Abstract
We reported previously that low-density lipoprotein (LDL)-containing immune complexes (LDL-IC) stimulated matrix metalloproteinase-1 (MMP-1) expression in U937 histiocytes through Fc gamma receptor (FcγR)-mediated extracellular signal-regulated kinase pathway. The present study has explored the transcriptional mechanisms involved in the stimulation. Deletion analysis showed that LDL-IC stimulated MMP-1 promoter activity in cells transfected with the Construct 1 that contained a 4,334-bp MMP-1 promoter fragment, but had no effect in cells transfected with other constructs that had shorter MMP-1 promoter (2685-bp or less), suggesting that cis-acting elements located between −4334 and −2685 are required for the promoter stimulation. The mutation study further indicated that the activator protein-1 (AP-1) (−3471) or Ets (−3836) motifs in this distal region were essential for the LDL-IC-stimulated MMP-1 expression. Moreover, although above deletion analysis showed that LDL-IC did not stimulate MMP-1 promoter activity in cells transfected with constructs that contained the proximal AP-1 (−72) and Ets (−88) in the promoter fragments that are 2685-bp or less, the mutations of the −72 AP-1 or the −88 Ets motif in the construct 1 abolished the stimulation of MMP-1 expression by LDL-IC, suggesting that a long promoter sequence is required for the −72 AP-1 and −88 Ets motifs to be involved in the stimulation. Finally, electrophoretic mobility shift assay showed that LDL-IC stimulated the activities of transcription factors AP-1 and Ets. In conclusion, the present study shows that both the distal and proximal AP-1 and Ets motifs are required for LDL-IC-stimulated MMP-1 expression in U937 histiocytes.
Introduction
In 1980, Hansson and coworkers reported that immunoglobulin G (IgG) was deposited in the subendothelial zone of the atherosclerotic lesions. Using electronic microscopy, they found that IgG was adhering to collagen fibres and coating the surfaces of foam cells.1 In 1981, Parums and Mitchinson also reported the diffuse positive IgG-staining in the amorphous parts of the atheromatous material.2 While these studies provided evidence that a relatively large quantity of IgG was present in atherosclerotic lesions, it was not clear whether these immunoglobulins were in free form or complexed with antigens and what the nature of the antigen was if immune complexes (IC) were formed. In 1994, an elegant study conducted by Witztum and coworkers showed that the IgG isolated from atherosclerotic lesions recognized epitopes characteristic of oxidized low-density lipoprotein (LDL) and that IC isolated from the lesions contained apoprotein B-100 and its fragments.3 Thus, this study clearly demonstrates that LDL-containing IC (LDL-IC) are present in atherosclerotic plaques.
In vitro studies have shown that the interaction of LDL-IC with macrophages leads to the engagement of Fc gamma receptors (FcγRs) and subsequent macrophage activation.4,5 Activated macrophages release cytokines, such as tumour necrosis factor-α and interleukin (IL)-1β, and free radicals that are believed to play crucial roles in atherogenesis.6 Moreover, studies have also demonstrated that Fc gamma receptor (FcγR)-mediated uptake of LDL-IC by macrophages leads to foam cell formation.7 All these observations suggest that LDL-IC in atherosclerotic lesions contribute to atherogenesis.
Recently, we have explored the potential role of LDL-IC in the macrophage-involved matrix degradation that contributes to the disruption of atherosclerotic plaques.8 We found that LDL-IC stimulated expression of matrix metalloproteinase-1 (MMP-1, interstitial collagenase), a proteinase responsible for the initial cleavage of fibrillar interstitial collagen, through FcγR-mediated extracellular signal-regulated kinase (ERK) pathway in both human monocyte-derived macrophages and U937 histiocytes. We also observed that both FcγRI and FcγRII were involved in the stimulation. Furthermore, our results showed that IC that contain human IgG and rabbit anti-human IgG also stimulated MMP-1 expression, suggesting that IC, regardless of their antigen contents, are capable of stimulating MMP-1 secretion from U937 histiocytes by crosslinking FcγRs. In a follow-up study, we demonstrated that pretreatment of macrophage-like U937 cells with interferon-γ (IFN-γ) augmented LDL-IC-induced MMP-1 expression.9 Because MMP-1 released by macrophages has been implicated in the destabilization of atherosclerotic plaques10–14 these studies suggest that LDL-IC may contribute to plaque rupture.
Although above studies have shown that LDL-IC up-regulate MMP-1 expression through FcγR-mediated ERK signalling pathway, the molecular mechanisms involved in the gene expression remain unknown. The current investigation was undertaken to determine the cis- and trans-acting elements required for LDL-IC-stimulated MMP-1 expression. Our results showed that the activator protein-1 (AP-1) and Ets motifs located in both the distal and the proximal regions of the MMP-1 promoter were involved in the stimulation and that LDL-IC stimulated transcription factors AP-1 and Ets.
Materials and methods
Materials
The human histiocytic cell line U93715, the plasmid clone of human MMP-1, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were purchased from the American Type Culture Collection (Rockville, MD). The cell culture media were obtained from Life Technologies, Inc (Rockville, MD). MMP enzyme-linked immunosorbent assay (ELISA) kits were obtained from Oncogene (Cambridge, MA). [α-32P]CTP, [γ-32P]ATP, and the chemiluminescence reagents were from NEN Life Science Products (Boston, MA). The Prime-a-Gene Labeling System was purchased from Promega (Madison, WI). The DNA assay kit was purchased from Molecular Probes, Inc. (Eugene, OR).
Cell culture
The histiocytic origin (resident macrophage) of U937 cells was shown by its capacity for lysozyme production and its strong esterase activity.15 U937 cells were cultured in a 5% CO2 atmosphere in Iscove's modified Dulbecco's medium supplemented with 10% fetal calf serum, and the medium was changed every 2–3 days.
Preparation of LDL-IC
LDL (d = 1·019 to 1·063 g/mol) was isolated from the plasma of normal volunteers. Insoluble LDL-IC were prepared with human native LDL and rabbit anti-LDL antiserum as described previously.16 The endotoxin level in LDL-IC preparations was measured using an endotoxin assay kit (Sigma, St Louis, MO) and the level was found to be below the lower limit of detection (0·015 units/ml).
ELISA of secreted MMP-1
Secreted MMP-1 from U937 cells was quantified using the MMP-1 ELISA kits (Oncogene), according to the protocol provided by the manufacturer. The Absorbance at 450 nm was measured by a spectrophotometric plate reader.
Northern blot analysis
Total cellular RNA was isolated from control and stimulated cells with the RNeasy minikit (Qiagen, Inc., Santa Clarita, CA) according to the instructions from the manufacturer. Northern blot of MMP-1 mRNA was performed as described previously.17
DNA assay
Cellular DNA was quantified with the CyQUANT cell proliferation assay kit (Molecular Probes), according to the procedures provided by the manufacturer.
Preparation of promoter–reporter constructs
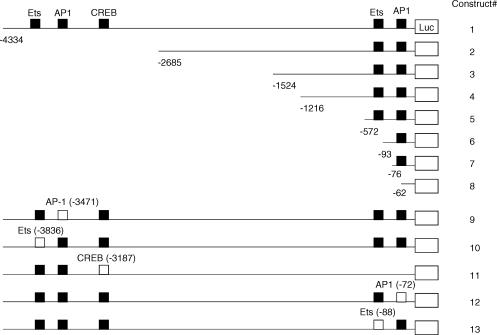
A promoter fragment of the human MMP-1 gene from −4334 to +5218 relative to the transcription start point was amplified from human DNA by polymerase chain reaction (PCR), using a pair of primers (agatgtaagagctgggaaaggacgg/tcagtgcaaggtaagtgatggcttc) and the Expand Long Template PCR System (Boehringer Mannheim Corp., Indianapolis, IN). The fragment was subcloned into the pCR-XL-TOPO vector (Invitrogen) for propagation in bacteria. The isolated plasmids were then digested with restriction enzymes MluI and XhoI to release the inserted fragment. The fragment was re-subcloned in the sense orientation into the promoter-free pGL3-Basic vector (Promega) at the MluI and XhoI sites in the 5′ flanking region of the luciferase sequence. This construct was designated as Construct 1.
Seven reporter constructs with successively 5′-deleted fragments of the MMP-1 promoter region were prepared (Fig. 1) similarly as described above, using the downstream primer used for Construct 1 and the following different upstream primers: Construct 2 (−2685/+52), agatgctcccagaggaaac; Construct 3 (−1524/+52), caggaatccataaggggagg; Construct 4 (−1216/+52), gggcaggagatgctaaataag; Construct 5 (−572/+52), tgcctggctctgagtaaag; Construct 6 (−93/+52), tcaagaggatgttataaagc; Construct 7 (−76/+52), agcatgagtcagacacctct; Construct 8 (−62/+52), acctctggctttctggaagg. All constructs were sequenced to ensure fidelity.
Figure 1.
Promoter–reporter gene constructs. Eight 5′-restricted (1–8) and five mutated human MMP-1 promoter–reporter constructs (9–13) were prepared as described in Materials and Methods. Normal binding sites for AP-1, Ets, and CREB (closed squares) and mutated binding sites (opened squares) are indicated.
Site-directed mutagenesis
Five reporter constructs designated as Construct 9, 10, 11, 12, and 13, which contain mutations in the AP-1, Ets, and cAMP-responsive element binding protein (CREB) motifs (Fig. 1), were prepared using the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Construct 1 was used as template and five oligonucleotides with the mutated AP-1, Ets and CREB motifs (Table 1) were synthesized to generate mutated constructs.
Table 1.
Oligonucleotides used for the site-directed mutagenesis
| Constructs # | Mutated motifs | Wild-type sequences | Mutation sequences |
|---|---|---|---|
| ″9 | AP-1 motif at −3471 | aaataagagagattgagtgacagcttgaagggg | aaataagagagattgCAtgacagcttgaagggg |
| 10 | PEA-3 motif at −3836 | gggagtctgaggcaggaagattgcttaagccca | gggagtctgaggcagTCagattgcttaagccca |
| 11 | CREB motif at −3187 | ggtaaccatgaggactgacggaaccagtgtgtacc | ggtaaccatgaggactTaTggaaccagtgtgtac |
| 12 | AP-1 motif at −72 | atgttataaagcatgagtcagacacctctggct | atgttataaagcatgCAtcagacacctctggct |
| 13 | PEA-3 motif at −88 | atagctaatcaagaggatgttataaagcatgag | atagctaatcaagagTCtgttataaagcatgag |
The motifs are underlined. The mutated nucleotides are indicated by capital letters.
Transient transfection
U937 histiocytes were transiently transfected with the reporter constructs, using FuGene 6 transfection reagent (Roche Molecular Biochemicals, Indianapolis, IN), according to the instructions provided by the manufacturer. For transfection, cells were seeded in 35-mm culture dishes (Costar Corp, Cambridge, MA.) at 5 × 105/ml. Transfection was performed in triplicate with 1 µg each of construct DNA and control pSVβGal vector (Promega) and 6 µl of FuGene 6 per dish for 24 hr. After the transfection, cells were incubated with or without 150 µg/ml of LDL-IC for 24 hr. Luciferase activity in cells was measured using a luciferase activity assay kit (Promega). The β-galactosidase activity was determined by a colorimetric assay (Promega) for the correction of transfection efficiency.
Electrophoretic mobility shift assay (EMSA)
U937 cells were seeded at 10 × 106 per 100-mm dish and incubated with or without 150 µg/ml of LDL-IC for 24 hr. Nuclear extracts were prepared using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Pierce, Rockford, IL) according to the instructions provided by the manufacturer. Nuclear extracts were quickly frozen at −70°.
The EMSA was performed using a gel shift assay system (Promega). DNA probes were synthesized according to the MMP-1 promoter sequence.18 The sequence for the AP-1 motif-containing oligonucleotides is gagagattgagtgacagcttg (−3478/−3458) and the one for the Ets motif-containing oligonucleotides is ctgaggcaggaagattgctta (−3844/−3824). DNA probes were radiolabelled with 32P in the presence of T4 polynucleotide kinase and purified by chromatography using a Sephadex G-25 spin column (Roche Molecular Biochemicals). An aliquot of the labelled probe (0·035 pmol/µl) was incubated with 3–5 µg of nuclear extract in 5× gel shift binding buffer containing 20% glycerol, 5 mm MgCl2, 2·5 mm ethylenediaminetetra-acetic acid (EDTA), 2·5 mm dithiothreitol (DTT), 250 mm NaCl, 50 mm Tris–HCl (pH 7·5) and 0·25 mg/ml poly (dI-dC)poly (dI-dC) at room temperature for 20 min After the incubation, an aliquot of the incubation mixture was loaded onto a 4% polyacrylamide non-denature gel or Novex precast gels (Invitrogen Life Technologies, Carlsbad, CA) and electrophoresis was performed at 350 V for about 10 min The gel was then dried and exposed to X-ray film for 4–12 hr. For supershift assay, the radiolabelled AP-1 oligonucleotides were incubated with nuclear extract in the presence or absence of 1 µg of antic-Jun antibody (Santa Cruz Biotechnology, Inc, Santa Cruz, CA) overnight at 4° and the mixture was electrophoresed as described above.
Statistical analysis
Data was presented as mean ± SEM. Comparison between treatments was performed using the one-way analysis of variance (anova). A value of P < 0·05 was considered significant.
Results
LDL-IC stimulate MMP-1 transcription
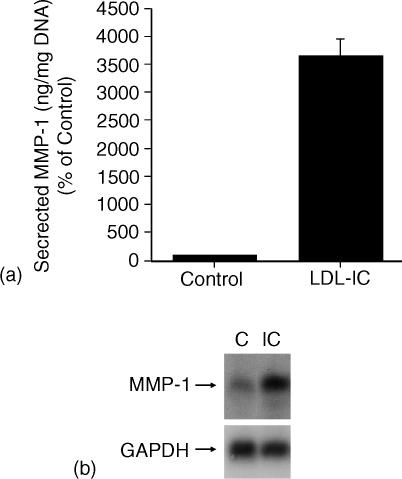
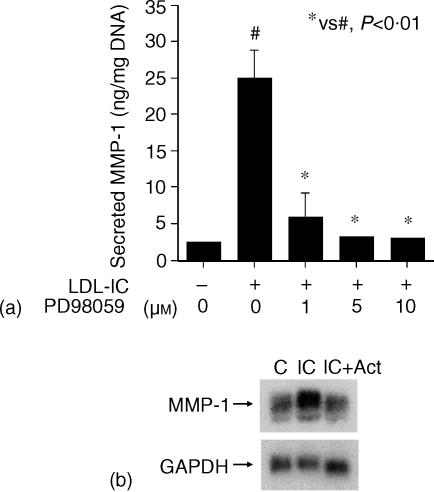
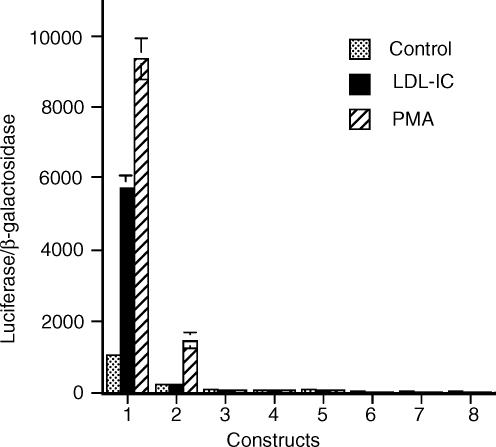
The effect of LDL-IC on MMP-1 expression was re-examined at the beginning of this study to ensure that the response of U937 cells to LDL-IC was similar to that we reported previously.8 As shown in Fig. 2, LDL-IC stimulated MMP-1 secretion and mRNA expression by U937 cells. MAPK/ERK kinase (MEK) inhibitor PD98059 abolished the stimulation of MMP-1 expression (Fig. 3a), suggesting that LDL-IC-stimulated MMP-1 expression is ERK-dependent. All these results are consistent with our previous reports.8 To determine whether the stimulation of MMP-1 mRNA expression by LDL-IC is caused by a transcriptional or post-transcriptional regulation, cells were pretreated with actinomycin D, a transcription inhibitor, before the stimulation with LDL-IC. Results showed that 5 µg/ml of actinomycin D abolished the stimulation (Fig. 3b), indicating that LDL-ICs up-regulate MMP-1 expression by activating the transcription of MMP-1 gene. The transcriptional activation by LDL-IC was also indicated by the observation that LDL-IC stimulated MMP-1 promoter activity in cells transfected with a MMP-1 promoter–luciferase reporter construct (Fig. 4).
Figure 2.
Stimulation of MMP-1 secretion and mRNA expression by LDL-IC. U937 cells were incubated with or without 150 µg/ml of LDL-IC for 24 hr. After the incubation, the conditioned medium was collected for quantitative analysis of secreted MMP-1 by ELISA (a) while the total RNA was isolated for Northern analysis of MMP-1 mRNA (b). The data are representatives of three experiments with similar results.
Figure 3.
Inhibition of LDL-IC-stimulated MMP-1 expression by PD98059 and actinomycin D. (a) U937 cells were incubated for 24 hr with 150 µg/ml of LDL-IC in the presence of increasing concentrations of PD98059 as indicated. After incubation, the conditioned medium was subjected to ELISA to quantify secreted MMP-1. Dimethyl sulphoxide, a vehicle for PD98059, was 0·1% of the medium volume and has been shown to have no effect on MMP-1 secretion.8 Data presented are the mean ± SEM of three different experiments run in duplicate. (b) U937 cells were preincubated in medium with actinomycin D (5 µg/ml) for 30 min The cells were then incubated with 150 µg/ml of LDL-IC for 24 hr. After the incubation, total RNA was isolated for Northern blot analysis of MMP-1 mRNA as described in Materials and Methods. (c) control; IC, LDL-IC; Act, actinomycin D.
Figure 4.
LDL-IC stimulate MMP-1 promoter activity in cells transfected with Construct 1. U937 cells were transiently transfected with MMP-1 promoter-reporter constructs and β-galactosidase vector for 24 hr, and then stimulated without (control) or with 150 µg/ml of LDL-IC or 100 nm PMA for 24 hr. The cellular luciferase and β-galactosidase activities were measured and the ratios were calculated for correction of the transfection efficiency. Data presented are mean ± SEM of three experiments run in triplicate.
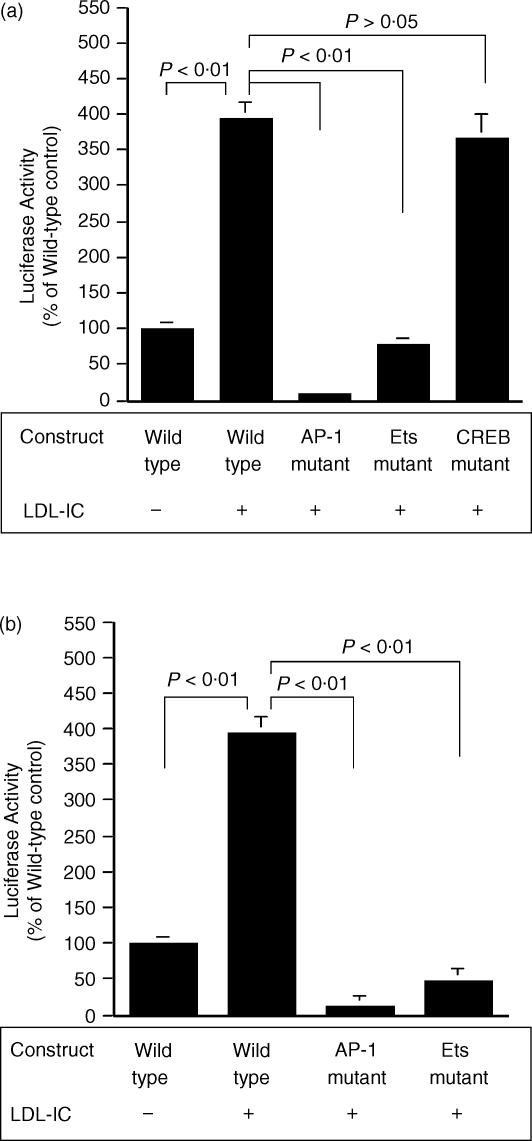
The −3471 AP-1 and −3836 Ets motifs Are essential for LDL-IC-stimulated MMP-1 expression
To explore the transcriptional mechanisms by which LDL-IC stimulate MMP-1 expression, deletion analysis was performed by transfecting U937 cells with eight luciferase reporter constructs, which contain successively 5′-deleted fragments of MMP-1 promoter region (Fig. 1), followed by treatment with LDL-IC or phorbol 12-myristate 13-acetate (PMA) as positive control. Results from luciferase activity assay showed that PMA stimulated luciferase activity in cells transfected with Construct 1 (−4334/+52) and 2 (−2685/+52) (Fig. 4). Interestingly, LDL-IC only stimulated luciferase activity in cells transfected with the Construct 1 (−4,334/+52), suggesting that the cis-acting elements that are responsive to LDL-IC are located between −4334 and −2685. By analysing the sequence of the region between −4334 and −2685, cis-acting elements AP-1 (−3471), Ets (−3836), and CREB (−3187) motifs were found. To determine whether these motifs are involved in the LDL-IC-stimulated MMP-1 expression, mutation analysis using Construct 9 (−3471 AP-1 mutant), Construct 10 (−3836 Ets mutant), and Construct 11 (−3187 CREB mutant)19 (Fig. 1) was performed. Results showed that the AP-1 mutation in Construct 9 and the Ets mutation in Construct 10, but not the CREB mutation in Construct 11, abolished LDL-IC-stimulated luciferase activity (Fig. 5a). The AP-1 mutation in Construct 9 also markedly inhibited the baseline level of luciferase activity (Fig. 5a), which is consistent with the deletion study showing that control cells transfected with Constructs 2–8, which do not have the −3471 AP-1 motif, had a substantial reduction of MMP-1 promoter activity as compared to those transfected with wild-type Construct 1 (Fig. 4). These results indicate that these ‘distal’ AP-1 and Ets motifs play an essential role in the LDL-IC-stimulated MMP-1 expression, and the former is also critical for the constitutive expression of MMP-1.
Figure 5.
Mutations in the distal AP-1 (−3471)/Ets (−3836) (a) and proximal AP-1 (−72)/Ets (−88) motifs (b) inhibit LDL-IC-stimulated MMP-1 promoter activity. (a). U937 cells were transiently transfected with Construct 1 (wild-type construct), Construct 9 (−3471 AP-1 mutant), Construct 10 (−3836 Ets mutant), or Construct 11 (−3187 CREB mutant) and then treated with 150 µg/ml of LDL-ICs for 24 hr. The cellular luciferase activity was measured and corrected for transfection efficiency. (b). U937 cells were transiently transfected with Construct 1 (wild-type), Construct 12 (−72 AP-1 mutant), and Construct 13 (−88 Ets mutant), and then treated with 150 µg/ml of LDL-ICs for 24 hr. The cellular luciferase activity was measured and corrected for transfection efficiency. Data presented are mean ± SEM of three experiments run in triplicate. The luciferase activity in cells transfected with wild-type Construct 1 and that had no exposure to LDL-IC was designated as 100%.
The −72 AP-1 and −88 Ets-1 motifs require a long promoter sequence for activation
The results from the deletion studies (Fig. 4) showed that LDL-ICs did not stimulate MMP-1 promoter activity in cells transfected with Constructs 2, 3, 4, 5, 6 and 7, which all contain the proximal AP-1 (−72) and Ets (−88) motifs that have been shown previously to be essential in MMP-1 expression.20 Two reasons may explain this observation. First, these motifs may need long promoter sequence for their activation. Second, the proximal AP-1 and Ets motifs may simply not respond to LDL-IC. To clarify this issue, mutation analysis, using Construct 12 and Construct 13, which have a long MMP-1 promoter sequence (4334-bp) and contain mutations in −72 AP-1 and −88 Ets motifs, respectively (Fig. 1), was conducted. Interestingly, results showed that the mutations of either the AP-1 or the Ets motif inhibited both the basal and the LDL-IC-stimulated luciferase activity (Fig. 5b), indicating that the activation of the proximal AP-1 and Ets motifs requires a long promoter sequence.
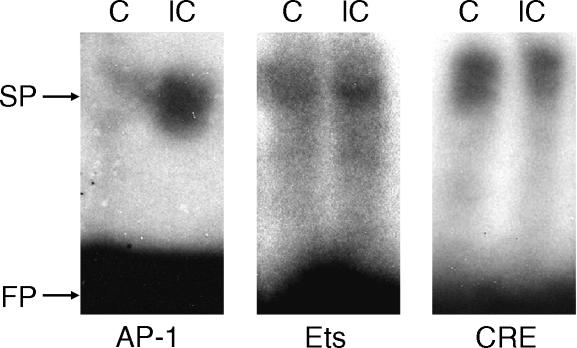
LDL-ICs stimulate transcription factor AP-1 and Ets-1
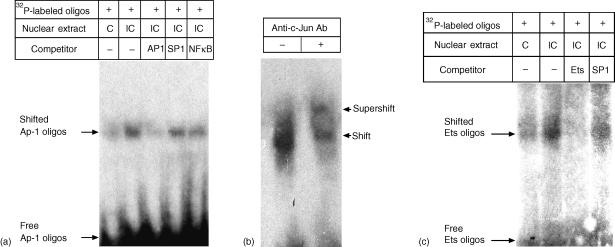
The above deletion and mutation studies showed that both the distal and the proximal AP-1 and Ets motifs were LDL-IC-responsive cis-acting elements. Based on these observations, we investigated the effects of LDL-IC on the trans-acting elements (transcription factor) AP-1 and Ets. Results from the EMSA (Fig. 6) showed that the nuclear AP-1 activity, which was very low in control cells, was markedly stimulated by LDL-IC. The Ets activity was also stimulated by LDL-IC. In contrast, LDL-IC had no effect on CREB activities, which is consistent with the mutation analysis (Fig. 5a) showing that Construct 11 (−3187 CREB mutant) did not affect LDL-IC-stimulated MMP-1 promoter activity. The binding of AP-1 to the AP-1 motif was confirmed by the competition studies showing that the unlabelled AP-1 probes competed effectively against the radiolabelled AP-1 probe while the unlabeled SP-1 and nuclear factor (NF)κB probes had no competition at all (Fig. 7a) and the supershift assay using anti-c-jun antibody (Fig. 7b). The unlabeled Ets-1 probe also effectively competed against the labeled Ets probe (Fig. 7c).
Figure 6.
LDL-IC stimulate transcription factor AP-1 and Ets activities. Nuclear proteins of U937 cells treated without (C, control) or with 150 µg/ml of LDL-IC (IC) were extracted and incubated with radiolabelled oligonucleotide probes containing AP-1, Ets, or CREB motifs. The binding of transcription factor in the nuclear extracts to their corresponding probe was detected by the EMSA as described under Materials and Methods. FP, free probes. SP, shifted probes. The blot presented is representative of three blots with similar results.
Figure 7.
The competition between the unlabeled and radiolabelled probes and probe supershift. (a) The radiolabelled AP-1 probes were incubated with nuclear extracts of U937 cells, which were treated without (control, C) or with 150 µg/ml of LDL-IC (IC), in the presence or absence of 50-fold unlabelled AP-1, SP-1 or NFκB probes. The protein–DNA complexes were detected by EMSA. (b) The radiolabelled AP-1 probes were incubated with nuclear extracts of U937 cells, which were treated with 150 µg/ml of LDL-IC, in the presence of absence of anti-c-jun antibodies. The protein–DNA–antibody complexes were detected by EMSA as supershift. (c) The radiolabelled Ets probes were incubated with nuclear extracts of U937 cells, which were treated without (control, C) or with 150 µg/ml of LDL-IC (IC), in the presence or absence of 50-fold unlabeled Ets and SP-1 probes. The protein–DNA complexes were detected by EMSA.
Discussion
It has been shown that the expression of human MMP-1 is stimulated by phorbol esters, cytokines, and growth factors at the transcription level21–23. Many studies have demonstrated that multiple promoter elements are involved in the transcriptional regulation of MMP-1 expression. For examples, transcriptional activation of MMP-1 by PMA is mediated by the proximal AP-1 (−72) and Ets (−88) as well as the ‘TTCA’ (−104) motifs.24,25 Stimulation of MMP-1 expression by insulin is also mediated by the proximal AP-1 and Ets.26 Besides the proximal AP-1 and Ets motifs, MMP-1 promoter sequence contains several other AP-1 and Ets motifs, and some of them are located in the ‘distal’ promoter region that is more than 3000-bp from the transcription start site.18 Although the potential involvement of the distal motifs in MMP-1 expression has been suggested by previous studies showing that higher levels of stimulation on the MMP-1 promoter activity can be induced with larger promoter fragments, the particular motifs in the distal promoter region that are involved in MMP-1 expression have not been identified. In the present study, our deletion and mutation analyses demonstrate that, in addition to the proximal AP-1 (−72) and Ets (−88) motifs, the distal AP-1 (−3471) and Ets (−3836) motifs are also LDL-IC-responsive elements. To our best knowledge, this is the first study that shows the role of the distal AP-1 and Ets motifs in the MMP-1 expression.
Another compelling finding from the present study is that the proximal AP-1 and Ets motifs require a long distal promoter sequence for their activation by LDL-IC. Our deletion analysis shows that LDL-IC do not stimulate MMP-1 promoter activity of Constructs 2, 3, 4, 5, 6 and 7 that all contain the proximal AP-1 and Ets motifs. However, the mutation analysis, using constructs containing 4334-bp promoter fragment, shows that the mutated proximal AP-1 or Ets motifs completely inhibited the stimulation of MMP-1 expression by LDL-IC, suggesting that the promoter sequence equal or less than 2685-bp is insufficient for the proximal AP-1 and Ets motifs to be activated by LDL-IC. Previous studies have shown that the regulation of MMP-1 expression in certain types of cells requires a long promoter sequence. For examples, Brinckerhoff and colleagues showed that in the breast cancer cell line BC-8701, a reporter construct containing 512-bp MMP-1 promoter fragment was sufficient for a fourfold stimulation of the MMP-1 promoter activity by IL-1β, whereas in human foreskin fibroblasts, a 3300-bp promoter sequence is required for a significant stimulation by the same cytokine.18 In another study by the same group, it was found that transcriptional repression of MMP-1 gene in MDA231 breast cancer cells by all-trans-retinoic acid requires distal regions of the MMP-1 promoter.27 These reports and our current study suggest that different types of cells may utilize different promoter elements, either proximal or distal to the transcription start site, for MMP-1 gene expression. It is likely that for U937 cells, both the distal and proximal AP-1 and Ets motifs are involved in the interaction with transcription factors AP-1 and Ets. Additionally, our studies also show that the proximal AP-1 and Ets as well as the distal AP-1 motifs are required for the baseline level of MMP-1 promoter activity, which agrees with the previous report that the AP-1 and Ets motifs are essential for the constitutive expression of the MMP-1 gene.23
In conclusion, we have provided evidence that LDL-IC up-regulate the MMP-1 expression through AP-1 and Ets-dependent transcriptional activation in U937 histiocytes. Thus, this study has elucidated a transcriptional mechanism involved in LDL-IC-stimulated MMP-1 expression.
Acknowledgments
This work was supported by a Merit Review Grant from the Research Service of the Department of Veterans Affairs and the Grant-in-Aid from the American Heart Association, Mid-Atlantic Affiliate. This study was presented as an abstract at the 75th American Heart Association Scientific Sessions, Chicago, IL on November 17, 2002.
Abbreviations
- LDL
low-density lipoprotein
- IC
immune complexes
- MMP
matrix metalloproteinase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- AP-1
activator protein-1
- CREB
cAMP-responsive element binding protein
- MAPK
mitogen-activated protein kinase
- ERK
extracellular signal-regulated kinase
References
- 1.Hansson GK, Bondjers G, Bylock A, Hjalmarsson L. Ultrastructural studies on the location of IgG in the aortic endothelium and subendothelial intima of atherosclerotic and non-atherosclerotic rabbits. Exp Mol Pathol. 1980;33:302–15. doi: 10.1016/0014-4800(80)90028-3. [DOI] [PubMed] [Google Scholar]
- 2.Parums O, Mitchinson MJ. Demonstration of immunoglobulin in the neighbourhood of advanced atherosclerotic plaques. Atherosclerosis. 1981;38:211–6. doi: 10.1016/0021-9150(81)90118-0. [DOI] [PubMed] [Google Scholar]
- 3.Yla-Herttuala S, Palinski W, Butler SW, Picard S, Steinberg D, Witztum JL. Rabbit and human atherosclerotic lesions contain IgG that recognizes epitopes of oxidized LDL. Arterioscler Thromb. 1994;14:32–40. doi: 10.1161/01.atv.14.1.32. [DOI] [PubMed] [Google Scholar]
- 4.Huang Y, Jaffa A, Koskinen S, Takei A, Lopes-Virella MF. Oxidized LDL-containing immune complexes induce Fc gamma receptor I-mediated mitogen-activated protein kinase activation in THP-1 macrophages. Arterioscler Thromb Vasc Biol. 1999;19:1600–7. doi: 10.1161/01.atv.19.7.1600. [DOI] [PubMed] [Google Scholar]
- 5.Virella G, Munoz JF, Galbraith GM, Gissinger C, Chassereau CH, Lopes-Virella MF. Activation of human monocyte-derived macrophage by immune complexes containing low density lipoprotein. Clin Immunol Immunopathol. 1995;75:179–89. doi: 10.1006/clin.1995.1069. [DOI] [PubMed] [Google Scholar]
- 6.Libby P. Inflammatory and immune mechanisms in atherogenesis. In: Leaf A, Weber PC, editors. Atherosclerosis Reviews. New York, NY: Raven Press; 1990. pp. 79–89. [Google Scholar]
- 7.Griffith RL, Virella GT, Stevenson HC, Lopes-Virella MF. Low density lipoprotein metabolism by human macrophages activated with low density lipoprotein immune complexes. J Exp Med. 1988;168:1041–59. doi: 10.1084/jem.168.3.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang Y, Wu S, Fleming AJ, Virella G, Lopes-Virella MF. Fc gamma receptor crosslinking by immune complexes induces matrix metalloproteinase-1 expression in human U937 histiocytes via mitogen-activated protein kinase cascade. Arterioscler Thromb Vasc Biol. 2000;20:2533–8. doi: 10.1161/01.atv.20.12.2533. [DOI] [PubMed] [Google Scholar]
- 9.Anderson F, Game BA, Atchley D, Xu M, Lopes-Virella MF, Huang Y. IFN-γ pretreatement augments immune complex-induced matrix metalloproteinase-1 expression in U937 histiocytes. Clin Immunol. 2002;102:200–7. doi: 10.1006/clim.2001.5161. [DOI] [PubMed] [Google Scholar]
- 10.Libby P. Molecular bases of the acute coronary syndromes. Circulation. 1995;91:2844–50. doi: 10.1161/01.cir.91.11.2844. [DOI] [PubMed] [Google Scholar]
- 11.Galis ZS, Suknova GK, Lark MW, Libby P. Increased expression of matrix metalloproteinases and matrix degrading activity in vulnerable regions of human atherosclerotic plaques. J Clin Invest. 1995;94:2493–503. doi: 10.1172/JCI117619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aikawa M, Mabkin E, Okada Y, Voglic SJ, Clinton SK, Brinckerhoff CE, Sukhova GK, Libby P. Lipid lowering by diet reduces matrix metalloproteinase activity and increases collagen content of rabbit atheroma: a potential mechanism of lesion stabilization. Circulation. 1998;97:2433–44. doi: 10.1161/01.cir.97.24.2433. [DOI] [PubMed] [Google Scholar]
- 13.Lee RT, Schoen FJ, Loree HM, Lark MW, Libby P. Circumferential stress and matrix metalloproteinase 1 in human coronary atherosclerosis. Implication for plaque rupture. Arterioscler Thromb Vasc Biol. 1996;16:1070–3. doi: 10.1161/01.atv.16.8.1070. [DOI] [PubMed] [Google Scholar]
- 14.Cheng GC, Loree HM, Kamm RD, Fishbein MC, Lee RT. Distribution of circumferential stress in ruptured and stable atherosclerotic lesions. A structural analysis with histopathological correlation. Circulation. 1993;87:1179–87. doi: 10.1161/01.cir.87.4.1179. [DOI] [PubMed] [Google Scholar]
- 15.Sundstrom C, Nilsson K. Establishment and characterization of a human histiocytic lymphoma cell line (U937) Int J Cancer. 1976;17:565–77. doi: 10.1002/ijc.2910170504. [DOI] [PubMed] [Google Scholar]
- 16.Lopes-Virella MF, Binzafar N, Rackley S, Takei A, LaVia M, Virella G. The uptake of LDL-containing immune complexes by human macrophages: Predominant involvement of the FcγRI receptor. Atherosclerosis. 1997;135:161–70. doi: 10.1016/s0021-9150(97)00157-3. [DOI] [PubMed] [Google Scholar]
- 17.Huang Y, Mironova M, Lopes-Virella MF. Oxidized LDL stimulates matrix metalloproteinase-1 expression in human vascular endothelial cells. Arterioscler Thromb Vasc Biol. 1999;19:2640–7. doi: 10.1161/01.atv.19.11.2640. [DOI] [PubMed] [Google Scholar]
- 18.Rutter JL, Benbow U, Coon CI, Brinckerhoff CE. Cell-type specific regulation of human interstitial collagenase-1 gene expression by interleukin-1β (IL-1β) in human fibroblasts and BC-8701 breast cancer cells. J Cell Biochem. 1997;66:322–36. [PubMed] [Google Scholar]
- 19.Illi B, Puri P, Morgante L, Capogrossi MC, Gaetano C. Nuclear factor-κB and cAMP response element binding protein mediate opposite transcriptional effects on the Flk-1/KDR gene promoter. Circ Res. 2000;86:e110–e117. [PubMed] [Google Scholar]
- 20.Borden P, Heller RA. Transcriptional control of matrix metalloproteinases and the tissue inhibitors of matrix metalloproteinases. Crit Rev. 1997;7:159–78. doi: 10.1615/critreveukargeneexpr.v7.i1-2.90. [DOI] [PubMed] [Google Scholar]
- 21.Harris ED., Jr Rheumatoid arthritis. Pathophysiology and implications for therapy. N Engl J Med. 1990;322:1277–89. doi: 10.1056/NEJM199005033221805. [DOI] [PubMed] [Google Scholar]
- 22.Brinckerhoff CE. Regulation of metalloproteinase gene expression: implication for osteoarthritis. Crit Rev Eukaryot Gene Expr. 1992;2:145–64. [PubMed] [Google Scholar]
- 23.Aho S, Rouda S, Kennedy SH, Qin H, Tan EML. Regulation of human interstitial collagenase (matrix metalloproteinase-1) promoter activity by fibroblast growth factor. Eur J Biochem. 1997;247:503–10. doi: 10.1111/j.1432-1033.1997.00503.x. [DOI] [PubMed] [Google Scholar]
- 24.Vincenti MP, White LA, Schroen DJ, Benbor U, Brinckerhoff CE. Regulating expression of the gene for matrix metalloproteinase-1 (collagenase). mechanisms that control enzyme activity, transcription, and mRNA stability. Crit Rev Eukaryot Gene Expr. 1996;6:391–411. doi: 10.1615/critreveukargeneexpr.v6.i4.40. [DOI] [PubMed] [Google Scholar]
- 25.Gutman A, Wasylyk B. The collagenase gene promoter contains a TPA and oncogene-responsive unit encompassing the PEA-3 and AP-1 binding sites. EMBO J. 1990;9:2241–6. doi: 10.1002/j.1460-2075.1990.tb07394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chapman SC, Ayala JE, Streeper RS, et al. Multiple promoter elements are required for the stimulatory effect of insulin on human collagenase-1 gene transcription. J Biol Chem. 1999;274:18625–34. doi: 10.1074/jbc.274.26.18625. [DOI] [PubMed] [Google Scholar]
- 27.Benbow U, Rutter JL, Lowrey CH, Brinckerhoff CE. Transcriptional repression of the human collagenase-1 (MMP-1) gene in MDA 231 breast cancer cells by all-trans-retinoic acid requires distal regions of the promoter. Br J Cancer. 1999;79:221–8. doi: 10.1038/sj.bjc.6690037. [DOI] [PMC free article] [PubMed] [Google Scholar]