Abstract
C-reactive protein (CRP), the prototypical human acute phase protein, is produced primarily by hepatocytes. Its expression is modestly induced by interleukin (IL)-6 in Hep3B cells while IL-1, which alone has no effect, synergistically enhances the effects of IL-6. In previous studies of the proximal CRP promoter, we found that signal transducer and activator of transcription-3 (STAT3) and C/EBPβ -mediated IL-6-induced transcription and that Rel p50 acted synergistically with C/EBPβ, in the absence of p65, to enhance CRP transcription. Neither a requirement nor a binding site for the classic nuclear factor (NF)-κB heterodimer p50/p65 were found. The current studies were undertaken to determine whether similar novel transcription factor interactions might regulate the endogenous CRP gene. Transiently overexpressed p50 or p65 induced CRP mRNA accumulation in Hep3B cells. The heterodimer p50/p65 was markedly more effective than p50 or p65 homodimers. Co-overexpression of p50 or p65 with C/EBPβ or STAT3 synergistically enhanced CRP expression. Maximal expression was observed with overexpression of all four transcription factors; comparable effects were observed with IL-1β treatment of cells overexpressing STAT3 + C/EBPβ. Data from the Human Genome Project revealed 13 potential κB sites in the first 4000 bases of the CRP promoter, only one of which, centred at −2652, bound nuclear p50/p65 heterodimer activated by IL-1β. Our findings indicate that classical NF-κB activation can participate in endogenous CRP induction, and that activated NF-κB may synergistically enhance the effects of C/EBPβ and STAT3. They raise the possibility, not as yet established, that NF-κB activation may be responsible for the synergistic effect of IL-1β on IL-6-induced CRP expression.
Introduction
C-reactive protein (CRP) is a major human acute-phase protein whose rate of synthesis may increase 1000-fold or more during inflammatory states.1,2 It is considered to be a host-defence protein and a component of the innate immune system.3,4 Although a single precise in vivo function of CRP has not been defined, CRP has been found to bind to phosphocholine-containing substances and thus has the ability to recognize some foreign pathogens as well as phospholipid constituents of damaged cells.5–7 Ligand-complexed or aggregated CRP binds to C1q and activates the classic complement pathway.8–10 CRP is primarily produced by hepatocytes11 and transcriptional induction of CRP expression, in model systems, requires interleukin (IL)-6.12–16 In the human hepatoma cell line Hep3B, CRP gene expression is modestly induced by IL-6, while IL-1, which alone has no effect on CRP expression, synergistically enhances the effects of IL-6.17 The mechanism by which IL-1 exerts its synergistic effect on IL-6-induced expression of CRP is still unclear.
In a number of other model systems18 including several acute phase proteins, many of the effects of IL-1 have been found to be mediated by NF-κB. The five known members of the mammalian Rel family: p50, p52, p65 (Rel A), Rel B and c-Rel can form homodimers or heterodimers with each other. Classic nuclear factor (NF)-κB, the heterodimer of p50 and p65, is an activator of gene transcription while p50 homodimers have been shown to have both activating and repressing activities on gene transcription.19 In Hep3B cells, we have found that IL-6 activates signal transducer and activator of transcription-3 (STAT3) and C/EBPβ/δ while IL-1β activates p50/p65 heterodimers and p50 homodimers.20–22
In transactivation studies of constructs containing the proximal 157 bp of the CRP promoter, we have found that overexpressed STAT3, C/EBPβ, and Rel p50 are capable of inducing transcription without a requirement for p65.20–22 IL-6 treatment resulted in binding of C/EBPβ and C/EBPδ to two sites on the CRP promoter centred at −53 and −219, and of STAT3 to a site centred at −108.20–24 A novel role for p50 was found in these studies. Rel p50 homodimers were found to bind to a nonconsensus κB site centred at −46, overlapping the proximal C/EBP site on the CRP promoter, with consequent enhanced transcription. Binding sites for the classic NF-κB heterodimer, p50/p65, could not be identified in the first 256 bp of the promoter.
The current studies, undertaken to determine whether similar novel transcription factor interactions might explain the synergistic effect of IL-1 on the endogenous CRP gene, demonstrated that the overexpressed NF-κB heterodimer p50/p65 induced CRP mRNA accumulation and acted synergistically with overexpressed STAT3 and C/EBPβ. IL-1β similarly synergistically enhanced the effects of overexpressed STAT3 and C/EBPβ. Electrophoretic mobility shift assay showed that IL-1β treatment led to the appearance of nuclear p50/p65 heterodimers capable of binding to a κB site centred at position −2652 in the 5′-flanking region of CRP. Our findings indicate that classic NF-κB activation can participate in endogenous CRP induction, and raise the possibility that such activation may be responsible for the synergistic effect of IL-1β on IL-6-induced CRP expression. Further experiments are required to test this hypothesis, and to determine whether such an effect results from binding of NF-κB to the κB site at −2652.
Materials and methods
Materials
Human hepatoma Hep3B cells were provided by Dr G. J. Darlington (Baylor College of Medicine, Houston, TX). Expression vectors encoding p50 and p65 were provided by Dr G. Nabel (National Institutes of Health, Bethesda, MD). Expression vector encoding STAT3-C was provided by Dr J. E. Darnell, Jr. (Rockefeller University, New York, NY). STAT3-C is a constitutively active form of STAT3 that dimerizes spontaneously, binds to DNA, and activates transcription.25 Expression vector encoding C/EBPβ was provided by Dr P. Johnson (National Cancer Institute, Frederick, MD). CRP cDNA probe pCRP5, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) cDNA probe pH 1 cGAP3, were provided by Dr H. R. Colten (Stanford University, Palo Alto, CA) and Dr R. Wu (Cornell University, Ithaca, NY), respectively. Recombinant human IL-1β was purchased from Biosource International (Camarillo, CA). Random-primed DNA labeling reagents were purchased from Boehringer-Mannheim (Indianapolis, IN). Synthetic oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA). Polyclonal antibodies to p50 (H119), p65 (C20) and p52 (C5) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Cell culture, transient transfection and cytokine treatment
Hep3B cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) at 37° in a humidified atmosphere containing 5% CO2. Transfections were performed using the FuGENE 6 reagent (Roche Molecular Laboratory, Indianapolis, IN). Cells were plated onto 100 mm dishes one day prior to transfection so as to achieve 50–80% confluency on the day of transfection. Medium in the dishes was changed with 6 ml of RPMI-1640 medium containing 10% FBS and 1 µm of dexamethasone, just before the transfections. The transfection solution comprising 20 µl of FuGENE 6 and 200 µl of serum-free RPMI-1640 was preincubated for 5 min prior to the addition of plasmids expressing various transcription factors for 15 min at room temperature. Unless otherwise noted, the plasmids were used at: 250 ng for C/EBPβ, 500 ng for p50, 500 ng for p65, and 1 µg for STAT3-C. The FuGENE 6-plasmid mixture was then applied to Hep3B cells and incubated at 37°. The transfection efficiency was found to be approximately 10%. At 16 hr post-transfection, cells were either treated with IL-1β (20 ng/ml) or left untreated. Cells were further incubated for 24 hr, harvested at 40 hr post-transfection and subsequently processed for isolation of RNA as detailed below.
RNA isolation and Northern blot analysis
Total cellular RNA was isolated from Hep3B cells using the RNeasy Mini Kit (Qiagen Inc., Valencia, CA). The isolated RNA was stored at −80° until use. Northern blot analysis was performed as described previously.26 RNA samples (20–25 µg) were denatured for 5 min at 65° in 2 m formaldehyde and fractionated on 0·9% agarose gels containing 2·2 m formaldehyde. RNA was transferred to Gene Screen membranes (NEN, Boston, MA) by capillary blotting and subsequently UV cross-linked. PstI-cut pCRP5 cDNA clone was radiolabelled with [α-32P]dicytosine triphosphate (dCTP) by random priming and used as a probe for hybridization, which was performed overnight at 65° in Church buffer (0·5 m phosphate buffer containing 7% sodium dodecyl sulphate (SDS), 1 mm ethylenediaminetetraacetic acid (EDTA) and 1% bovine serum albumin (BSA)). After hybridization, the membranes were washed with 2× standard sodium citrate (SSC) containing 0·1% SDS at 25° for 15 min and then with 0·2× SSC containing 0·1% SDS at 65° for 45 min. To correct for variations in RNA loading, the blots were stripped (15 min at 85° in water) and rehybridized with a [32P]-labelled PstI-cut pH 1cGAP3 clone, coding for the constitutively expressed GAPDH mRNA. Gels were analysed in a phosphorimager using ImageQuant software (Molecular Dynamics, Sunnyvale, CA). The results are expressed as ratios of CRP to GAPDH (arbitrary units) mRNA accumulation and were termed ‘relative expression of CRP mRNA’ for purposes of comparison. In every set of transfections and Northern blots, overexpression of NF-κB was included to account for any variability between experiments.
Preparation of nuclear extracts
Hep3B cells were grown to 80–90% confluence in 100 mm dishes and treated with 20 ng/ml IL-1β for 15 min Cells were harvested and nuclear extracts were prepared as described previously.27 Briefly, cells were rinsed with ice-cold PBS, harvested by scraping in PBS, and pelleted by centrifugation at 12000 g for 1 min. Pellets were resuspended in 400 µl of buffer A (10 mm Hepes, pH 7·9, 10 mm KCl, 0·1 mm EDTA, 0·1 mm egtazic acid (EGTA), 1 mm dithiothreitol (DTT), 1 mm phenylmethylsulphony fluoride (PMSF), and 0·1 mm sodium orthovanadate) and allowed to swell at 4° for 15 min. Twenty-five µl of 10% nonidet P-40 was added and tubes were vortexed. Nuclei were pelleted by centrifugation at 12000 g at 4° for 1 min, and resuspended in 40 µl of ice-cold buffer B (20 mm Hepes, pH 7·9, 10 mm KCl, 1 mm EDTA, 1 mm EGTA, 420 mm NaCl, 20% glycerol, 1 mm DTT, 1 mm PMSF). The suspension was rocked gently at 4° for 30 min followed by centrifugation at 12000 g at 4° for 10 min. The supernatants (nuclear extracts) were collected and stored in aliquots at −80°. Protein concentration was determined in an aliquot using the Bio-Rad protein assay kit (Bio-Rad, Richmond, CA).
Electrophoretic mobility shift assay (EMSA)
The sequences of the double-stranded oligonucleotides (oligo) used for EMSAs were derived from the CRP promoter; sequences of the top strands are shown in Table 1. The NF-κB consensus oligo was designed according to a published sequence.28 Complementary oligos were annealed and end-labelled with [γ-32P]ATP using T4 polynucleotide kinase. EMSAs were carried out as described previously29 with some modifications. Nuclear extract (6 µg protein/2 µl extract) was incubated with about 0·5 ng of 32P-labelled double-stranded oligo probe (specific activity approximately 5·0 × 106 c.p.m./ng) in gel shift incubation buffer (40 mm KCl, 16 mm Hepes, pH 7·9, 1 mm EDTA, 2·5 mm DTT, 0·25% nonidet P-40, 8% Ficoll, 1 µg of poly dI-dC) at room temperature for 20 min. In supershift experiments, antibodies (2 µg) were added to the reaction mixture and incubated on ice for 15 min before addition of the probe. In competition experiments, 200-fold excess of unlabeled oligo was added to the binding reactions. DNA-protein complexes were resolved by 5% native polyacrylamide gel electrophoresis (PAGE) in 0·25× TBE (1 × TBE = 89 mm Tris-HCl, 89 mm boric acid and 2 mm EDTA) at 10 V/cm. The gels were dried and analysed in a phosphorimager using ImageQuant software.
Table 1.
Putative NF-κB binding sites present within the first 4000 bp of the CRP promoter
| κB-like sites centred at position | Nucleotide sequences* |
|---|---|
| −3920 | −3927 AGTAGAGGTTTTCTGTGAGGG −3907 |
| −3906 | −3920 GTTTTCTGTGAGGGCTCCATGCCTG −3896 |
| −3834 | −3845 CTAGACAGAAACTCCCAAAGC −3825 |
| −3290 | −3300 GTGCTTAGAAATTTCTTAAGCAC −3278 |
| −2955 | −2966 ATTTTTAGGAATCTTTATAGCAG −2944 |
| −2652 | −2662 ATGGGGGAAACCGCTCCTATG −2642 |
| −1700 | −1708 ACAGGGAGAGCTCATCCCTTTTAATAG −1682 |
| −1572 | −1581 AAGAGGAGAGTATTTCTCCTTTA −1559 |
| −1135 | −1145 GATACGGTGGTTCTTTTCTGATT −1123 |
| −966 | −975 TGTGGAGGGATTACTTGAATC −955 |
| −780 | −791 ATGTCTAGAGAGTTCTTAATAAG −769 |
| −643 | −653 ACTCTGGGGACTGTTGTGGGGTG −631 |
| −222 | −236 TGGAGCCCTGAGAGATTTCTT −216 |
| κB consensus | CCAAGGGGACTTTCCATG |
The sequences of the coding strands of 13 possible κB sites on the CRP promoter, that closely match the κB consensus sequence, are shown. These oligonucleotides were used in the EMSAs shown in Fig. 7. The κB-site-like sequences are in bold.
Results
Overexpressed homodimers and heterodimers of p50 and p65 induce endogenous CRP expression
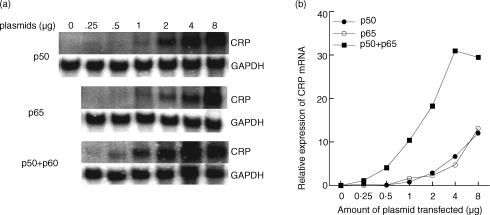
Employing Northern blot analyses, we measured the accumulation of CRP mRNA in Hep3B cells in response to overexpressed transcription factors known to be activated by IL-6 or IL-1. Transient transfection of Hep3B cells with increasing amounts of plasmids (0·25–8·0 µg) expressing the NF-κB proteins p50 or p65 induced CRP mRNA accumulation in a dose-dependent manner. A representative set of Northern blots is shown in Fig. 1a and quantitated in Fig. 1(b). Overexpressed p50 and overexpressed p65 had equivalent activities on induction of CRP expression, while their combination was markedly (approximately sixfold between 1 and 3 µg) more effective. The effect of the combination of overexpressed p50 and p65 was not caused by the double amount of the plasmids since the effect was not merely additive. No CRP mRNA was detected in mock-transfected cells.
Figure 1.
Overexpressed NF-κB induces endogenous CRP expression. (a) Dose-dependent induction of CRP mRNA accumulation in Hep3B cells in response to overexpressed p50, p65, and their combination. The cells were transiently transfected with increasing amounts of plasmids expressing the indicated transcription factors. The amounts of plasmids shown are the amounts used for each plasmid; thus the cells overexpressing both p50 and p65 received double amount of total DNA. After 40 hr, total RNA was isolated and subjected to Northern blot analysis. The blots were probed for CRP and GAPDH mRNA. One set of Northern blots, representative of three independent experiments, is shown. (b) CRP mRNA accumulation was analysed in a phosphorimager and normalized to GAPDH expression. The relative expression of CRP mRNA indicates the ratio of CRP to GAPDH mRNA at each dose relative to the other doses of overexpressed proteins. The numbers are arbitrary.
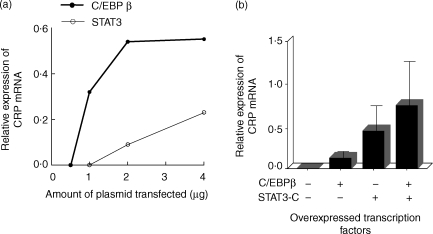
In contrast, overexpressed C/EBPβ and overexpressed STAT3, at concentrations of transfected plasmids varying from 0·5 to 4 µg, induced CRP mRNA accumulation only minimally and variably and the effect of their combination was additive (Fig. 2). In subsequent experiments, concentrations of C/EBPβ (250 ng) and STAT3-C plasmids (1 µg) were selected which generated minimal CRP mRNA responses.
Figure 2.
Overexpressed C/EBPβ and STAT3 induce endogenous CRP expression minimally. Hep3B cells were transiently transfected with plasmids expressing the indicated transcription factors. After 40 hr, total RNA was isolated and subjected to Northern blot analysis. The blots were probed for CRP and GAPDH mRNA. CRP mRNA accumulation was analysed in a phosphorimager and normalized to GAPDH expression. (a) Dose–response curves of the effects of increasing amounts of the plasmids expressing C/EBPβ and STAT3 on CRP mRNA accumulation. A representative of three experiments is shown. (b) Cells were tranfected with 250 ng of the plasmid expressing C/EBPβ and 1 µg of the plasmid expressing STAT3-C. The values on the y-axis are the normalized means + SD expression from four separate experiments.
NF-κB acts synergistically with C/EBPβ and STAT3 to induce endogenous CRP expression
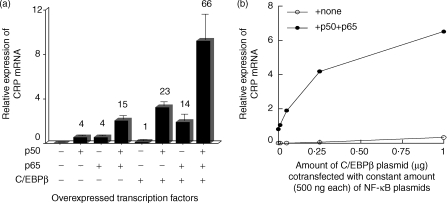
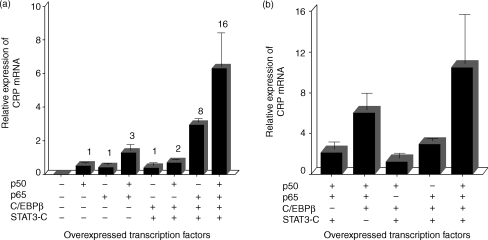
When NF-κB proteins and C/EBPβ were overexpressed together (co-overexpression), both p50 and p65 (500 ng of each plasmid) acted synergistically with C/EBPβ in inducing CRP mRNA accumulation: co-overexpressing p50 enhanced the inducing effect of overexpressed C/EBPβ by 23-fold while co-overexpressing p65 enhanced the inducing effect of overexpressed C/EBPβ by 14-fold (Fig. 3a). The effect of the heterodimer was striking: co-overexpression of both p50 and p65 enhanced the inducing effect of C/EBPβ by 66-fold. The effect of overexpressed NF-κB proteins was four- to sixfold greater in the presence of overexpressed C/EBPβ than in its absence. Synergism between C/EBPβ and NF-κB was dose related: CRP expression was increased when increasing amounts of a plasmid expressing C/EBPβ were cotransfected with constant amounts (500 ng) of the plasmids expressing p50 and p65 (Fig. 3b).
Figure 3.
NF-κB acts synergistically with C/EBPβ to induce endogenous CRP expression. Hep3B cells were transiently transfected with the plasmids expressing the indicated transcription factors. After 40 hr, total RNA was isolated and subjected to Northern blot analysis. The blots were probed for CRP and GAPDH mRNA. CRP mRNA accumulation was analysed in a phosphorimager and normalized to GAPDH expression. (a) Cells were transfected with 250 ng of the plasmid expressing C/EBPβ and 500 ng of the individual plasmids expressing p50 and p65. The values on the y-axis are the normalized means + SD expression from five separate experiments. The numbers above each bar represent fold induction compared to C/EBPβ alone, which was taken as 1. (b) Cells were cotransfected with increasing amounts of the plasmid expressing C/EBPβ with a constant amount (500 ng) of the individual plasmids expressing p50 and p65.
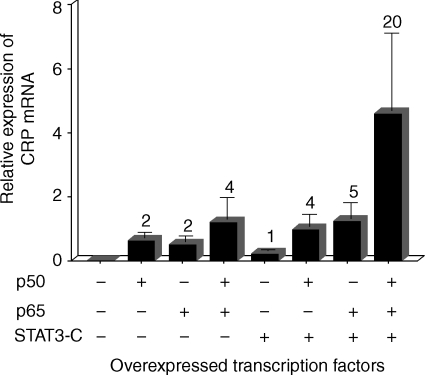
Similarly, both p50 and p65 enhanced the effect of overexpressed STAT3 on CRP mRNA accumulation (Fig. 4). Co-overexpressing p50 or p65 enhanced the effect of overexpressed STAT3 by fourfold and fivefold, respectively. Again, the effect of the heterodimer was strikingly greater; co-overexpression of p50/p65 with STAT3 enhanced the effect of STAT3 by 20-fold. In the presence of overexpressed STAT3, the effect of overexpressed NF-κB proteins was two- to fivefold greater than in the absence of overexpressed STAT3.
Figure 4.
NF-κB acts synergistically with STAT3 to induce endogenous CRP expression. Hep3B cells were transiently transfected with 1 µg of the plasmid expressing STAT3-C and 500 ng of the plasmids expressing p50 and p65. After 40 hr, total RNA was isolated and subjected to Northern blot analysis. The blots were probed for CRP and GAPDH mRNA. CRP mRNA accumulation was analysed in a phosphorimager and normalized to GAPDH expression. The values on the y-axis are the normalized means + SD expression from two separate experiments. The numbers above each bar represent fold induction compared to STAT3 alone, which was taken as 1.
Maximal CRP expression is induced by co-overexpressing p50, p65, C/EBPβ and STAT3
The effect of co-overexpression of NF-κB with the combination STAT3 + C/EBPβ was found to be substantially greater than that of co-overexpression of NF-κB with either of the two other transcription factors alone. Co-overexpressing p50 or p65 enhanced the inducing effect of overexpressed [STAT3 + C/EBPβ] by twofold and eightfold, respectively (Fig. 5a). However, co-overexpression of both the Rel proteins synergistically enhanced the inducing effect of overexpressed [STAT3 + C/EBPβ] by 16-fold. In the presence of overexpressed [STAT3 + C/EBPβ], the effect of overexpressed NF-κB proteins was two- to eightfold greater than in the absence of [STAT3 + C/EBPβ]. The effects of co-overexpressing four transcription factors (STAT3, C/EBPβ, p50 and p65) on endogenous CRP expression were compared with the effects of any three of the four proteins (Fig. 5b). CRP mRNA accumulation was induced maximally when all four transcription factors were over-expressed.
Figure 5.
Maximal endogenous CRP expression is induced by co-overexpressing NF-κB, STAT3 and C/EBPβ. Hep3B cells were transiently transfected with 250 ng of plasmid expressing C/EBPβ, 1 µg of the plasmid expressing STAT3-C, and 500 ng of each plasmid expressing p50 and p65. After 40 hr, total RNA was isolated and subjected to Northern blot analysis. The blots were probed for CRP and GAPDH mRNA. CRP mRNA accumulation was analysed in a phosphorimager and normalized to GAPDH expression. The values on the y-axis are the normalized means + SD expression from three separate experiments. (a) Overexpressed NF-κB induces CRP mRNA accumulation synergistically with overexpressed C/EBPβ + STAT3. The numbers above each bar represent fold induction compared to C/EBPβ + STAT3 alone, which was taken as 1. (b)Maximal CRP expression was induced when all four proteins, p50, p65, STAT3 and C/EBPβ, were co-overexpressed.
Both IL-1β and NF-κB overexpression greatly enhance endogenous CRP expression in cells overexpressing STAT3 + C/EBPβ
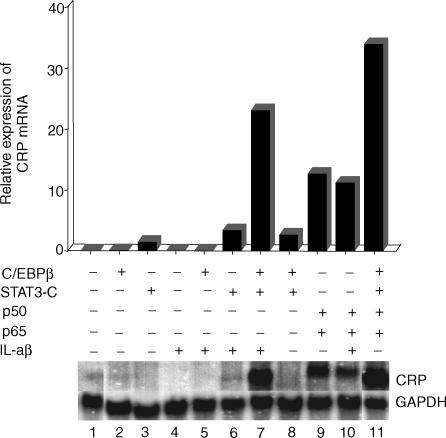
We next compared the effects of IL-1β treatment with that of NF-κB overexpression on CRP mRNA accumulation in cells transiently overexpressing C/EBPβ, STAT3, or their combination (Fig. 6). Overexpressed C/EBPβ, overexpressed STAT3, and IL-1β alone induced CRP mRNA accumulation minimally or not at all (lanes 2–4). Treatment with IL-1β did not affect CRP expression in cells overexpressing C/EBPβ and only modestly enhanced CRP expression in cells overexpressing STAT3 (lanes 5 and 6), the latter to a level comparable to that seen in cells co-overexpressing C/EBPβ and STAT3 (lane 8). However, IL-1β enhanced the inducing effect of overexpressed STAT3 + C/EBPβ ninefold (comparison of lanes 7 and 8), as did overexpressed NF-κB, which enhanced the inducing effect of overexpressed STAT3 + C/EBPβ by 13-fold (comparison of lanes 8 and 11). IL-1β treatment had no effect on CRP mRNA accumulation in cells overexpressing p50 and p65 (comparison of lanes 9 and 10). In a separate experiment, the effects of overexpressed STAT3 + C/EBPβ + p50 + p65 on the induction of CRP mRNA accumulation were compared in cells with or without IL-1β-treatment. IL-1β treatment had no enhancing effect on the induction of CRP mRNA accumulation in cells overexpressing all four transcription factors (data not shown).
Figure 6.
IL-1β greatly enhances endogenous CRP expression in cells overexpressing STAT3 and C/EBPβ. Hep3B cells were transiently transfected with 250 ng of the plasmid expressing C/EBPβ, 1 µg of the plasmid expressing STAT3-C, and 500 ng each for expressing p50 and p65. After 16 hr, indicated cells were treated with IL-1β. At 40 hr post-transfection, total RNA was isolated and subjected to Northern blot analysis. The blots were probed for CRP and GAPDH mRNA. CRP mRNA accumulation was analysed in a phosphorimager and normalized to GAPDH expression. A representative of two experiments is shown.
IL-1β-activated NF-κB binds to a κB site centred at −2652 on the CRP promoter
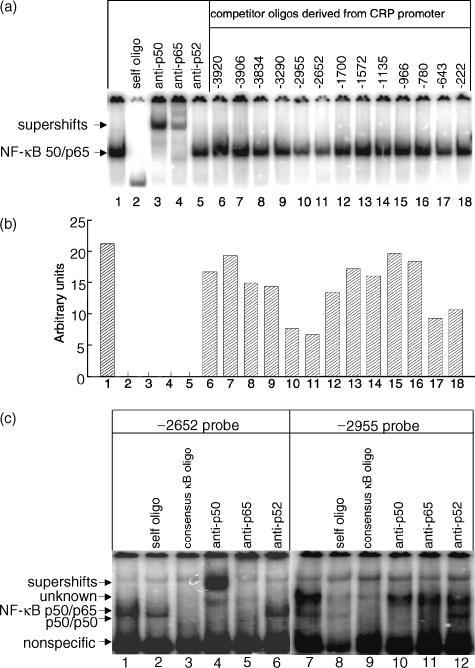
Data generated by the Human Genome Project30 permitted us to analyse 4000 bp upstream of the transcriptional start site of the CRP promoter. Visual inspection of this sequence revealed 13 potential κB sites on the CRP promoter (Table 1). To determine whether any of these sequences were capable of binding the classical NF-κB heterodimer, p50/p65, we synthesized 13 pairs of complementary oligos derived from these sites and used them as competitors in an EMSA experiment. The assay used the consensus κB oligo as a probe and nuclear extracts of Hep3B cells activated by IL-1β treatment for 15 min as a source of p50/p65 heterodimers (Fig. 7). The NF-κB in this extract is shown by the single complex formed on the consensus κB oligo (Fig. 7a, lane 1). This complex was specific and contained the NF-κB p50/p65 heterodimer as determined by competition and supershift analyses (lanes 2–5). Determination of the ability of the 13 oligos we synthesized to compete for binding of NF-κB to the consensus κB probe (lanes 6–18) revealed competition with four of the 13 oligos, those centred at position −2955, −2652, −643 and −222 (lanes 10, 11, 17 and 18). Although employed in 200-fold excess, competition was only partial (Fig. 7b, lanes 10, 11, 17 and 18) indicating that binding of NF-κB to these four oligos was weaker than the binding of NF-κB to its consensus sequence.
Figure 7.
IL-1β-activated NF-κB binds to a κB site centred at −2652 on the CRP promoter. The EMSAs utilized nuclear extracts from Hep3B cells treated with IL-1β for 15 min Antibodies recognizing p50, p65 and p52 were added to the nuclear extract before the addition of the labelled probe. DNA-protein complexes were separated by 5% native PAGE and the results were analysed by phosphorimager. The mobility of the free probe is not shown. (a) The consensus κB oligo was used as the radiolabelled probe (lanes 1–18). Unlabeled self (lane 2) and various oligos derived from the CRP promoter (lanes 6–18) were used as competitors and were added in 200-fold excess. The numbers above lanes 6–18 are the positions where the oligos are centred in the CRP promoter. A representative of two EMSAs is shown. (b) The intensities of the NF-κB complex in each lane were quantitated using ImageQuant software and are expressed as arbitrary densitometric units on the y-axis against the lane number on the x-axis. (c) The oligos centred at position −2652 (lanes 1–6) and at position −2955 (lanes 7–12) both derived from the CRP promoter were used as radiolabelled probes. Unlabelled self oligos and the consensus κB oligo were used as competitors and were added in 200-fold excess. A representative of two EMSAs is shown.
We therefore radiolabelled these four oligos and performed EMSA to determine whether they bound directly to NF-κB, employing nuclear extracts from IL-1β-treated cells. The analysis for −2652 and −2955 oligos is shown in Fig. 7(c). The single complex, formed on the −2652 probe, was competed by excess unlabelled self or excess unlabelled consensus κB oligos (lanes 1–3). This complex could be supershifted by antibodies to p50 and abolished by antibodies to p65 (lanes 4, and 5), indicating that the classical NF-κB p50/p65 bound to this site. Antibodies to p52 had no effect. Two complexes were formed on the −2955 probe. The faster migrating faint complex was affected only by anti-p50 antibodies and not affected by other anti-Rel antibodies, possibly indicating that few p50 homodimers may be capable of binding to this site. In separate experiments, NF-κB was not found to bind to the oligos centred at positions −222 and −643 either (data not shown). Thus, only the site centred at −2652 among the 13 possible κB sites we studied bound NF-κB in nuclear extracts from IL-1β-treated Hep3B cells.
Discussion
In Hep3B cells, exposure to IL-6 modestly induces CRP expression and results in binding of C/EBPβ and C/EBPδ to two sites on the CRP promoter, centred at −53 and −219, and binding of STAT3 to a site centred at −108, with consequent activation of gene expression.20–24 IL-1, which alone has no effect on CRP expression, markedly and synergistically enhances the inducing effect of IL-6 by unknown mechanisms. This series of studies was undertaken to evaluate the possible role of NF-κB, a transcription factor commonly activated by IL-1β in Hep3B cells. Our previous studies of the proximal 256 nucleotides of the CRP promoter failed to indicate a role for classical NF-κB, the heterodimer of p50 and p65 (henceforth referred to as NF-κB) but revealed a novel finding: Rel p50 homodimers were found to bind to a non-consensus κB site overlapping the proximal C/EBP site on the CRP promoter, with consequent enhanced transcription. Binding sites for NF-κB could not be identified in the first 256 bp of the promoter.22
We evaluated the possible role of Rel proteins in induction of the endogenous CRP gene. In contrast to our findings in the proximal promoter, the major findings in this study were: (1) Overexpression of either Rel 50 or Rel 65 induced expression of the endogenous CRP gene and their combination was markedly more effective than either Rel protein alone. (2) NF-κB acted synergistically with both C/EBPβ and STAT3 to induce endogenous CRP expression; maximal CRP expression was observed when all four transcription factors p50 + p65 + C/EBPβ + STAT3 were overexpressed. (3) IL-1β treatment also greatly enhanced endogenous CRP expression in cells overexpressing both STAT3 + C/EBPβ, to a degree roughly comparable to that observed with NF-κB overexpression. (4) IL-1β-activated NF-κB was found to bind to a κB site centred at −2652 on the CRP promoter.
Whether the synergistic effect of IL-1β on IL-6-induced CRP expression is in fact mediated through NF-κB activation is not clear. However, many of the effects of IL-1 have been found to be mediated by the NF-κB system. An example is serum amyloid-A (SAA), which exhibits great similarity to CRP in magnitude and kinetics of induction and the cytokines required for its optimal induction in Hep3B cells.31 IL-1 has been shown to induce SAA expression by activation of NF-κB, with consequent physical interaction between p65 and activated C/EBPβ.32 Besides SAA, NF-κB has been shown to play a role in expression of many other inflammation associated genes including IL-6,33 IL-1,34 intercellular adhesion molecule-1,35 lipopolysaccharide-binding protein,36 angiotensinogen37 and granulocyte colony-stimulating factor.38 The synergism between NF-κB and C/EBPβ on induction of CRP expression that we observed on induction of CRP expression has also been shown for a number of other gene promoters.39–41
The most obvious explanation for the discrepancy between the findings in the proximal promoter and the larger region we have surveyed here is, of course, the location of the κB site, which is not present on the proximal promoter, but rather is present 2·6 kb upstream. Such a conclusion is consistent with studies in transgenic mice indicating that extended 5′- and 3′-flanking regions are capable of influencing CRP gene expression.42 In addition, in contrast to the current studies of the endogenous gene, studies of the proximal CRP promoter relied heavily on study of transiently transfected promoters, which may not be subject to the same chromatin assembly and subsequent regulatory mechanisms that operate on their integrated counterparts.43
Induction of CRP expression by overexpressed NF-κB alone was unexpected, since IL-1β does not itself induce CRP expression in Hep3B cells although it activates NF-κB. A possible explanation may reside in the kinetics of transcription factor activation. We have shown that IL-1β activates NF-κB rapidly within 15 min, but only p50 homodimers are present in these cells at 18 hr.22 In contrast, overexpressed NF-κB might persist for a considerably longer period. IL-1, in addition to activating NF-κB, could conceivably activate other transcription factors capable of repressing CRP expression in the absence of IL-6-activated transcription factors. Our finding of induction of CRP expression by overexpressed p50 alone, which does not contain a transactivation domain, may be due to synergy with the basal levels of C/EBPβ present in Hep3B cells, as found in our studies of the proximal promoter.21 EMSA of Hep3B nuclear extracts did not reveal any endogenous p65, although a low level of p50 was always found to be present in these cells.21 Thus, the results obtained from overexpressed p50 were not due in part to endogenous p65. Indeed the results obtained with overexpressed p65 might involve endogenous p50.
Our observation that co-overexpressed NF-κB and STAT3 synergistically induce CRP expression is supported by the recent report that proteolysis-inducing factor induces CRP expression in primary hepatocyte cultures by activation of NF-κB and STAT3.44 Synergy between NF-κB and STAT proteins has also been reported for the promoters of interferon regulatory factor-1 and toll-like receptor-2, and for IL-4-induced transcription.45–47 Our data strongly indicate that NF-κB acts synergistically with both STAT3 and C/EBPβ to induce CRP expression. This conclusion is arrived at with a degree of caution since transcription factor overexpression may not accurately reflect physiologic conditions.
We have found that in IL-6 + IL-1β-treated Hep3B cells, NF-κB and STAT3 are activated after only 15 min of treatment, while C/EBPβ activation is not apparent until 4 hr.19,20 Substantial IL-1 induction of CRP expression occurred only in cells overexpressing both STAT3 and C/EBPβ; IL-1 caused no induction of CRP expression in cells overexpressing C/EBPβ and only minimal enhancement of CRP induction in cells overexpressing STAT3. Taken together with our finding that maximal CRP expression was similarly observed in cells overexpressing p50, p65, STAT3, and C/EBPβ, this finding suggests that the initiation of CRP expression starts with the interaction between STAT3 and NF-κB and is sustained through the interaction between C/EBPβ and p50. The effects of cytokine treatment and overexpressed transcription factors were not compared in more detail because transfection efficiency in our hands is low while cytokine treatment affects virtually all cells.
Until now, an NF-κB binding site had not been identified in the 5′-flanking region of the CRP promoter. The NF-κB binding sequence we have identified, centred at −2652, 5′-GGGAAACCGCTCC-3′, is similar to κB sites present on promoters of other genes, including tumour necrosis factor-α and IL-2·48 The subtle difference between the −2652 κB site and the consensus κB site probably explains our observation of relatively weaker binding of NF-κB to the −2652 κB site on the CRP promoter. Nevertheless such variant κB sites have been found to be important in gene transcription49,50 because protein–protein interactions with other transcription factors are believed to stabilize their binding to DNA. Thus, interactions between NF-κB and IL-6-activated transcription factors could enhance the stability of binding of these transcription factors to the CRP promoter.
Taken together, our findings indicate that NF-κB activation can participate in endogenous CRP induction. We are currently mutating the κB site at −2652 and performing transactivation assays to document the function of this κB site for the regulation of CRP gene expression. In addition, we are employing inhibitors of NF-κB activation to test the hypothesis that the synergistic effect of IL-1β on IL-6-induced CRP expression is mediated through activation of NF-κB and its subsequent binding to the κB site at −2652.
Acknowledgments
We thank Dr Gary Nabel, Dr Peter Johnson, Dr J. E. Darnell, Dr H. R. Colten, Dr X. H. Sun and R. Wu for various plasmid constructs and Dr G.J. Darlington for providing Hep3B cells. The work was supported by National Institutes of Health.
Abbreviations
- CRP
C-reactive protein
- IL
interleukin
- EMSA
electrophoretic mobility shift assay
- Oligo
oligonucleotides
References
- 1.Tillett WS, Francis T., Jr Serological reactions in pneumonia with a non-protein somatic fraction of pneumococcus. J Exp Med. 1930;52:561–71. doi: 10.1084/jem.52.4.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kushner I. The phenomenon of the acute phase response. Ann NY Acad Sci USA. 1982;389:39–48. doi: 10.1111/j.1749-6632.1982.tb22124.x. [DOI] [PubMed] [Google Scholar]
- 3.Gabay C, Kushner I. Acute-phase proteins and other systemic responses to inflammation. N Engl J Med. 1999;340:448–54. doi: 10.1056/NEJM199902113400607. [DOI] [PubMed] [Google Scholar]
- 4.Volanakis JE. Human C-reactive protein: expression, structure and function. Mol Immunol. 2001;38:189–97. doi: 10.1016/s0161-5890(01)00042-6. [DOI] [PubMed] [Google Scholar]
- 5.Kushner I, Kaplan MH. Studies of acute phase protein. I. an immunohistochemical method for the localization of Cx-reactive protein in rabbits. association with necrosis in local inflammatory lesions. J Exp Med. 1961;114:961–73. doi: 10.1084/jem.114.6.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Volanakis JE, Kaplan MH. Specificity of C-reactive protein for choline phosphate residues of pneumococcal C-polysaccharide. Proc Soc Exp Biol Med. 1971;136:612–4. doi: 10.3181/00379727-136-35323. [DOI] [PubMed] [Google Scholar]
- 7.Agrawal A, Simpson MJ, Black S, Carey MP, Samols D. A C-reactive protein mutant that does not bind to phosphocholine and pneumococcal C-polysaccharide. J Immunol. 2002;169:3217–22. doi: 10.4049/jimmunol.169.6.3217. [DOI] [PubMed] [Google Scholar]
- 8.Kaplan MH, Volanakis JE. Interaction of C-reactive protein complexes with the complement system. I. Consumption of human complement associated with the reaction of C-reactive protein with pneumococcal C-polysaccharide and with the choline phosphatides, lecithin and sphingomyelin. J Immunol. 1974;112:2135–47. [PubMed] [Google Scholar]
- 9.Jiang H, Lint TF, Gewurz H. Defined chemically cross-linked oligomers of human C-reactive protein: characterization and reactivity with the complement system. Immunology. 1991;74:725–31. [PMC free article] [PubMed] [Google Scholar]
- 10.Agrawal A, Shrive AK, Greenhough TJ, Volanakis JE. Topology and structure of the C1q-binding site on C-reactive protein. J Immunol. 2001;166:3998–4004. doi: 10.4049/jimmunol.166.6.3998. [DOI] [PubMed] [Google Scholar]
- 11.Kushner I, Feldmann G. Control of the acute phase response. Demonstration of C-reactive protein synthesis and secretion by hepatocytes during acute inflammation in the rabbit. J Exp Med. 1978;148:466–77. doi: 10.1084/jem.148.2.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganapathi MK, May LT, Schultz D, Brabenec A, Weinstein J, Sehgal PB, Kushner I. Role of interleukin-6 in regulating synthesis of C-reactive protein and serum amyloid A in human hepatoma cell lines. Biochem Biophys Res Comm. 1988;157:271–7. doi: 10.1016/s0006-291x(88)80043-3. [DOI] [PubMed] [Google Scholar]
- 13.Ganter U, Arcone R, Toniatti C, Morrone G, Ciliberto G. Dual control of C-reactive protein gene expression by interleukin-1 and interleukin-6. EMBO J. 1989;8:3773–9. doi: 10.1002/j.1460-2075.1989.tb08554.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Steel DM, Whitehead AS. Heterogeneous modulation of acute-phase-reactant mRNA levels by interleukin-1β and interleukin-6 in the human hepatoma cell line PLC/PRF/5. Biochem J. 1991;277:477–82. doi: 10.1042/bj2770477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang D, Jiang S, Rzewnicki D, Samols D, Kushner I. The effect of interleukin-1 on C-reactive protein expression in Hep3B cells is exerted at the transcriptional level. Biochem J. 1995;310:143–8. doi: 10.1042/bj3100143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinhold B, Bader A, Poli V, Rüther U. Interleukin-6 is necessary, but not sufficient, for induction of the human C-reactive protein gene in vivo. Biochem J. 1997;325:617–21. doi: 10.1042/bj3250617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ganapathi MK, Rzewnicki D, Samols D, Jiang S, Kushner I. Effect of combinations of cytokines and hormones on synthesis of serum amyloid A and C-reactive protein in Hep3B cells. J Immunol. 1991;147:1261–5. [PubMed] [Google Scholar]
- 18.Dinarello CA. Interleukin-1. Cytokine Growth Factor Rev. 1997;8:253–165. doi: 10.1016/s1359-6101(97)00023-3. [DOI] [PubMed] [Google Scholar]
- 19.Baeuerle PA, Baltimore D. NF-κB. ten years after. Cell. 1996;87:13–20. doi: 10.1016/s0092-8674(00)81318-5. [DOI] [PubMed] [Google Scholar]
- 20.Zhang D, Sun M, Samols D, Kushner I. STAT3 participates in transcriptional activation of the C-reactive protein gene by interleukin-6. J Biol Chem. 1996;271:9503–9. doi: 10.1074/jbc.271.16.9503. [DOI] [PubMed] [Google Scholar]
- 21.Agrawal A, Cha-Molstad H, Samols D, Kushner I. Transactivation of C-reactive protein by IL−6 requires synergistic interactions of CCAAT/enhancer binding protein β (C/EBPβ) and Rel p50. J Immunol. 2001;166:2378–84. doi: 10.4049/jimmunol.166.4.2378. [DOI] [PubMed] [Google Scholar]
- 22.Cha-Molstad H, Agrawal A, Zhang D, Samols D, Kushner I. The Rel family member p50 mediates cytokine-induced C-reactive protein expression by a novel mechanism. J Immunol. 2000;165:4592–7. doi: 10.4049/jimmunol.165.8.4592. [DOI] [PubMed] [Google Scholar]
- 23.Li S, Goldman ND. Regulation of human C-reactive protein gene expression by two synergistic IL-6 responsive elements. Biochemistry. 1996;35:9060–8. doi: 10.1021/bi953033d. [DOI] [PubMed] [Google Scholar]
- 24.Majello B, Arcone R, Toniatti C, Ciliberto G. Constitutive and IL-6-induced nuclear factors that interact with the human C-reactive protein promoter. EMBO J. 1990;9:457–65. doi: 10.1002/j.1460-2075.1990.tb08131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr STAT3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- 26.Jiang S, Samols D, Rzewnicki D, Macintyre SS, Greber I, Sipe J, Kushner I. Kinetic modeling and mathematical analysis indicate that acute phase gene expression in Hep3B cells is regulated by both transcriptional and posttranscriptional mechanisms. J Clin Invest. 1995;95:1253–61. doi: 10.1172/JCI117775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schreiber E, Matthias P, Müller MM, Schaffner W. Rapid detection of octamer binding proteins with ‘mini-extracts’, prepared from a small number of cells. Nucl Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sen R, Baltimore D. Multiple nuclear factors interact with the immunoglobulin enhancer sequences. Cell. 1986;46:705–16. doi: 10.1016/0092-8674(86)90346-6. [DOI] [PubMed] [Google Scholar]
- 29.Levy D, Kessler DS, Pine R, Darnell JE., Jr Cytoplasmic activation of ISGF3, the positive regulator of interferon-α-stimulated transcription, reconstituted in vitro. Genes Dev. 1989;3:1362–71. doi: 10.1101/gad.3.9.1362. [DOI] [PubMed] [Google Scholar]
- 30.International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 31.Ganapathi MK, Schultz D, Mackiewicz A, Samols D, Hu S, Brabenec A, Macintyre SS, Kushner I. Heterogeneous nature of the acute phase response: differential regulation of human serum amyloid A, C-reactive protein, and other acute phase proteins by cytokines in Hep3B cells. J Immunol. 1988;141:564–9. [PubMed] [Google Scholar]
- 32.Betts JC, Cheshire JK, Akira S, Kishimoto T, Woo P. The role of NF-κB and NF-IL-6 transactivating factors in the synergistic activation of human serum amyloid A gene expression by interleukin-1 and interleukin-6. J Biol Chem. 1993;268:25624–31. [PubMed] [Google Scholar]
- 33.Matsusaka T, Fujikawa K, Nishio Y, Mukaida N, Matsushima K, Kishimoto T, Akira S. Transcription factor NF-IL-6 and NF-κB synergistically activate transcription of the inflammatory cytokines, interleukin-6 and interleukin-8. Proc Natl Acad Sci USA. 1993;90:10193–7. doi: 10.1073/pnas.90.21.10193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Rom WN. Regulation of the interleukin-β (IL-1β) gene by mycobacterial components and lipopolysaccharide is mediated by two nuclear factor-IL-6 motifs. Mol Cell Biol. 1993;13:3831–7. doi: 10.1128/mcb.13.6.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Catron KM, Brickwood JR, Shang C, Li Y, Shannon MF, Parks TP. Cooperative binding and synergistic activation by RelA and C/EBPβ on the intercellular adhesion molecule-1 promoter. Cell Growth Differ. 1998;9:949–59. [PubMed] [Google Scholar]
- 36.Schumann RR, Kirschning CJ, Unbehaun A, Aberle HP, Knope HP, Lamping N, Ulevitch RJ, Herrmann F. The lipopolysaccharide-binding protein is a secretory class 1 acute phase protein whose gene is transcriptionally activated by APRF/STAT3 and other cytokine-inducible nuclear proteins. Mol Cell Biol. 1996;16:3490–503. doi: 10.1128/mcb.16.7.3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brasier AR, Ron D, Tate JE, Habener JF. A family of constitutive C/EBP-like DNA binding proteins attenuate the IL-1α-induced, NF-κB mediated trans-activation of the angiotensinogen gene acute phase response element. EMBO J. 1990;9:3933–44. doi: 10.1002/j.1460-2075.1990.tb07614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunn SM, Coles LS, Lang RK, Gerondokis S, Vadas MA, Shannon MF. Requirement for nuclear factor (NF)-κB p65 and NF-interleukin-6 binding elements in the tumor necrosis factor response region of the granulocyte colony stimulating factor promoter. Blood. 1994;83:2469–79. [PubMed] [Google Scholar]
- 39.LeClair KP, Blanar MA, Sharp PA. The p50 subunit of NF-κB associates with the NF-IL6 transcription factor. Proc Natl Acad Sci USA. 1992;89:8145–9. doi: 10.1073/pnas.89.17.8145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stein B, Cogswell PC, Baldwin AS., Jr Functional and physical associations between NF-κB and C/EBP family members: a Rel domain–bZIP interaction. Mol Cell Biol. 1993;13:3964–74. doi: 10.1128/mcb.13.7.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia C, Cheshire JK, Patel H, Woo P. Cross-talk between transcription factors NF-κB and C/EBP in the transcriptional regulation of genes. Int J Biochem Cell Biol. 1997;29:1525–39. doi: 10.1016/s1357-2725(97)00083-6. [DOI] [PubMed] [Google Scholar]
- 42.Murphy C, Beckers J, Rüther U. Regulation of the human C-reactive protein gene in transgenic mice. J Biol Chem. 1995;270:704–8. doi: 10.1074/jbc.270.2.704. [DOI] [PubMed] [Google Scholar]
- 43.Smith CL, Hager GL. Transcriptional regulation of mammalian genes in vivo. A tale of two templates. J Biol Chem. 1997;272:27493–6. doi: 10.1074/jbc.272.44.27493. [DOI] [PubMed] [Google Scholar]
- 44.Watchorn TM, Waddell ID, Dowidar N, Ross JA. Proteolysis-inducing factor regulates hepatic gene expression via the transcription factors NF-κB and STAT3. FASEB J. 2001;15:562–564. doi: 10.1096/fj.00-0534fje. [DOI] [PubMed] [Google Scholar]
- 45.Ohmori Y, Schreiber RD, Hamilton TA. Synergy between interferon-γ and tumor necrosis factor-α in transcriptional activation is mediated by cooperation between signal transducer and activator of transcription 1 and nuclear factor κB. J Biol Chem. 1997;272:14899–907. doi: 10.1074/jbc.272.23.14899. [DOI] [PubMed] [Google Scholar]
- 46.Musikacharoen T, Matsuguchi T, Kikuchi T, Yoshikai Y. NF-κB and STAT5 play important roles in the regulation of mouse toll-like receptor 2 gene expression. J Immunol. 2001;166:4516–24. doi: 10.4049/jimmunol.166.7.4516. [DOI] [PubMed] [Google Scholar]
- 47.Shen C, Stavnezer J. Interaction of STAT6 and NF–κB: direct association and synergistic activation of interleukin-4-induced transcription. Mol Cell Biol. 1998;18:3395–404. doi: 10.1128/mcb.18.6.3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen FE, Ghosh G. Regulation of DNA binding by Rel/NF-κB transcription factors: structural views. Oncogene. 1999;18:6845–52. doi: 10.1038/sj.onc.1203224. [DOI] [PubMed] [Google Scholar]
- 19.Lewis H, Kaszubska W, DeLamarter JF, Whelan J. Cooperativity between two NF-κB complexes, mediated by high-mobility-group protein I (Y), is essential for cytokine-induced expression of the E-selectin promoter. Mol Cell Biol. 1994;14:5701–9. doi: 10.1128/mcb.14.9.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brouillard F, Bouthier M, Leclerc T, Clement A, Baudouin-Legros M, Edelman A. NF-κB mediates up-regulation of CFTR gene expression in Calu-3 cells by interleukin-1β. J Biol Chem. 2001;276:9486–91. doi: 10.1074/jbc.M006636200. [DOI] [PubMed] [Google Scholar]