Abstract
Interleukin-5 (IL-5) is a T helper type 2 cytokine, which is implicated in the pathogenesis of eosinophilic diseases such as asthma. Both peripheral blood mononuclear cells (PBMC) and primary human T cells display similar patterns of IL-5 expression when stimulated with both phorbol-12-myristate 13-acetate and phytohaemagglutinin. The expression of IL-5 stimulated by these agents was shown to require de novo transcription and translation. However, although dexamethasone was a potent inhibitor of both IL-5 release and messenger RNA accumulation from PBMC and T cells, dexamethasone had no effect on the luciferase activity of a reporter construct under the control of an IL-5 promoter region transiently transfected into primary human T cells. Furthermore, dexamethasone appeared to decrease the stability of IL-5 messenger RNA and this effect was dependent upon de novo transcription. Taken together, the results presented here suggest that, whilst transcriptional processes predominantly regulate IL-5 release, the mechanism by which dexamethasone inhibits IL-5 is post-transcriptional.
Introduction
Asthma is a complex inflammatory disease characterized by eosinophil infiltration into the airway.1 Interleukin-5 (IL-5) is an important regulator of eosinophil differentiation, proliferation and survival and thus, is thought to be a major cytokine involved in the pathogenesis of asthma.2–4 IL-5 is predominantly produced by T cells of the T helper type 2 (Th2) phenotype,5,6 although it can also be produced by eosinophils, mast cells and epithelial cells.7–9
Inhaled glucocorticoids are the drugs of choice for treatment of the underlying inflammation in all but the mildest cases of asthma.10 The effectiveness of glucocorticoids in controlling the inflammation of the asthmatic airways is attributed to the ability of these drugs to inhibit the release of pro-inflammatory factors, such as cytokines, released by multiple cell types.11 The mechanism by which most, if not all, of the effects of glucocorticoids on cells are considered to be mediated, is by binding to the glucocorticoid receptor.12 Binding of the glucocorticoid receptor to a glucocorticoid response element on the DNA has been shown to be essential for the transcriptional activation (transactivation) of glucocorticoid-responsive genes.13 However, the anti-inflammatory abilities of glucocorticoids are generally considered to be the result of the negative regulation (transrepression) of pro-inflammatory gene transcription.14
Although IL-5 can be inhibited by glucocorticoids both in vivo and in vitro15,16 the IL-5 promoter does not contain a consensus glucocorticoid response element sequence. Mechanisms have therefore been proposed by which glucocorticoids may affect the activity of the nuclear factor of activated T cells (NF-AT) and activator protein-1 (AP-1) binding sites that are present in the promoter regions of many pro-inflammatory cytokines, including IL-5.17–19 However, gene expression is not simply a matter of the up- or down-regulation of gene transcription, as post-transcriptional, translational and even post-translational events are also required for the release of pro-inflammatory cytokines. Indeed, the glucocorticoid, dexamethasone has been shown to inhibit release of several pro-inflammatory cytokines, such as, interferon-β and tumour necrosis factor-α, by affecting post-transcriptional and even translational mechanisms.20,21
We have previously shown evidence that IL-10 may inhibit IL-5 by post-transcriptional mechanisms.22 In the present study, we have used both peripheral blood mononuclear cells (PBMC) and primary human T cells to evaluate the hypothesis that the glucocorticoid, dexamethasone, inhibits IL-5 release by post-transcriptional mechanisms.
Materials and methods
Reagents
Phorbol 12-myristate 13-acetate (PMA), phytohaemagglutinin (PHA), dexamethasone, actinomycin D and cycloheximide were purchased from Sigma (Poole, UK)
Isolation and treatment of human PBMC
Mononuclear cells were prepared from the peripheral blood of healthy human volunteers and cultured at a density of 3 × 106 cells/ml for all experiments as previously described.22 Cells were stimulated by the addition of PMA (50 nm) and PHA (5 μg/ml) as previously described.22
Isolation of primary human T cells
T cells were isolated from PBMC using a MACS™ Pan T cell isolation kit on a magnetic depletion column according to the manufacturer's instructions (Miltenyi Biotec, Bergisch Gladbach, Germany). The flow through from this column typically contained >93% T cells as determined by fluorescence-activated cell sorter (FACS) analysis following labelling with fluorescein isothiocyanate-labelled anti-CD3. T cells were then resuspended at 2 × 106 cells/ml in RPMI-1640 medium supplemented with 10% fetal calf serum, 2 mm l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin and 2·5 μg/ml amphotericin B (all Sigma). Cells were then cultured for 72 hr in a humidified atmosphere at 37°, 5% CO2, with 10 ng/ml human recombinant IL-2 (R & D Systems, Abingdon, UK). This treatment increased the percentage of CD3+ T cells to >97% as measured by FACS.
Enzyme-linked immunosorbent assay (ELISA)
Supernatants from 3 × 106 cells were harvested 24 hr after treatment and ELISA was performed as described by the manufacturer (BD Pharmingen, Cambridge, UK). Human recombinant IL-5 was used as a standard (R & D Systems). For intracellular IL-5 measurements, PBMCs (6 × 106 cells/treatment) were harvested after the indicated time-points and spun down. Supernatants were collected for standard ELISA analysis and cells were resuspended in 100 μl 1× reporter lysis buffer (RLB – Promega, Southampton, UK). ELISA was carried out as above except that cytokine standards were made up in 1× RLB and only 50 μl of sample was added per well. RLB had no effect on the detection of cytokines by ELISA.
Semi-quantitative reverse transcription–polymerase chain reaction (RT-PCR)
Total RNA was extracted from 6 × 106 cells using a one-step guanidine thiocyanate–phenol–chloroform method.23 RT-PCR for IL-5 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was carried out using the primers and conditions previously described.22 To confirm the semi-quantitative nature of this technique, cycle profiles were established for each set of different RT reactions by performing PCR with various cycle numbers on an ‘average’ sample created by combining aliquots of all cDNA samples in that particular experiment as previously published.24 This ensured that the amplification was in the linear phase, where the signal is proportional to the starting amount of template. Using this method, cycle numbers for IL-5 ranged between 32 and 36 cycles and for GAPDH between 24 and 28 cycles. Quantification of PCR products was carried out using gelworks 1d intermediate software (Ultra Violet Products Limited, Cambridge, UK). Dilutions of cDNA as well as negative controls for both the RT reaction and the PCR were used for each set of PCR reactions to ensure linearity of the PCR (Fig. 1).
Figure 1.
RT-PCR is a semi-quantiative method for measuring IL-5 mRNA expression. PBMC were incubated with or without PMA (50 nm) and PHA (5 μg/ml) and RNA was harvested at 6 hr for semi-quantitative RT-PCR analysis of IL-5 and GAPDH. Complementary DNA samples from the reverse transcriptase reaction were pooled to create an average sample. Dilutions of that average sample (1, ½, ¼) as well as negative controls for both the reverse transcriptase reaction (No RNA) and the PCR (No cDNA) were always used for every PCR and subsequent gel run. A representative agarose gel is shown with corresponding optical density data to demonstrate linearity of the technique. λ = IL-5 (R2 = 0·97), ν = GAPDH (R2 = 0·99).
Cloning of promoter fragments into luciferase reporter vectors
IL-5 promoter fragments were amplified by PCR, supplemented with 1 U Taq extender, from human genomic DNA using the following primers: 0·5-kilobase (kb) promoter 5′-AAG CCT ATC CTA ATC AAG ACC CCA GTG-3′ forward, 5′-AAG CTT CTC TGA AAC GTT CTG CGT TTG-3′ reverse; 1·5-kb promoter 5′-GTC TGA AGA TCT CTC TGA AAC GTT CTG CGT TT-3′ forward, 5′-GTC TGA AGA TCT TGC AGT CTC AAG GAA ACA TT-3′ reverse. Amplification products were agarose gel separated and purified using the GFX™ kit according to the manufacturer's instructions (Amersham Pharmacia, Little Chalfont, UK). Products were ligated overnight at 15° into pGEM-T vector using DNA ligase according to the manufacturer's instructions (Promega). Ligation mixes were transformed into JM109 competent cells and grown on l-agar plates supplemented with 50 μg/ml ampicillin. Colonies containing the plasmid were detected by blue/white screening using 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) and isopropyl-β-d-thiogalactopyranoside (IPTG). Colonies were then grown in l-broth supplemented with 50 μg/ml ampicillin prior to Wizard® plus SV miniprep DNA extraction kit (Promega). Sequence identity as determined by Cambridge Bioscience (Cambridge, UK) was >99%. Isolated plasmids were then incubated with the appropriate restriction enzyme (HindIII for 0·5 kb, BglII for 1·5 kb) and run on a low-melt agarose gel. Promoter fragments were excised from the gel and purified by GFX™ kit. Purified fragments were subcloned into pGL3basic plasmid following the manufacturer's instructions (Promega). Orientation was then confirmed by manual sequencing using the usb sequenase version 2·0, DNA sequencing kit (Amersham Pharmacia).
Transient transfection of primary human T cells
After washing, primary human T cells were resuspended in supplemented RPMI-1640 medium at 2 × 107 cells/ml and aliquots of 250 μl were incubated with 10 μg plasmid DNA for 5 min. Electroporation was carried out on a BioRad Gene Pulser II (Hemel Hempstead, UK) at 260 V and 960 μF in 0·4-cm cuvettes. After electroporation, cells were washed and resuspended in 400 μl of serum-free RPMI-1640 medium. Experiments were carried out in a 96-well, round-bottom culture plate (Costar, High Wycombe, UK) with 100-μl aliquots of cells.
Luciferase assay
Transfected cells were treated as indicated and harvested after 12 hr. Supernatants were collected for ELISA analysis and cells were resuspended in 30 μl 1× RLB. Luciferase activity was measured on a luminometer (Turner Design, Steptech, Stevenage, UK) by adding 20 μl supernatant to 40 μl luciferin substrate (Promega). Results were normalized to total protein concentration as determined by Bradford assay.
Statistical analysis
Data were analysed using Wilcoxon's signed rank test or analysis of variance (anova) as appropriate. Results were considered significant when P < 0·05.
Results
IL-5 release requires de novo transcription and translation
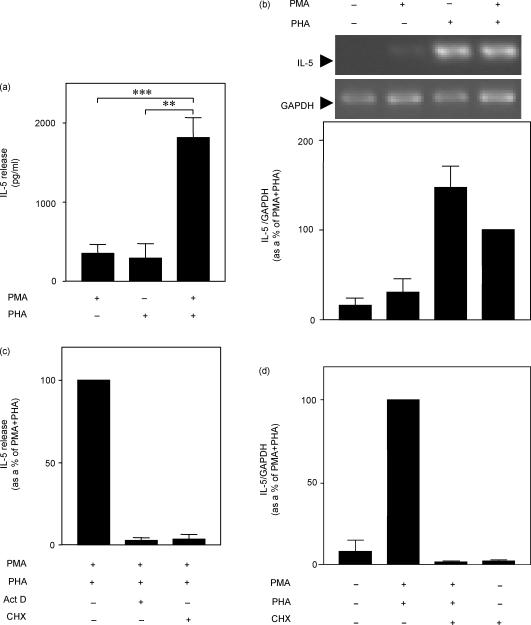
After 24 hr, basal levels of IL-5 were below the limit of detection (<16 pg/ml) and treatment of PBMC with PMA, PHA, or a combination of both resulted in increased release of IL-5, with PMA and PHA causing similar levels of release when added independently (Fig. 2a). When added simultaneously, a marked synergy was observed (Fig. 2a). In contrast, PMA had little effect on IL-5 mRNA accumulation, whereas stimulation of cells with PHA induced similar levels of IL-5 mRNA to stimulation with PMA + PHA (Fig. 2b,c). Similar results were seen when PBMC were stimulated with activating antibodies to the CD3 and CD28 cell surface markers with CD28 being analogous to PMA (data not shown).
Figure 2.
IL-5 expression requires de novo transcription and translation. (a) Cells were incubated with PMA (50 nm) and PHA (5 μg/ml), as indicated, for 24 hr. Supernatants were harvested and IL-5 release was measured. Data from at least five independent experiments are expressed as pg/ml means ± SEM. (b) Cells were incubated with PMA + PHA and RNA was harvested at 6 hr for semi-quantitative RT-PCR analysis of IL-5 and GAPDH. Agarose gels representative of six such experiments are shown. Data from six independent experiments are expressed as means ± SEM of percentage of the PMA + PHA stimulation. (c) PBMC were incubated with PMA + PHA as above in the presence or absence of actinomycin D (10 μg/ml) or cycloheximide (10 μg/ml). Supernatants were harvested at 24 hr and IL-5 release measured by ELISA. (d) Cells were treated with PMA + PHA and cycloheximide (10 μg/ml) as indicated and RNA was harvested after 6 hr for semi-quantitative RT-PCR analysis of IL-5 and GAPDH. Data (n = 3) were normalized to GAPDH. In (c) and (d) data from at least three independent experiments are expressed as percentage of stimulated as means ± SEM. ***P < 0·001, **P < 0·01.
PBMC were stimulated with PMA + PHA in the presence of the RNA polymerase II inhibitor, actinomycin D or the translation inhibitor, cycloheximide and in each case IL-5 release was suppressed to near basal levels (Fig. 2c). Furthermore, cycloheximide could also inhibit IL-5 mRNA accumulation, suggesting that new translation is necessary for IL-5 gene transcription (Fig. 2d).
Effect of dexamethasone on IL-5 protein and mRNA production
In agreement with previous studies, dexamethasone dose-dependently inhibited IL-5 release (IC50 = 0·66 nm) when added simultaneously with the PMA + PHA stimulus (Fig. 3a).15 Dexamethasone (1 μm) was also able to abolish both the PMA + PHA-induced release and the intracellular formation of IL-5 from PBMCs over 24 hr (Fig. 3b,e). Furthermore, dexamethasone inhibited steady-state IL-5 mRNA levels over 24 hr. However, dexamethasone incompletely suppressed IL-5 mRNA at 2 hr, suggesting that there is a time lag between the addition of dexamethasone and the onset of inhibition (Fig. 3c,d).
Figure 3.
Characterization of the effect of dexamethasone on IL-5 expression. (a) PBMCs were incubated with PMA (50 nm) + PHA (5 μg/ml) and co-incubated with various concentrations of dexamethasone as indicated. Supernatants were harvested at 24 hr and IL-5 release was measured. Data are expressed as means ± SEM of four independent experiments. (b) PBMC were stimulated with PMA + PHA, as above, with or without dexamethasone (1 μm). Supernatants were harvested at the times indicated and IL-5 release was measured. Data are expressed as means ± SEM of nine independent experiments. (c) Cells were incubated with PMA + PHA as previously described with or without dexamethasone (1 μm) and RNA was harvested at the time-points indicated for semi-quantitative RT-PCR analysis of IL-5 and GAPDH. NS = not stimulated. Agarose gels representative of five such experiments are shown. (d) Data were normalized to GAPDH and expressed as a percentage of the 6 hr PMA + PHA response. Data from five independent experiments are expressed as means ± SEM. (e) PBMC were stimulated with PMA + PHA, as above, with or without dexamethasone (1 μm). Supernatants were removed and the cells were lysed in 1 × RLB and harvested at the times indicated and intracellular IL-5 was measured by ELISA. Data are expressed as means ± SEM of at least four independent experiments. **P < 0·01, *P < 0·05.
Effect of dexamethasone on IL-5 promoter constructs
Previous studies have identified a 500-base-pair (bp) region in the proximal part of the IL-5 promoter, which includes binding sites for the transcription factors NF-AT, GATA-3 and Ets, which have been demonstrated to be important for the induction of IL-5 gene transcription.19,25–27 To investigate whether dexamethasone could affect IL-5 promoter activity, IL-5 promoter fragments were cloned into a luciferase reporter plasmid. One promoter construct spanned the proximal promoter region (−508/+41 bp). The second construct, which spanned from −1553 bp to +41 bp, was used to investigate the role of any previously unidentified regions that may also be involved in the transcriptional activation of the IL-5 gene. These constructs were transiently transfected into primary human T cells isolated by negative magnetic selection (Fig. 4a). Transfected T cells were treated with PMA + PHA with or without addition of dexamethasone (1 μm) and cell lysates were assayed for luciferase activity after 12 hr. Treatment of transfected cells with PMA + PHA induced luciferase activity from both IL-5 promoter constructs (Fig. 4a). However, addition of dexamethasone had no effect on the inducibility of either of these promoter constructs (Fig. 4b). In contrast, addition of dexamethasone significantly inhibited the IL-5 release from transfected T cells induced by PMA + PHA (Fig. 4c). These results confirm that dexamethasone was still actively inhibiting IL-5 release from the same cells, even though there was no effect on promoter activity and suggests that dexamethasone does not inhibit IL-5 gene transcription.
Figure 4.
Effect of dexamethasone on IL-5 promoter constructs. (a) Primary human T cells, transiently transfected with the constructs indicated, were incubated with PMA (50 nm) + PHA (5 μg/ml). Cells were harvested at 12 hr and luciferase activity was measured. Data are expressed as means ± SEM of at least four independent experiments. (b) Primary human T cells were transiently transfected with either of the two IL-5 promoter constructs and stimulated as in (a) and co-incubated with dexamethasone (1 μm) and harvested as above. Data are expressed as fold induction as means ± SEM of at least four independent experiments. (c) Supernatants from the above experiments were harvested and IL-5 release was measured. Data are expressed as means ± SEM of 13 independent experiments. ***P < 0·001.
Actinomycin D blocks dexamethasone-mediated inhibition of IL-5 mRNA
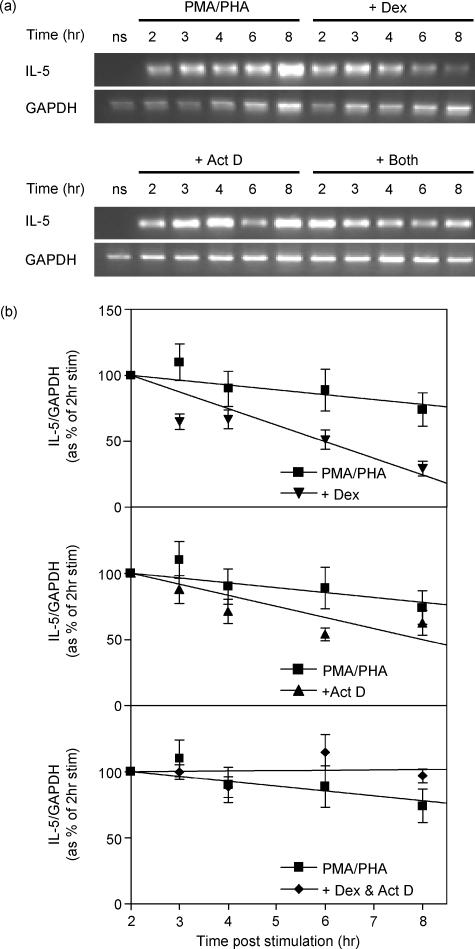
As dexamethasone appears to inhibit IL-5 release by post-transcriptional mechanisms, it was necessary to investigate the effect of dexamethasone on IL-5 mRNA accumulation once transcription had been initiated. Initial studies revealed that, whereas steady-state IL-5 mRNA levels remained reasonably constant between 2 and 8 hr post-PMA + PHA stimulation, the addition of dexamethasone at 2 hr resulted in ∼70% repression of IL-5 mRNA over the following 6 hr (Fig. 5b, upper panel). When the transcriptional inhibitor, actinomycin D was added 2 hr after PMA + PHA stimulation, the half-life of IL-5 mRNA was approximately 6 hr (Fig. 5a,b, middle panel). This contrasts with the apparent half-life of 4 hr when dexamethasone was added 2 hr after stimulation (Fig. 5). The difference between these two results suggests that inhibition of transcription alone cannot fully account for the inhibition of IL-5 observed upon treatment with dexamethasone. Furthermore, when added together 2 hr after stimulation, actinomycin D totally reversed the dexamethasone-dependent reduction of IL-5 mRNA (Fig. 5). This result suggests that the inhibition of IL-5 mRNA by dexamethasone is dependent upon de novo transcription. Taken together with the promoter data, these observations indicate that dexamethasone may inhibit IL-5 by post-transcriptional mechanisms.
Figure 5.
Effect of dexamethasone and actinomycin D on IL-5 mRNA stability. PBMCs were incubated with PMA (50 nm) + PHA (5 μg/ml) for 2 hr then dexamethasone (1 μm) and/or actinomycin D (10 μg/ml) were added and the cells were harvested at the time-points indicated for semi-quantitative RT-PCR analysis of IL-5 and GAPDH. (a) Agarose gels representative of five such experiments are shown. (b) Data were normalized to GAPDH and expressed as a percentage of the response after 2 hr of stimulation. Data from five independent experiments are expressed as means ± SEM.
Effects of dexamethasone on IL-5 protein release
From the previous sections, it appears that whilst dexamethasone may have some effect on the induction of IL-5 mRNA transcription, dexamethasone may decrease IL-5 mRNA stability. To examine whether these changes in mRNA levels were reflected at the level of protein release, PBMCs were stimulated with PMA + PHA, then dexamethasone was added at the time-points indicated (Fig. 6). In common with the effects observed at the mRNA level, dexamethasone could inhibit IL-5 release when added 2 hr after PMA + PHA stimulation. Furthermore, dexamethasone could still inhibit IL-5 release from PBMC by ∼40% when added up to 12 hr after stimulation (Fig. 6). Similar results were obtained when cycloheximide and actinomycin D were added at various time-points after stimulation (Fig. 6). These results are consistent with the data presented in Fig. 2 demonstrating that IL-5 release was dependent upon ongoing transcription and translation.
Figure 6.
Reverse time–course effect of dexamethasone, actinomycin D and cycloheximide on IL-5. PBMCs were incubated with PMA (50 nm) + PHA (5 μg/ml) and dexamethasone (1 μm), actinomycin D (10 μg/ml), or cycloheximide (10 μg/ml) were added at the time-points indicated. Supernatants were harvested at 24 hr and IL-5 release was measured. Data are expressed as percentage of stimulus at 24 hr as means ± SEM of four independent experiments.
Discussion
From the data presented here, expression of IL-5 certainly requires a transcriptional component, since de novo transcription is necessary for the release of IL-5 (Fig. 2). These observations are further supported by the ability of PMA + PHA to induce activity of the IL-5 proximal promoter transiently transfected into human T cells. These data are consistent with previous studies that have shown transcription to be important in the regulation of IL-5 expression.28–30 Indeed, stimulation of PBMCs with PMA + ionomycin has been shown to increase the rate of IL-5 gene transcription.28 Furthermore, this increase in transcription rate was abolished upon addition of the inhibitors of NF-AT activation, cyclosporin A and FK506.28 Several other transcription factors, including AP-1, GATA, Ets and Oct have also been reported to control IL-5 gene expression.26,31–34 These previous investigations studied the role of transcription in IL-5 gene expression using IL-5 promoter constructs transiently transfected into murine and human T-cell clones and T-cell lines.25,31,35,36 Both of the IL-5 promoter constructs used in this study included the 500-bp region previously identified as being essential for an increase in IL-5 gene expression.25 Thus, the up-regulation of IL-5 induced by PMA + PHA appears to be predominantly via a transcriptional mechanism. However, several observations presented here, support a role for post-transcriptional mechanisms in IL-5 gene expression.
There are few data in the literature on the regulation of IL-5 by post-transcriptional processes. However, the ability of PMA to increase IL-5 release by PHA synergistically without a concomitant increase in steady-state IL-5 mRNA levels (Fig. 2b), suggests that stimulation of T cells with PMA does cause IL-5 release by post-transcriptional mechanisms. Similarly, we have observed that stimulation of T cells with an activating antibody to the CD28 receptor could synergistically increase IL-5 release by an activating αCD3 antibody without any effect on steady-state IL-5 mRNA levels (data not shown), raising the possibility that PHA and αCD28 share a common post-transciptional/translational mechanism of action. Stimulation of T cells via the CD28 receptor can up-regulate the expression of several cytokines, including granulocyte–macrophage colony-stimulating factor (GM-CSF), by increasing mRNA stability.37 In contrast, Umland and colleagues have shown that CD28 only has a negligible effect on IL-5 mRNA stability.38 However, a recent study demonstrated no role for either the 5′- or 3′-untranslated region (UTR) of IL-5 in the regulation of this cytokine, both of which have been shown to be involved in the post-transcriptional regulation of other cytokines (e.g. GM-CSF).39–41 However, this study used IL-5 constructs under the control of an SV40 promoter, thus leading to a constitutive, not inducible, expression of IL-5.39 Since any factors that regulate IL-5 mRNA stability or translation may require up-regulation by a stimulus, the fact that these promoters were constitutively active may explain the lack of effect observed.39 For example, in an inducible system, deletion of the IL-5 3′-UTR was shown to decrease production of IL-5.42 Thus, the nature of the roles of the 5′- and 3′-UTR in the control of IL-5 gene expression is currently equivocal.
Further evidence for the regulation of IL-5 by post-transcriptional mechanisms is provided by studying the effects of dexamethasone on IL-5 expression since dexamethasone could not inhibit the inducibility of either the 0·5-kb or 1·5-kb IL-5 promoter constructs. These observations are in contrast to a previous report by Mori and colleagues. which demonstrated that dexamethasone could inhibit the activity of a similar 0·5-kb IL-5 promoter construct stimulated by αCD3 or IL-2.31 One explanation for the differences observed between this study and the results presented by Mori and associates may be the different stimuli used.31 However, dexamethasone appears to inhibit both PMA + PHA and αCD3 + αCD28-stimulated IL-5 release equally well (data not shown). Furthermore, unlike cAMP-elevating agents,22 dexamethasone can inhibit IL-5 release in a stimulus-independent manner (data not shown). An alternative explanation may be that the IL-5 promoter is regulated differently in the T-cell clones used by Mori and associates and the primary human T cells used in this study.31 However, it should be noted that these are transient transfection studies and therefore may not accurately reflect the situation at the level of the native chromosomal DNA. This is especially pertinent in the light of recent data that suggest that some of dexamethasone's inhibitory effects may be mediated by changes in the profile of histone acetylation.43 Thus, we cannot completely exclude the possibility that dexamethasone has some effects on IL-5 mRNA transcription.
The ability of dexamethasone to inhibit IL-5 mRNA significantly more than actinomycin D when added after stimulation, suggests that dexamethasone may also activate mRNA decay pathways. Furthermore, the ability of actinomycin D to reverse the dexamethasone-mediated repression of IL-5 implies that dexamethasone may inhibit IL-5 mRNA by de novo synthesis of an inhibitory factor. Taken together with the inability of dexamethasone to inhibit IL-5 promoter activation, these data imply that dexamethasone may inhibit IL-5 solely by post-transcriptional mechanisms, such as activation of mRNA destabilizing factors. Several destabilizing factors, including ARE/poly(U)-binding/degradation factor 1 and tristetraprolin, have so far been characterized and at least one of these, tristetraprolin, requires de novo transcription and translation to function.44–46 However, AUF-1 and tristetraprolin exert their destabilizing effects by binding to 3′-UTR Au-rich elements (AREs) and whilst this has been demonstrated using the GM-CSF 3′-UTR,45 no studies have examined the binding of such proteins to the IL-5 3′-UTR. As new protein synthesis is an absolute requirement for induction of IL-5 mRNA (Fig. 2d), this suggests that the regulation of IL-5 mRNA stability may be a dynamic process, involving competitive binding between mRNA stabilizing and destabilizing factors. Thus, newly translated stabilizing factors, such as HuR,47 may be responsible for the initial accumulation of IL-5 mRNA upon treatment of PBMCs with PMA + PHA. Addition of dexamethasone may therefore cause an increase in destabilizing factors, and may also inhibit production of stabilizing proteins, thereby displacing stabilizing factors and inhibiting IL-5 mRNA accumulation. Indeed, tristetraprolin and a murine homologue of HuR, HuA, have recently been demonstrated to have overlapping binding specificities to ARE-sequences in both IL-3 and GM-CSF RNA sequences.48 Furthermore, the authors of this study speculated that this overlap in binding activities may follow a similar pattern to that which we have outlined above,48 with HuA being responsible for RNA-stabilization and tristetraprolin for RNA-degradation. However, the products of these dexamethasone-inducible genes do not have to be RNA-binding proteins. Alternative mechanisms of inhibition by dexamethasone-inducible genes, may include up-regulation of factors that inhibit stabilizing factors binding to IL-5 mRNA or activation of enzymes that increase the turnover of stabilizing proteins. These scenarios are purely hypothetical and further studies are required to identify the factors induced by dexamethasone and the roles of these species in the degradation of IL-5 mRNA.
In summary, the work presented here, in agreement with previous studies, shows that dexamethasone is a potent inhibitor of IL-5 release from human PBMCs and T cells.15,16 The data also indicate that a glucocorticoid-inducible gene(s) may be responsible for the inhibition of IL-5 and provide further evidence for the inhibitory action of dexamethasone occurring at a post-transcriptional level.
Acknowledgments
This work was supported by a grant from Boehringer Ingelheim. M. W. Bergmann held a Deutsche Forschungsgemeinschaft scholarship.
References
- 1.Djukanovic R, Roche WR, Wilson JW, Beasley CR, Twentyman OP, Howarth RH, Holgate ST. Mucosal inflammation in asthma. Am Rev Respir Dis. 1990;142:434–57. doi: 10.1164/ajrccm/142.2.434. [DOI] [PubMed] [Google Scholar]
- 2.Clutterbuck EJ, Hirst EM, Sanderson CJ. Human interleukin-5 (IL-5) regulates the production of eosinophils in human bone marrow cultures: comparison and interaction with IL-1, IL-3, IL-6, and GMCSF. Blood. 1989;73:1504–12. [PubMed] [Google Scholar]
- 3.Ema H, Suda T, Nagayoshi K, Miura Y, Civin CI, Nakauchi H. Target cells for granulocyte colony-stimulating factor, interleukin-3, and interleukin-5 in differentiation pathways of neutrophils and eosinophils. Blood. 1990;76:1956–61. [PubMed] [Google Scholar]
- 4.Humbert M, Corrigan CJ, Kimmitt P, Till SJ, Kay AB, Durham SR. Relationship between IL-4 and IL-5 mRNA expression and disease severity in atopic asthma. Am J Respir Crit Care Med. 1997;156:704–8. doi: 10.1164/ajrccm.156.3.9610033. [DOI] [PubMed] [Google Scholar]
- 5.Hamid Q, Azzawi M, Ying S, et al. Expression of mRNA for interleukin-5 in mucosal bronchial biopsies from asthma. J Clin Invest. 1991;87:1541–6. doi: 10.1172/JCI115166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ying S, Durham SR, Corrigan CJ, Hamid Q, Kay AB. Phenotype of cells expressing mRNA for TH2-type (interleukin 4 and interleukin 5) and TH1-type (interleukin 2 and interferon gamma) cytokines in bronchoalveolar lavage and bronchial biopsies from atopic asthmatic and normal control subjects. Am J Respir Cell Mol Biol. 1995;12:477–87. doi: 10.1165/ajrcmb.12.5.7742012. [DOI] [PubMed] [Google Scholar]
- 7.Giembycz MA, Lindsay MA. Pharmacology of the eosinophil. Pharmacol Rev. 1999;51:213–340. [PubMed] [Google Scholar]
- 8.Shichijo M, Inagaki N, Kimata M, Serizawa I, Saito H, Nagai H. Role of cyclic 3′,5′-adenosine monophosphate in the regulation of chemical mediator release and cytokine production from cultured human mast cells. J Allergy Clin Immunol. 1999;103:S421–S428. doi: 10.1016/s0091-6749(99)70157-0. [DOI] [PubMed] [Google Scholar]
- 9.Salvi S, Semper A, Blomberg A, et al. Interleukin-5 production by human airway epithelial cells. Am J Respir Cell Mol Biol. 1999;20:984–91. doi: 10.1165/ajrcmb.20.5.3463. [DOI] [PubMed] [Google Scholar]
- 10.National Asthma Education and Prevention Program. Expert Panel Report 2. Bethesda MD: National Institutes of Health, National Heart, Lung and Blood Institute; 1997. NIH Publication 97–4051. Guidelines for the Diagnosis and Management of Asthma. [Google Scholar]
- 11.Barnes PJ, Pedersen S, Busse WW. Efficacy and safety of inhaled corticosteroids. New developments. Am J Respir Crit Care Med. 1998;157:S1–53. doi: 10.1164/ajrccm.157.3.157315. [DOI] [PubMed] [Google Scholar]
- 12.Beato M. Gene regulation by steroid hormones. Cell. 1989;56:335–44. doi: 10.1016/0092-8674(89)90237-7. [DOI] [PubMed] [Google Scholar]
- 13.Dahlman-Wright K, Wright A, Gustafsson JA, Carlstedt-Duke J. Interaction of the glucocorticoid receptor DNA-binding domain with DNA as a dimer is mediated by a short segment of five amino acids. J Biol Chem. 1991;266:3107–12. [PubMed] [Google Scholar]
- 14.Newton R. Molecular mechanisms of glucocorticoid action: what is important? Thorax. 2000;55:603–13. doi: 10.1136/thorax.55.7.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rolfe FG, Hughes JM, Armour CL, Sewell WA. Inhibition of interleukin-5 gene expression by dexamethasone. Immunology. 1992;77:494–9. [PMC free article] [PubMed] [Google Scholar]
- 16.Corrigan CJ, Haczku A, Gemou-Engesaeth V, Doi S, Kikuchi Y, Takatsu K, Durham SR, Kay AB. CD4 T-lymphocyte activation in asthma is accompanied by increased serum concentrations of interleukin-5. Effect of glucocorticoid therapy. Am Rev Respir Dis. 1993;147:540–7. doi: 10.1164/ajrccm/147.3.540. [DOI] [PubMed] [Google Scholar]
- 17.Cato AC, Wade E. Molecular mechanisms of anti-inflammatory action of glucocorticoids. Bioessays. 1996;18:371–8. doi: 10.1002/bies.950180507. [DOI] [PubMed] [Google Scholar]
- 18.Barnes PJ, Karin M. Nuclear factor-kB – a pivotal transcription factor in chronic inflammatory diseases. N Engl J Med. 1997;336:1066–71. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 19.Karlen S, D'Ercole M, Sanderson CJ. Two pathways can activate the interleukin-5 gene and induce binding to the conserved lymphokine element 0. Blood. 1996;88:211–21. [PubMed] [Google Scholar]
- 20.Peppel K, Vinci JM, Baglioni C. The AU-rich sequences in the 3′ untranslated region mediate the increased turnover of interferon mRNA induced by glucocorticoids. J Exp Med. 1991;173:349–55. doi: 10.1084/jem.173.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han J, Brown T, Beutler B. Endotoxin-responsive sequences control cachectin/tumor necrosis factor biosynthesis at the translational level [published erratum appears in J Exp Med 1990; 171:971–2] J Exp Med. 1990;171:465–75. doi: 10.1084/jem.171.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Staples KJ, Bergmann M, Barnes PJ, Newton R. Stimulus-specific inhibition of IL-5 by cAMP-elevating agents and IL-10 reveals differential mechanisms of action. Biochem Biophys Res Commun. 2000;273:811–15. doi: 10.1006/bbrc.2000.3023. [DOI] [PubMed] [Google Scholar]
- 23.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 24.Newton R, Kuitert LM, Slater DM, Adcock I, Barnes PJ. Cytokine induction of cytosolic phospholipase A2 and cyclooxygenase-2 mRNA is suppressed by glucocorticoids in human epithelial cells. Life Sci. 1997;60:67–78. doi: 10.1016/s0024-3205(96)00590-5. [DOI] [PubMed] [Google Scholar]
- 25.Bourke PFLB, Campbell HD, Young IG. Localization of the inducible enhancer in the mouse interleukin-5 gene that is responsive to T-cell receptor stimulation. Blood. 1995;85:2069–77. [PubMed] [Google Scholar]
- 26.Stranick KS, Zambas DN, Uss AS, Egan RW, Billah MM, Umland SP. Identification of transcription factor binding sites important in the regulation of the human interleukin-5 gene. J Biol Chem. 1997;272:16453–65. doi: 10.1074/jbc.272.26.16453. [DOI] [PubMed] [Google Scholar]
- 27.Mori A, Kaminuma O, Mikami T, Hoshino A, Ohmura T, Miyazawa K, Suko M, Okudaira H. Dissection of human IL-5 promoter – essential role of CLEO element in human IL-5 gene transcription. Int Arch Allergy Immunol. 1997;113:272–4. doi: 10.1159/000237570. [DOI] [PubMed] [Google Scholar]
- 28.Rolfe FG, Valentine JE, Sewell WA. Cyclosporin A and FK506 reduce interleukin-5 mRNA abundance by inhibiting gene transcription. Am J Respir Cell Mol Biol. 1997;17:243–50. doi: 10.1165/ajrcmb.17.2.2819. [DOI] [PubMed] [Google Scholar]
- 29.Mori A, Kaminuma O, Mikami T, Inoue S, Okumura Y, Akiyama K, Okudaira H. Transcriptional control of the IL-5 gene by human helper T cells: IL-5 synthesis is regulated independently from IL-2 or IL-4 synthesis. J Allergy Clin Immunol. 1999;103:S429–36. doi: 10.1016/s0091-6749(99)70158-2. [DOI] [PubMed] [Google Scholar]
- 30.Ogawa K, Kaminuma O, Kikkawa H, Akiyama K, Mori A. IL-5 synthesis by T cells of allergic subjects is regulated at the transcriptional level. Int Arch Allergy Immunol. 2000;122(Suppl. 1):63–6. doi: 10.1159/000053636. [DOI] [PubMed] [Google Scholar]
- 31.Mori A, Kaminuma O, Suko M, et al. Two distinct pathways of interleukin-5 synthesis in allergen-specific human T-cell clones are suppressed by glucocorticoids. Blood. 1997;89:2891–900. [PubMed] [Google Scholar]
- 32.Blumenthal SG, Aichele G, Wirth T, Czernilofsky AP, Nordheim A, Dittmer J. Regulation of the human interleukin-5 promoter by ets transcription factors. Ets1 and Ets2, but not Elf-1, cooperate with GATA3 and HTLV-I Tax1. J Biol Chem. 1999;274:12910–16. doi: 10.1074/jbc.274.18.12910. [DOI] [PubMed] [Google Scholar]
- 33.Lee HJ, O'Garra A, Arai K, Arai N. Characterization of cis-regulatory elements and nuclear factors conferring Th2-specific expression of the IL-5 gene: a role for a GATA-binding protein. J Immunol. 1998;160:2343–52. [PubMed] [Google Scholar]
- 34.Gruart-Gouilleux V, Engels P, Sullivan M. Characterization of the human interleukin-5 gene promoter: involvement of octamer binding sites in the gene promoter activity. Eur J Immunol. 1995;25:1431–5. doi: 10.1002/eji.1830250544. [DOI] [PubMed] [Google Scholar]
- 35.Stranick KS, Uss AS, Zambas DN, Egan RW, Billah MM, Umland SP. Characterization of the mouse interleukin-5 promoter in a mouse TH2 T cell clone. Biochem Biophys Res Commun. 1998;252:56–62. doi: 10.1006/bbrc.1998.9594. [DOI] [PubMed] [Google Scholar]
- 36.Tsuruta L, Lee HJ, Masuda ES, Yokota T, Arai N, Arai K. Regulation of expression of the IL-2 and IL-5 genes and the role of proteins related to nuclear factor of activated T cells. J Allergy Clin Immunol. 1995;96:1126–35. doi: 10.1016/s0091-6749(95)70197-4. [DOI] [PubMed] [Google Scholar]
- 37.Lindstein T, June CH, Ledbetter JA, Stella G, Thompson CB. Regulation of lymphokine messenger RNA stability by a surface-mediated T cell activation pathway. Science. 1989;244:339–43. doi: 10.1126/science.2540528. [DOI] [PubMed] [Google Scholar]
- 38.Umland SP, Razac S, Shah H, Nahrebne DK, Egan RW, Billah MM. Interleukin-5 mRNA stability in human T cells is regulated differently than interleukin-2, interleukin-3, interleukin-4, granulocyte/macrophage colony-stimulating factor, and interferon-gamma. Am J Respir Cell Mol Biol. 1998;18:631–42. doi: 10.1165/ajrcmb.18.5.3046. [DOI] [PubMed] [Google Scholar]
- 39.Thomas MA, Karlen S, D'Ercole M, Sanderson CJ. Analysis of the 5′ and 3′UTRs in the post-transcriptional regulation of the interleukin-5 gene. Biochim Biophys Acta. 1999;1444:61–8. doi: 10.1016/s0167-4781(98)00268-1. [DOI] [PubMed] [Google Scholar]
- 40.Shaw G, Kamen R. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–67. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 41.Jarzembowski JA, Rajagopalan LE, Shin HC, Malter JS. The 5′-untranslated region of GM-CSF mRNA suppresses translational repression mediated by the 3′ adenosine-uridine-rich element and the poly (A) tail. Nucleic Acids Res. 1999;27:3660–6. doi: 10.1093/nar/27.18.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Westhuizen FH, Pretorius PJ, de Wet WJ. Contribution of regions 3′ and 5′ to the hIL-5 gene on the expression of rhIL-5 in CHO-cells. Biochem Biophys Res Commun. 1996;227:576–80. doi: 10.1006/bbrc.1996.1548. [DOI] [PubMed] [Google Scholar]
- 43.Ito K, Barnes PJ, Adcock IM. Glucocorticoid receptor recruitment of histone deacetylase 4 inhibits interleukin-1beta-induced histone H4 acetylation on lysines 8 and 12. Mol Cell Biol. 2000;20:6891–903. doi: 10.1128/mcb.20.18.6891-6903.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buzby JS, Brewer G, Nugent DJ. Developmental regulation of RNA transcript destabilization by A + U-rich elements is AUF1-dependent. J Biol Chem. 1999;274:33973–8. doi: 10.1074/jbc.274.48.33973. [DOI] [PubMed] [Google Scholar]
- 45.Carballo E, Lai WS, Blackshear PJ. Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood. 2000;95:1891–9. [PubMed] [Google Scholar]
- 46.Lai WS, Stumpo DJ, Blackshear PJ. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J Biol Chem. 1990;265:16556–63. [PubMed] [Google Scholar]
- 47.Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–70. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raghavan A, Robison RL, McNabb J, Miller CR, Williams DA, Bohjanen PR. HuA and Tristetraprolin are induced following T cell activation and display distinct but overlapping RNA binding specificities. J Biol Chem. 2001;276:47958–65. doi: 10.1074/jbc.M109511200. [DOI] [PubMed] [Google Scholar]