However, restimulation of PBMC from this animal with rFPV-F induced little or no IFN-γ production by CD8+ T cells (Fig. 4e). Therefore, CD8+ T cells from a BoLA A20 individual, previously infected with BRSV, recognized the N and the M2 proteins, but not the F protein of BRSV.
After restimulation of PBMC from the 14 BRSV-primed cattle with rFPV expressing different BRSV proteins, F protein-specific responses were observed in CD8+ T cells from 10 animals, N protein-specific responses were observed in seven animals and M2 protein-specific responses were observed in 10 animals (Fig. 5b, 5c, 5d). The frequencies of F, N and M2 protein-specific CD8+ T cells ranged from 0·07 to 2·55%, 0·05 to 2·09% and 0·05 to 0·92%, respectively. BRSV protein-specific CD8+ IFN-γ+ T cells in the three naïve calves were < 0·01% (Fig. 5).
Although the numbers of cattle in the different groups were small, there was some evidence that vaccination may have influenced the antigenic specificity of the CD8+ T-cell response. Thus, the N and the M2 proteins were recognized by CD8+ T cells from three of the four BRSV-infected animals, whereas the F protein was only recognized by one individual (Fig. 5). In contrast, the F protein was recognized by CD8+ T cells from all of four cattle that had been vaccinated with rVV-F and subsequently infected with BRSV, and in all of three cattle that had been vaccinated with an inactivated vaccine and subsequently exposed to BRSV. Although CD8+ T cells from two of the three animals that had been vaccinated with rVV-G and infected with BRSV recognized the F protein and two recognized the G protein, there appeared to be little or no recognition of the N and M2 protein.
Discussion
The pathogenesis of bronchiolitis and pneumonia induced by RS viruses in the natural host is still not clearly understood. However, cattle with impaired CD8+ T-cell responses suffer more prolonged BRSV infection than control calves,12 indicating that the CD8+ T-cell response plays a critical role in controlling BRSV infection. MHC class I-restricted CD8+ T cells can mediate antiviral protection via two pathways: direct cytotoxicity; or via release of cytokines such as IFN-γ and tumour necrosis factor-α (TNF-α).42 Functionally active BRSV-specific CTL have been demonstrated in the peripheral blood and lungs of calves following primary infection,14 and the secretion of IFN-γ by CD8+ T cells during a primary infection has also been demonstrated (R. Gaddum and G. Taylor, unpublished observations). In this study we have extended our previous findings by identifying some of the proteins recognized by BRSV-specific bovine CTLs generated during a primary response. In addition, the BRSV proteins recognized by memory CD8+ T cells in cattle that have either been infected with BRSV on a number of occasions or had been primed by vaccination and subsequently infected with BRSV, were analysed by flow cytometry to detect IFN-γ production. Using rVV and rFPV expressing four of the 10 BRSV proteins of the Snook or 391–2 strains of BRSV, the majority of infected and/or vaccinated cattle recognized the F, N and M2 proteins. In addition, the G protein of BRSV was strongly recognized in cattle that had been vaccinated with rVV-G and subsequently infected with BRSV.
Whereas MHC restriction of BRSV-specific CTL was demonstrated in the present study, MHC restriction of IFN-γ production by CD8+ T cells following restimulation with BRSV, rVV or rFPV was not analysed. However, the IFN-γ response observed was unlikely to be caused by natural killer (NK)-like CD8+ cells. Therefore, in the present study, activation of CD8+ T cells was analysed in cattle older than 12 months of age, and putative NK cells that have been identified in older cattle do not appear to express CD8.43 Furthermore, CD3– CD8+ NK-like cells from naïve neonatal calves that produce IFN-γ in response to Mycobacterium bovis bacille Calmette–Guérin (BCG)-infected dendritic cells do not express high levels of CD8.43 CD8 low cells are composed of CD3+ and CD3– populations, whereas cells expressing high levels of CD8, which were analysed in the present study, are predominantly43 CD3+ (R. Cook and G. Taylor, unpublished observations).
Differences in the activation of bovine CD8+ T cells by rFPV expressing individual BRSV proteins compared with that induced by rVV may have been a result of differences in the duration of culture. Analysis of the kinetics of intracellular synthesis of IFN-γ by bovine CD8+ T cells following activation with BRSV or rFPV demonstrated an increase in the antigen-specific IFN-γ synthesis at 22 hr compared with that after 8 hr of culture. However, there was poor survival of viable cells after 22 hr of culture when rVV was used to activate T cells. Therefore, cultures in which rVV was used to activate CD8+ T cells were only of 8 hr duration.
There did not appear to be any correlation between bovine MHC class I phenotype and recognition of BRSV proteins by CD8+ T cells. Furthermore, there was no evidence that cattle which are homozygous for A18 (i.e. express a single MHC class I gene) displayed a more restricted antigen-recognition repertoire than those which express A31 or A14 haplotypes. During a primary response, CTL assays demonstrated that the HD6 gene product could present BRSV to CD8+ T cells and that BRSV-specific, MHC class I HD6-restricted CTL recognized the M2 and N proteins. In addition, memory CD8+ T cells from a vaccinated animal (no. 14), which was homozygous for A18 and therefore only expressed the HD6 gene product, recognized the F, M2 and N proteins. These findings were similar to those observed in cattle that express two or more genes. For example, cattle 8 and 10, which were both A31/A31 homozygous and therefore expressed the HD1 and HD7 MHC class I gene products, recognized the F, N and M2 proteins or the F and G proteins, respectively. However, it has not been demonstrated that both HD1 and HD7 were presenting peptides from any or all of these proteins. Further studies using additional L-cell transfectants would be required to clarify the situation, and to compare the presentation capabilities of these class I alleles. For example, at present it is not known how many different peptides may be presented by each allele. The peptide-binding motifs for each of the three alleles HD6, HD1 and HD7 are known.44 The motif and the range of peptides eluted from HD6 predict that this allele would have a broad binding capability, which may in part explain the results seen here.
Although the number of animals in the different treatment groups was small, there was some evidence that the specificity of BRSV-specific CD8+ T cells was influenced by previous vaccination. Therefore, the F protein was recognized less frequently in cattle that had been primed with BRSV alone than in those that had been primed by vaccination prior to BRSV infection. This observation may reflect differences in the duration of F protein-specific memory CD8+ T cells in animals primed in different ways. Studies of HRSV-specific human CTL suggest that recognition of the F protein may be associated with recent HRSV infection.24 Taken together, these findings suggest that CD8+ T-cell recognition of the F protein may be short-lived. Further studies are required to determine whether differences in the duration of CD8+ memory T cells specific for different BRSV proteins correlates with susceptibility to reinfection.
The number of cattle recognizing the N and M2 proteins was similar in each of the treatment groups, apart from those animals vaccinated with rVV-G (group III) prior to BRSV infection. Although there was little or no recognition of the N or M2 proteins in these cattle, they did recognize the F and G proteins. Furthermore, CD8+ T cells from one of these animals (no. 10) consistently failed to respond to BRSV. Vaccination of BALB/c mice with rVV-G does not prime CD8+ T cells,45 and following HRSV challenge, the HRSV-specific pulmonary CTL response is reduced compared with that seen in mice undergoing a primary HRSV infection (L. J. Mackay, G. Bembridge & G. Taylor, manuscript in preparation). Taken together, these findings suggest that priming of CD8+ T cells to certain RSV proteins may not be particularly effective in animals that have been previously vaccinated with rVV-G, possibly because prior vaccination with rVV-G restricts the replication of RSV following subsequent challenge. These findings contrast with those seen in cattle vaccinated with glutaraldehyde-fixed BRSV-infected nasal mucosal cells, which are also effective at restricting replication of BRSV following challenge.33 Therefore, CD8+ T cells from cattle vaccinated with this inactivated vaccine and subsequently exposed to BRSV recognized the F, N and M2 proteins. However, as vaccination with glutaraldehyde-fixed BRSV-infected nasal mucosal cells primes RSV-specific CTLs in BALB/c mice,46 it seems probable that this vaccine also primes CD8+ T cells in cattle.
In conclusion, this study has demonstrated that the F, N and M2 proteins are recognized by BRSV-specific bovine CD8+ T cells. Knowledge of the BRSV proteins recognized by bovine CD8+ T cells will facilitate the identification of MHC class I-restricted epitopes from these proteins. This will enable the development of certain methodologies, such as the use of bovine MHC class I–peptide tetramers, to allow a detailed quantitative and qualitative analysis to be carried out of the CD8+ T-cell response to BRSV. Such studies will provide an insight into the relationship between BRSV-specific CD8+ T-cell memory and frequency of reinfection, and will help to ensure that potential vaccines induce a qualitatively appropriate CTL response.
Acknowledgments
This work was supported by MAFF (UK) and the Biotechnology and Biological Sciences Research Council. We thank Dr Lewis Thomas and other members of the IAH staff for care of the cattle used within this study.
References
- 1.Stott EJ, Taylor G. Respiratory syncytial virus. Brief review. Arch Virol. 1985;84:1–52. doi: 10.1007/BF01310552. [DOI] [PubMed] [Google Scholar]
- 2.Elvander M. Severe respiratory disease in dairy cows caused by infection with bovine respiratory syncytial virus. Vet Rec. 1996;138:101–5. doi: 10.1136/vr.138.5.101. [DOI] [PubMed] [Google Scholar]
- 3.Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol. 1969;89:422–34. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 4.Schreiber P, Matheise JP, Dessy F, Heimann M, Letesson JJ, Coppe P, Collard A. High mortality rate associated with bovine respiratory syncytial virus (BRSV) infection in Belgian white blue calves previously vaccinated with an inactivated vaccine. J Vet Med B, Infectious Dis Vet Public Health. 2000;47:535–50. doi: 10.1046/j.1439-0450.2000.00380.x. [DOI] [PubMed] [Google Scholar]
- 5.Anderson LJ, Heilman CA. Protective and disease-enhancing immune responses to respiratory syncytial virus. J Infect Dis. 1995;171:1–7. doi: 10.1093/infdis/171.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Alwan WH, Record FM, Openshaw PJM. CD4+ T-cells clear virus but augment disease in mice infected with respiratory syncytial virus. Comparison with the effects of CD8+ T cells. Clin Exp Immunol. 1992;88:527–36. doi: 10.1111/j.1365-2249.1992.tb06482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alwan WH, Kozlowski WJ, Openshaw PJM. Distinct types of lung disease caused by functional subsets of antiviral T cells. J Exp Med. 1994;179:81–9. doi: 10.1084/jem.179.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cannon MJ, Openshaw PJM, Askonas BA. Cytotoxic T cells clear virus but augment lung pathology in mice infected with respiratory syncytial virus. J Exp Med. 1988;168:1163–8. doi: 10.1084/jem.168.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham BS, Bunton LA, Wright PF, Karzon DT. Role of T-lymphocyte subsets in the pathogenesis of primary infection and rechallenge with respiratory syncytial virus in mice. J Clin Invest. 1991;88:1026–33. doi: 10.1172/JCI115362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor G, Thomas LH, Furze JM, Cook RS, Wyld SG, Lerch R, Hardy R, Wertz GW. Recombinant vaccinia viruses expressing the F, G or N, but not the M2, protein of bovine respiratory syncytial virus (BRSV) induce resistance to BRSV challenge in the calf and protect against the development of pneumonic lesions. J Gen Virol. 1997;78:3195–206. doi: 10.1099/0022-1317-78-12-3195. [DOI] [PubMed] [Google Scholar]
- 11.Thomas LH, Cook RS, Wyld SG, Furze JM, Taylor G. Passive protection of gnotobiotic calves using monoclonal antibodies directed at different epitopes on the fusion protein of bovine respiratory syncytial virus. J Infect Dis. 1998;177:874–80. doi: 10.1086/515234. [DOI] [PubMed] [Google Scholar]
- 12.Taylor G, Thomas LH, Wyld SG, Furze J, Sopp P, Howard CJ. Role of T-lymphocyte subsets in recovery from respiratory syncytial virus infection in calves. J Virol. 1995;69:6658–64. doi: 10.1128/jvi.69.11.6658-6664.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hall CB, Powell KR, MacDonald NE, Gala CL, Mengus ME, Suffin SC, Cohen HJ. Respiratory syncytial virus infection in children with compromised immune function. N Engl J Med. 1986;315:77–81. doi: 10.1056/NEJM198607103150201. [DOI] [PubMed] [Google Scholar]
- 14.Gaddum RM, Cook RS, Thomas LH, Taylor G. Primary cytotoxic T cell responses to bovine respiratory syncytial virus in calves. Immunology. 1996;88:421–7. doi: 10.1046/j.1365-2567.1996.d01-667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McInnes E, Sopp P, Howard CJ, Taylor G. Phenotypic analysis of local cellular responses in calves infected with bovine respiratory syncytial virus. Immunology. 1999;96:396–403. doi: 10.1046/j.1365-2567.1999.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Openshaw PJ, Anderson K, Wertz GW, Askonas BA. The 22,000-kilodalton protein of respiratory syncytial virus is a major target for Kd-restricted cytotoxic T lymphocytes from mice primed by infection. J Virol. 1990;64:1683–9. doi: 10.1128/jvi.64.4.1683-1689.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connors M, Kulkarni AB, Collins PL, Firestone C-Y, Holmes KL, Morse HC, III, Murphy BR., III Resistance to respiratory syncytial virus (RSV) challenge induced by infection with a vaccinia virus recombinant expressing the RSV M2 protein is mediated by CD8+ T cells, while that induced by Vac-F or Vac-G recombinants is mediated by antibodies. J Virol. 1992;66:1277–81. doi: 10.1128/jvi.66.2.1277-1281.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulkarni AB, Connors M, Firestone CY, Morse HCD, Murphy BR. The cytolytic activity of pulmonary CD8+ lymphocytes, induced by infection with a vaccinia virus recombinant expressing the M2 protein of respiratory syncytial virus (RSV), correlates with resistance to RSV infection in mice. J Virol. 1993;67:1044–9. doi: 10.1128/jvi.67.2.1044-1049.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Connors M, Collins PL, Firestone C-Y, Murphy BR. Respiratory syncytial virus (RSV) F, G, M2 (22K) and N proteins each induce resistance to RSV challenge, but resistance induced by the M2 and N proteins is short-lived. J Virol. 1991;65:1634–7. doi: 10.1128/jvi.65.3.1634-1637.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kulkarni AB, Collins PL, Bacik I, Yewdell JW, Bennink JR, Crowe JE, Jr, Murphy BR. Cytotoxic T cells specific for a single peptide on the M2 protein of respiratory syncytial virus are the sole mediators of resistance induced by immunization with M2 encoded by a recombinant vaccinia virus. J Virol. 1995;69:1261–4. doi: 10.1128/jvi.69.2.1261-1264.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chang J, Srikiatkhachorn A, Braciale TJ. Visualization and characterization of respiratory syncytial virus F-specific CD8+ T cells during experimental virus infection. J Immunol. 2001;167:4254–60. doi: 10.4049/jimmunol.167.8.4254. [DOI] [PubMed] [Google Scholar]
- 22.Chang J, Braciale TJ. Respiratory syncytial virus infection suppresses lung CD8+ T-cell effector activity and peripheral CD8+ T-cell memory in the respiratory tract. Nat Med. 2002;8:54–60. doi: 10.1038/nm0102-54. [DOI] [PubMed] [Google Scholar]
- 23.Bangham CRM, Openshaw PJM, Ball LA, King AMQ, Wertz GW, Askonas BA. Human and murine cytotoxic T-cells specific to respiratory syncytial virus recognise the viral nucleoprotein (N) but not the major glycoprotein (G), expressed by vaccinia virus recombinants. J Immunol. 1986;137:3973–7. [PubMed] [Google Scholar]
- 24.Cherrie AH, Anderson K, Wertz GW, Openshaw PJ. Human cytotoxic T cells stimulated by antigen on dendritic cells recognize the N, SH, F, M, 22K, and 1b proteins of respiratory syncytial virus. J Virol. 1992;66:2102–10. doi: 10.1128/jvi.66.4.2102-2110.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goulder PJR, Lechner F, Klenerman P, McIntosh K, Walker BD. Characterization of a novel respiratory syncytial virus-specific human cytotoxic T-lymphocyte epitope. J Virol. 2000;74:7694–7. doi: 10.1128/jvi.74.16.7694-7697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brandenburg AH, de Waal L, Timmerman HH, Hoogerhout P, de Swart RL, Osterhaus ADME. HLA class I-restricted cytotoxic T-cell epitopes of the respiratory syncytial virus fusion protein. J Virol. 2000;74:10240–4. doi: 10.1128/jvi.74.21.10240-10244.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Davies CJ, Joosten I, Bernoco D, Arriens MA. Polymorphism of bovine MHC class I genes. Joint report of the fifth International BoLA workshop. Eur J Immunogenet. 1994;21:239–58. doi: 10.1111/j.1744-313x.1994.tb00197.x. [DOI] [PubMed] [Google Scholar]
- 28.Ellis SA, Holmes EC, Staines KA, Smith KB, Stear MJ, McKeever DJ, MacHugh ND, Morrison WI. Variation in the number of expressed MHC genes in different cattle class I haplotypes. Immunogenetics. 1999;50:319–28. doi: 10.1007/s002510050608. [DOI] [PubMed] [Google Scholar]
- 29.Ellis SA, Staines KA, Stear MJ, Hensen EJ, Morrison WI. DNA typing for BoLA class I using sequence specific primers (PCR–SSP) Eur J Immunogenet. 1999;25:365–70. doi: 10.1046/j.1365-2370.1998.00112.x. [DOI] [PubMed] [Google Scholar]
- 30.Thomas LH, Gourlay RN, Stott EJ, Howard CJ, Bridger JC. A search for new microorganisms in calf pneumonia by the inoculation of gnotobiotic calves. Res Vet Sci. 1982;33:170–82. doi: 10.1016/S0034-5288(18)32331-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lerch RA, Anderson K, Wertz GW. Nucleotide sequence analysis and expression from recombinant vectors demonstrate that the attachment protein G of bovine respiratory syncytial virus is distinct from that of human respiratory syncytial virus. J Virol. 1990;64:5559–69. doi: 10.1128/jvi.64.11.5559-5569.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lerch RA, Anderson K, Amann VL, Wertz GW. Nucleotide sequence analysis of the bovine respiratory syncytial virus fusion protein mRNA and expression from a recombinant vaccinia virus. Virology. 1991;181:118–31. doi: 10.1016/0042-6822(91)90476-r. [DOI] [PubMed] [Google Scholar]
- 33.Stott EJ, Thomas LH, Taylor G, Collins AP, Jebbett J, Crouch S. A comparison of three vaccines against respiratory syncytial virus in calves. J Hyg Camb. 1984;93:251–61. doi: 10.1017/s0022172400064779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qingzhong Y, Barrett T, Brown TDK, Cook JKA, Green P, Skinner MA, Cavanagh D. Protection against turkey rhinotracheitis pneumovirus (TRTV) induced by a fowlpox virus recombinant expressing the TRTV fusion glycoprotein. Vaccine. 1994;12:569–73. doi: 10.1016/0264-410x(94)90319-0. [DOI] [PubMed] [Google Scholar]
- 35.Boulanger D, Green P, Smith T, Czerny C-P, Skinner MA. The 131-amine-acid repeat region of the essential 39-kilodatlon core protein of fowlpox virus FP9, equivalent to vaccinia virus A4L protein, is nonessential and highly immunogenic. J Virol. 1998;72:170–9. doi: 10.1128/jvi.72.1.170-179.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taylor G, Stott EJ, Bew M, Fernie BF, Cote PJ, Collins AP, Hughes M, Jebbett J. Monoclonal antibodies protect against respiratory syncytial virus infection in mice. Immunology. 1984;52:137–42. [PMC free article] [PubMed] [Google Scholar]
- 37.Taylor G, Stott EJ, Furze J, Ford J, Sopp P. Protective epitopes on the fusion protein of respiratory syncytial virus recognized by murine and bovine monoclonal antibodies. J Gen Virol. 1992;73:2217–23. doi: 10.1099/0022-1317-73-9-2217. [DOI] [PubMed] [Google Scholar]
- 38.Lerch RA, Stott EJ, Wertz GW. Characterisation of bovine respiratory syncytial virus proteins and mRNAs and generation of cDNA clones to the viral mRNAs. J Virol. 1989;63:833–40. doi: 10.1128/jvi.63.2.833-840.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaia JA, Oxman MN. Antibody to varicella-zoster virus-induced membrane antigen: immunofluorescence assay using monodisperse glutaradehyde fixed target cells. J Infect Dis. 1977;136:519–30. doi: 10.1093/infdis/136.4.519. [DOI] [PubMed] [Google Scholar]
- 40.MacHugh ND, Bensaid A, Howard CJ, Davis WC, Morrison WI. Analysis of the reactivity of anti-bovine CD8 monoclonal antibodies with cloned T-cell lines and mouse 1-cells transfected with bovine CD8. Vet Immunol Immunopathol. 1991;27:167–72. doi: 10.1016/0165-2427(91)90096-u. [DOI] [PubMed] [Google Scholar]
- 41.Collins RA, Howard CJ, Duggan SE, Werling D. Bovine interleukin-12 and modulation of IFN-γ. Vet Immunol Immunopathol. 1999;68:193–207. doi: 10.1016/s0165-2427(99)00020-3. [DOI] [PubMed] [Google Scholar]
- 42.Kagi D, Hengartner H. Different roles for cytotoxic T cells in the control of infections with cytopathic versus non-cytopathic viruses. Curr Opin Immunol. 1996;8:472–7. doi: 10.1016/s0952-7915(96)80033-1. [DOI] [PubMed] [Google Scholar]
- 43.Hope JC, Sopp P, Howard CJ. NK-like CD8+ cells in immunologically naive neonatal calves that respond to dendritic cells infected with Mycobacterium bovis BCG. J Leukoc Biol. 2002;71:184–94. [PubMed] [Google Scholar]
- 44.Gaddum G, Willis AC, Ellis SA. Peptide motifs from three cattle MHC (BoLA) class I antigens. Immunogenetics. 1996;43:238–9. doi: 10.1007/BF00587307. [DOI] [PubMed] [Google Scholar]
- 45.Pemberton RM, Cannon MJ, Openshaw PJ, Ball LA, Wertz GW, Askonas BA. Cytotoxic T cell specificity for respiratory syncytial virus proteins: fusion protein is an important target antigen. J Gen Virol. 1987;68:2177–82. doi: 10.1099/0022-1317-68-8-2177. [DOI] [PubMed] [Google Scholar]
- 46.Bangham CRM, Askonas BA. Murine cytotoxic T-cells specific to respiratory syncytial virus recognise different antigenic subtypes of the virus. J Gen Virol. 1986;67:623–9. doi: 10.1099/0022-1317-67-4-623. [DOI] [PubMed] [Google Scholar]