Abstract
The catalytic activities of eukaryotic protein kinases (EPKs) are regulated by movement of the C-helix, movement of the N and C lobes upon ATP binding, and movement of the activation loop upon phosphorylation. Statistical analysis of the selective constraints associated with AGC kinase functional divergence reveals conserved interactions between these regulatory regions and three regions of the C-terminal tail (C-tail): the N-lobe tether (NLT), the active-site tether (AST), and the C-lobe tether (CLT). The NLT serves as a docking site for an upstream kinase PDK1 and, upon activation, positions the C-helix within the ATP binding pocket. The AST directly interacts with the ATP binding pocket, and the CLT interacts with the interlobe linker and the αC–β4 loop, which appears to serve as a hinge for C-helix movement. The C-tail is a hallmark of AGC functional divergence inasmuch as most of the conserved core residues that distinguish AGC kinases from other EPKs are associated with the NLT, AST, or CLT. Moreover, several AGC catalytic core conserved residues that interact with the C-tail strikingly diverge from the canonical residues observed at corresponding positions in nearly all other EPKs, suggesting that the catalytic core may have coevolved with the C-tail in AGC kinases. These observations, along with the fact that the C-tail is needed for catalytic activity suggests that the C-tail is a cis-acting regulatory module that can also serve as a regulatory “handle,” to which trans-acting cellular components can bind to modulate activity.
Keywords: kinase mechanisms, phosphorylation, signaling
Protein kinases A, B, and C (designated PKA, PKB, and PKC, respectively) belong to the AGC group within the eukaryotic protein kinase (EPK) superfamily (1, 2). Because AGC kinases control critical cellular processes, such as cell growth, differentiation, and cell survival, they are subject to tight spatial and temporal regulation, which is achieved, in part, through activation loop phosphorylation and subsequent repositioning of key catalytic and substrate binding regions (3–5). In PKA, PKB, and PKC, phosphorylation of the activation loop is mediated by an upstream kinase called PDK1, which also is a member of the AGC group (6–9). It is believed that most AGC kinases are phosphorylated and activated by PDK1 because nearly all of them conserve a consensus PDK1 recognition site (10).
Translocation to the plasma membrane is a critical part of AGC kinase biological function (11) and occurs via distinct mechanisms in different AGC kinases. For example, PKB and PDK1 are translocated via their N- and C-terminal (respectively) pleckstrin homology domains (12), which bind with high affinity to membrane-associated phosphatidylinositol 3,4,5 triphosphate (13, 14). Plasma membrane association is proposed to induce conformational changes in the catalytic domain of these proteins (13). Conventional PKCs are translocated upon binding to diacylglycerol and Ca2+-triggered phospholipids by regulatory C1 and C2 domains, respectively (15). PKA recruitment to the membrane is mediated by interaction of A-kinase anchoring proteins with a PKA regulatory subunit (16) and myristylation of a glycine residue in a PKA N-terminal helix that is outside of the catalytic core (17).
Structural and biochemical studies suggest that activation of AGC kinases also involves conformational changes in the C-helix, a key regulatory helix within EPKs. Upon EPK activation, a conserved glutamate within the C-helix is repositioned to form a salt bridge with a conserved lysine that is within the β3 strand and coordinates with the α and β phosphates of ATP (4). Various regulatory proteins modulate kinase activity by altering the conformation of the C-helix. In cyclin-dependent kinase, for instance, cyclin binding repositions the C-helix into an active conformation, whereas in the absence of cyclin the C-helix swings away from the ATP-binding site, thereby disrupting the salt bridge interaction (18).
In AGC kinases, repositioning of the C-helix is mediated by the C-terminal tail (termed the C-tail), near the end of which is a hydrophobic motif (the HF motif) that is well conserved across AGC kinases. In the inactive state of PKB, both the HF motif and the C-helix are disordered (19), but, in the active state, the HF motif stabilizes the C-helix by docking to a hydrophobic groove between the C-helix and the β4 strand (19, 20), suggesting a regulatory role for this interaction (19). In the inactive state, the AGC HF motif also serves as a docking site for PDK1 (21–23), which phosphorylates the activation loop of and thereby activates the AGC kinase. In Akt/PKB, a serine or threonine adjacent to the HF motif gets phosphorylated by the mTor–Rictor complex (24, 25). This phosphorylation of the HF motif provides an additional level of regulation inasmuch as it increases binding affinity to PDK1 (23, 26).
Presumably there are other modes of AGC kinase regulation that involve interactions with regulatory proteins or the plasma membrane and that might be coupled to binding and positioning of ATP, substrate, or catalytically critical regions. Thus fully understanding AGC regulatory mechanisms will require many carefully designed experiments based on structural analyses of AGC kinases bound to various regulators and activators. Designing such experiments requires, however, that one first formulate the right hypotheses based on preliminary observations. One source of empirical information in this regard is to characterize the evolutionary constraints distinguishing AGC kinases from other, functionally divergent protein kinases through Bayesian inference (27) (reviewed in ref. 28). Using this approach, we recently compared EPKs to distantly related EPK-like kinases (ELKs) (29). This comparison revealed that, whereas the ELK C-helix appears to be held in a constitutively active conformation, the EPK C-helix, which is conformationally flexible, typically is associated with an elaborate network of conserved interactions involving the αC–β4 loop, which anchors the C-helix to the catalytic core and acts as a hinge that mediates C-helix conformational changes (29). However, the AGC kinases diverge from these canonical αC–β4 interactions, suggesting that they use alternative modes of regulation (29). Here, we examine the divergent nature of AGC kinases more carefully and present evidence that the C-tail is the hallmark of AGC kinases (Fig. 1) and plays a critical regulatory role, in which it can serve as a docking site for other cellular components. It is thus a cis regulatory element essential for activity and allostery. Furthermore, we propose that features distinguishing the AGC catalytic domain from other EPKs have coevolved with the C-tail to create an extended allosteric network that links the C-tail to the active site. Finally, the last subdomain of the catalytic core, from which the C-tail emanates, contains a short AGC kinase-specific insert segment that is allosterically linked to the substrate-binding site.
Fig. 1.
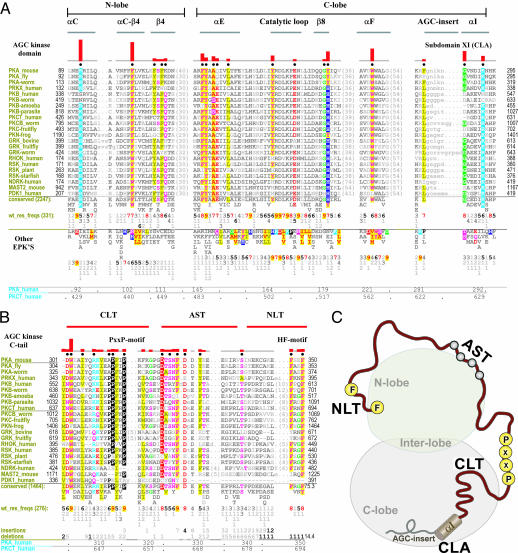
AGC kinase evolutionary constraints. (A) Constraints most distinguishing AGC kinases from other EPKs. The alignment shows representative AGC kinases (see SI Appendix 3 for a complete description of the analysis and alignments). Directly below this alignment are the patterns of conserved residues observed in AGC kinases as a whole with the residue frequencies corresponding to these patterns shown directly below conserved residues in integer tenths where, for example, a 5 indicates that the corresponding residue above it occurs in 50–60% of the (weighted) sequences. Below this are shown the patterns of conserved residues observed in other EPKs along with the corresponding (weighted) residue frequencies (wt_res_freqs). (Such sequence weighting adjusts for overrepresented families in the alignment.) Residue positions within human PKA and PKC-theta are indicated at the bottom. The histogram above the alignment plots the strength of the selective pressure shifting residues at each position in the AGC kinases away from the residue composition observed at those positions in other EPKs. Residue positions subject to the strongest constraints are highlighted with chemically similar amino acids colored similarly; very weakly conserved positions and nonconserved positions are shown in dark and light gray, respectively. Dots below the histograms indicate those residues positions that most strikingly distinguish AGC kinases from EPKs, as selected by our statistical procedure (27). Key secondary structural elements are indicated above the histogram with β strands indicated by their number designations (i.e., 6–9 corresponds to the β6–β9 strands, respectively) and helices by their letter designations (i.e., F and G correspond to the F helix and G helix, respectively). Regions corresponding to EPK lobes are indicated at the top. See SI Appendix 3 for sequence identifiers. (B) Alignment of AGC kinase C tail highlighting conserved residues. The NLT, AST, and CLT regions are indicated above the alignment. (C) Cartoon representation of the AGC kinase domain showing the key regions of the C-tail interacting with the two lobes (N and C lobes) of the catalytic domain. Some of the conserved motifs in the C tail such as the PxxP and hydrophobic motif residues are shown as yellow circles.
Results and Discussion
To determine which residues and corresponding structural features have most contributed to AGC kinase functional divergence we performed a statistical analysis (27) (see Methods) of the selective constraints distinguishing AGC kinases from other EPKs. These constraints generally correspond to residues that are highly conserved in AGC kinases but strikingly different in kinases outside of the AGC group [K92PKA, F102PKA, F108PKA (Y in some AGCs), F110PKA, K111PKA (typically Q), F145PKA, Y146PKA, A148PKA, Q149PKA (typically E), Y164PKA, Y179PKA (H in some AGCs), W221PKA, F281PKA (typically L), G287PKA, and K292PKA in Fig. 1A]. These AGC conserved residues, although widely dispersed in the sequence, spatially interact either with conserved residues within the AGC C-tail (Fig. 1 B and C) or conserved residues near the end of the C lobe, to which the C-tail is attached (Fig. 1 A and C). Thus the C-tail appears to be the hallmark of AGC kinase functional divergence.
The AGC prototypic features identified here are generally highly conserved across AGC families and across diverse phyla (Fig. 1 A and B). However, metazoan PKBs diverge from other AGC kinases because of the presence of an N-terminal pleckstrin homology domain [see supporting information (SI) Appendix 1]. Likewise, metazoan PKAs, which serve as a reference and prototype when discussing other protein kinases, diverge from the canonical AGC kinase residues at certain positions (Fig. 1A). The loss of some of these canonical features may be caused by the presence of an N-terminal helix (the A-helix) that occurs in metazoan PKAs but is absent from nonmetazoan PKAs and other AGC kinases: regions of the catalytic core that contact the A-helix lack the canonical AGC residues typically found there. Thus, in addition to referring in the following discussions to residues within mammalian PKA, which serves as the standard prototype, we will also refer to residue positions in the PKC family (human PKC-theta), because it conserves nearly all of the canonical AGC features.
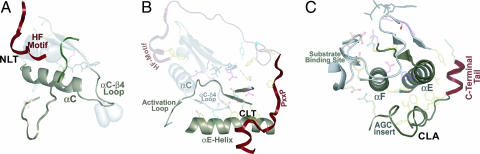
The AGC C-tail can be divided into three segments based on its conserved interactions with the catalytic domain (Fig. 2A and B): (i) At the C-terminal end of the tail is the N-lobe tether (NLT) (residues 680–695 in PKC; residues 340–350 in PKA) that includes the HF motif and interacts with the B-helix, C-helix, and β4 strand of the N lobe. (ii) At the beginning of the tail is the C-lobe tether (CLT) (residues 300–319 in PKA) that interacts with the E-helix of the C lobe and with the αC–β4 loop, β8 strand, and interlobe linker that spans the N and C lobes. (iii) In the region of the tail that joins the NLT and CLT is the active-site tether (AST) that serves as a gate for ATP and substrate binding and peptide substrate recruitment (30).
Fig. 2.
Structural location of AGC conserved features in a prototypic AGC kinase structure. (A) NLT interactions with the N lobe. (B) CLT interactions with the C lobe and interlobe region formed between the αC–β4 loop, β8 strand, and E-helix. (C) CLA interactions with F-helix. Secondary structural elements of the kinase domain and C-tail are colored gray and maroon, respectively.
In addition to the C-tail, subdomain XI of the catalytic core, to which the C-tail is attached, conserves features that distinguish AGC kinases from other EPKs (Fig. 1A). This subdomain is termed the C-lobe anchor (CLA) (residues 623–639 in PKC; residues 282–299 in PKA), because it is anchored to the F-helix of the C lobe by AGC-conserved interactions (Fig. 2C).
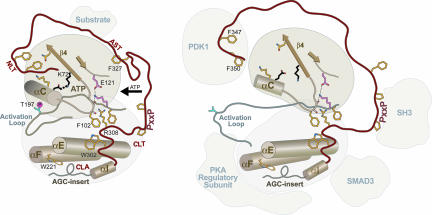
Whereas the role of the NLT interactions with the catalytic core is fairly well understood, little is known about the role of the CLT and CLA interactions. The role of AST in PKA is well understood (30), but not in other AGC kinases. The residues involved in the CLT and CLA interactions are conserved across diverse AGC families and across major eukaryotic phyla within each family. These regions presumably are not required to form the AGC structural fold, considering that the non-AGC Aurora kinase, which lacks nearly all of these AGC features, superimposes with PKA with a rmsd of ≈1 Å. The C-tail is clearly required for function, however, in that mutations in or deletion of the tail are inactivating (31). Thus one may view the C-tail either as a cis-interacting regulatory element that directly influences AGC activity or as a scaffold for transinteracting regulatory components that indirectly influence AGC activity by docking to the C-tail (Fig. 3). Because the role of the NLT and its interaction with the catalytic domain have been described (reviewed in SI Appendix 1), here we focus on AGC functional constraints imposed on the AST, CLT, and CLA regions.
Fig. 3.
A mechanistic model of a prototypical AGC kinase in the active closed state (Left) and other alternative states (Right). The C-tail is shown in red, and some of the AGC-conserved residue side chains are shown in stick representation (yellow) with their functional role. Binding of regulatory protein to the NLT, AST, or CLA may regulate activity by modulating some of the canonical AGC-conserved interactions. For example, a tryptophan at the beginning of the CLT within PKB is important for interaction with Smad3 (43) (see SI Appendix 2).
The AST Region Is an Integral Part of the ATP Binding Pocket in PKA.
The AST region, unlike the CLT and NLT regions, is disordered in most AGC kinase structures except PKA. In PKA, a conserved phenylalanine (F327PKA) within the AST region protrudes into the ATP binding pocket and mediates van der Waals interactions with the bound nucleotide (30). This phenylalanine and most of the AST become disordered in the absence of nucleotide, suggesting a role for the AST in nucleotide entry and exit (30).
Interactions Between the CLT and the αC–β4 Loop.
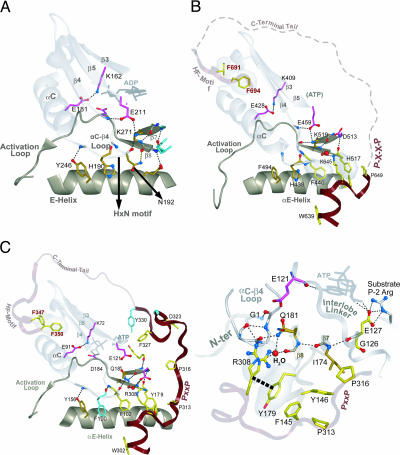
A previous analysis of EPKs (29) suggested that the αC–β4 loop plays a critical functional role by serving as a hinge point for C-helix movement (32). Functionally, it is the only secondary structure that is firmly anchored as a rigid body to the large lobe (33). Unlike distantly related eukaryotic-like kinases, EPKs characteristically harbor conserved residues within the αC–β4 loop and within the nearby β7 and β8 strands that form mutual, conserved interactions, indicating that in EPKs these regions perform a specialized function. One characteristic EPK feature within the αC–β4 loop is a conserved HxN motif (see residue patterns characteristic of other EPKs below the AGC alignment in Fig. 1A) that adopts a type I β turn and facilitates a “pseudo-β-sheeted interaction” with the β8 strand of the large lobe (29). In other words, both the backbone and the side chain of the HxN–asparagine (N192Aurora in Fig. 4 A) form a hydrogen bond to the β8 strand thereby extending a nearby β-sheet to the edge of the αC–β4 loop (Fig. 4A). The absence of the HxN motif in distantly related eukaryotic-like kinases, in which the C-helix is held in a constitutively active conformation, led to a suggested role for these EPK interactions in facilitating conformational transitions in the C-helix (29).
Fig. 4.
Comparison of interlobe interactions in non-AGC kinases (Aurora, A) and AGC kinases [PKC (B) and PKA (C)]. Main-chain traces of key regions are colored as indicated at the top of Fig. 1). Main-chain traces of phosphate moieties use the standard CPK color scheme). Oxygen, nitrogen, and hydrogen atoms establishing hydrogen bonds are red, blue, and white, respectively). Side chains of kinase-shared residues are pale magenta. Side chains of residues shared between Eukaryotic and Eukaryotic-like kinases are shown in gold (29). AGC-specific residues are in light yellow. Hydrogen bonds are depicted as dotted lines. CH–π interactions are depicted as dotted lines into dot clouds. (A) Canonical EPK interactions in the interlobe region of Aurora kinase, which does not belong to the AGC group. (B and C) Conserved interactions between the CLT and the interlobe region of PKC (B) and PKA (C) that belong to the AGC group. A close-in view of PKA interactions are shown in C Right. The role of only one of the conserved residues in the AST is known to play a role in PKA function, a phenylalanine (F327PKA), that acts as a gate for and assists in ATP binding (31). Much of the PKC AST region is disordered in the crystal structure; this region is represented by a dotted line. Water molecule is shown in red spheres representation (B and C) and labeled H2O in C.
The AGC kinases fail to conserve some of the canonical EPK residues in these regions, however, and, instead, conserve other residues that interact with each other and the CLT region of the C-tail. The HxN–asparagine within the αC–β4 loop, for instance, is replaced by a conserved phenylalanine (F440PKC in Figs. 1A and 4B; F102 PKA in Figs. 1A and 4C) that forms alternative interactions with a conserved basic residue within the CLT (K645PKC in Fig. 4 B; R308PKA in Fig. 4 C) and with a conserved buried water molecule (Fig. 4 B and C). This water molecule forms hydrogen bonds with both this CLT basic residue and the same carbonyl backbone oxygen of the β8 strand that forms a hydrogen bond with the HxN–asparagine in non-AGC kinases (Fig. 4A). The side chain of this CLT basic residue also mediates a CH–π interaction with a canonical AGC histidine within the β8 strand (H517 in Fig. 4 B; Y179PKA in Fig. 4 C). Taken together, these interactions appear to closely integrate the CLT with the αC–β4 loop, perhaps to coordinate C-helix movement with NLT movement during AGC kinase activation.
The CLT and αC–β4 loop are also coupled to the ATP binding site by a canonical EPK salt bridge that was proposed to facilitate interlobe movement (32, 34). Specifically, the conserved water molecule, which hydrogen-bonds to the CLT basic residue, also hydrogen-bonds to the side chain of a conserved lysine (K519PKC in Fig. 4 B; Q181PKA in Fig. 4 C) in the β8 strand. Within EPKs in general, this side chain forms a salt bridge with a conserved glutamate (E459PKC in Fig. 4 B; E121PKA in Fig. 4 C) at the beginning of the interlobe linker region, the backbone of which hydrogen-bonds to ATP (Fig. 4C). This salt bridge may couple ATP binding with the αC–β4 loop and thereby help coordinate C-helix movement with ATP binding and release (29). The interaction of the CLT with both the αC–β4 loop and the interlobe salt bridge in AGC kinases may allow conformational changes in the C-tail to modulate ATP binding and C-helix movement.
The AGC kinase GRK2 lacks some of the canonical interactions in the CLT–interlobe interface and thus appears to use an alternative mechanism that could involve a regulatory RGS homology domain, within which the GRK2 kinase domain and C-tail are inserted (35) (see SI Appendix 2).
The CLT PxxP Motif Interacts with the Interlobe Linker.
The CLT region contains a conserved PxxP motif (Fig. 1B) that, within PKB, was proposed to interact with the Src homology 3 (SH3) domain of Src kinase (36). The PxxP motif is present in nearly all AGC kinases, except for PDK1, NDRK, and a few AGC kinases from lower eukaryotes (Fig. 1B). This motif both interacts with the linker that joins the N and C lobes and docks to a hydrophobic groove created between the E-helix and the β7 and β8 strands, three structural elements also located at the interface between the N and C lobes (Fig. 2B). This region of the catalytic domain serves as an allosteric site in the MAP kinase Fus3. Binding of a peptide from the scaffold protein Ste5 to this region activates Fus3 by modulating the relative orientation of the N and C lobes, thereby inducing conformational changes in the distal activation loop (37, 38).
Interaction of the PxxP motif with the interlobe linker is critical for interlobe movement.
The PxxP motif is a potential site for allosteric regulation in AGC kinases inasmuch as it forms conserved interactions with the E-helix and the β7 and β8 strands (Fig. 4C), all of which, together with the D-helix, form a region of the catalytic domain that is critical for interlobe movement and, consequently, for ATP and substrate binding. The first PxxP proline (P650PKC, disordered in Fig. 4 B; P313PKA in Fig. 4 C Right) packs up against an AGC conserved phenylalanine (F145PKA in Fig. 4 C) that is within the E-helix and, in turn, interacts with an AGC-conserved histidine or tyrosine within the β8 strand (Y179PKA in Fig. 4 C). The second PxxP proline (P316PKA in Fig. 4 C Right) typically packs up against a conserved hydrophobic residue in the β7 strand (I174PKA in Fig. 4 C Right). The backbone of this hydrophobic residue hydrogen bonds to a glycine (G126PKA in Fig. 4 C) that is conserved within the interlobe linker (Fig. 3 C and D) and undergoes a backbone torsion angle change upon interlobe movement (39). This glycine is generally conserved in all EPKs, but within the non-AGC kinase GCN2 an arginine occurs at this position and mediates an autoinhibitory conformation, preventing interlobe movement (40). However, mutation of this arginine to glycine restores conformational flexibility in the linker and movement of the N and C lobes relative to each other, resulting in GCN2 activation (40). Thus within AGC kinases, the interaction between the PxxP motif and this glycine (G126PKA) likewise might modulate interlobe movement, perhaps upon binding to an SH3 domain, which recognize proline-rich regions (36).
For SH3 binding, however, the prolines need to be oriented in a polyproline type II conformation (41, 42) and the proline N-substituted backbone amides need to be solvent exposed, neither of which is the case for the PxxP motif in available AGC kinase structures. Thus, upon SH3 binding (as for PKB) (36), the PxxP motif presumably undergoes a conformational change (Fig. 3), a plausible event given the high-temperature factor of the PxxP motif in crystal structures and the dynamic nature of the C-tail (30). This finding suggests an allosteric mechanism for coupling SH3 binding to catalytic function.
The PxxP motif interaction with the interlobe linker might also modulate substrate binding inasmuch as adjacent to the AGC-conserved glycine (G126PKA in Fig. 4 C Right) in the sequence is a conserved glutamate or aspartate (E127PKA in Fig. 4 C Right) that, in PKA, hydrogen-bonds to a peptide substrate (Fig. 4C Right).
Smad3 interacts with a conserved tryptophan in the CLT region of Akt.
At the beginning of the CLT is a tryptophan (W639PKC in Fig. 4 B; W302PKA in Fig. 4 C), which in PKB (W414PKB in SI Appendix 2) is important for interaction with Smad3 (43). This tryptophan, like the PxxP motif, is also not solvent-exposed for interaction with Smad3, but instead is buried in the hydrophobic interface formed between the CLT and interlobe regions (Fig. 4 B and C).
CLA Is Coupled to the Substrate-Binding Regions of the Core.
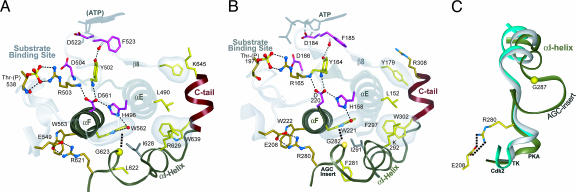
Subdomain XI of the kinase core, referred to here as the CLA, serves as an anchor to the rest of the core. It is anchored primarily through hydrophobic contacts with the exception of a single conserved electrostatic contact between a conserved arginine (R621PKC in Fig. 5 A; R280PKA in Fig. 5 B) that interacts with the glutamate of the APE motif at the end of the activation loop. This subdomain, which is characteristically missing in EPK-like kinases, serves to mediate the complex allosteric regulation that is a unique feature of the EPKs. In AGC kinases subdomain XI is attached to the C-tail and contains conserved features and interactions that distinguish AGC kinases from other EPKs (Fig. 1A). One of the distinguishing features of AGC kinases in this region is a short insert segment (AGC insert) connecting the H- and I-helices (Fig. 5C).
Fig. 5.
Coupling of CLA to substrate binding regions. (A and B) Conserved interactions coupling the CLA region with ATP- and substrate-binding regions of the catalytic core in PKC (A) and PKA (B). (C) Shown is the superposition of the αH-αI region in PKA (dark gray), Cdk2 (white), and insulin receptor tyrosine kinase (turquoise). The C-tail, which emanates from the CLA region (subdomain XI), is shown in red. The AGC insert is absent in some AGC kinases, including PKC-theta and therefore is not labeled in A.
One of the most distinguishing features of AGC kinases is a conserved tryptophan, termed the AGC tryptophan (W562PKC in Fig. 5 A; W221 in Fig. 5 B), which is located in the signal integrating F-helix (29, 44) and mediates specific interactions with AGC-conserved residues in the CLA. The AGC tryptophan forms a CH–π interaction with the Cα of the conserved glycine (G623PKC in Fig. 5 A; G282PKA in Fig. 5 B), which is located at the beginning of a short insert segment (AGC insert in Fig. 1A) in the CLA region. This insert was recently found to interact with the regulatory subunit (B domain) in PKA (unpublished work). Thus, this positioning of the insert by the AGC tryptophan may be functionally significant in that it may facilitate an allosteric coupling between the CLA and substrate binding regions of the catalytic core.
Both the AGC insert and the AGC tryptophan are strategically located relative to key kinase-conserved residues that are implicated in coupling distal ATP and substrate-binding regions. Specifically, a conserved arginine (R621PKC in Fig. 5 A; R280PKA in Fig. 5 B), which recently was proposed to be important for substrate binding and release (45), is two residues N-terminal of the AGC insert and forms a salt-bridge interaction with the APE-glutamate (E549PKC in Fig. 5 A; E208PKA in Fig. 5 B) that follows the P+1 loop in the substrate-binding region. The proline within the APE motif forms a CH–π interaction with a kinase-conserved tryptophan (W562PKC in Fig. 5 A; W221PKA in Fig. 5 B), which is sequence adjacent to the AGC trytophan in the F-helix. Also, the indole group of the AGC-tryptophan hydrogen-bonds (via a water molecule) to a kinase-conserved histidine (H496PKC in Fig. 5 A; H158PKA in Fig. 5 B) in the E-helix that is shared by both EPKs and distantly related EPK-like kinases and that is part of a hydrogen-bonding network that couples distal ATP and substrate-binding regions (29). The strategic location of the AGC tryptophan relative to this network suggests a role for this tryptophan in coupling CLA-associated regulatory functions with catalytic functions of the core. Whether or not this coupling is important for C-tail functions is an open question, but considering that these residues are located near the base of the C-tail suggests that conformational changes associated with the tail might be propagated to the substrate-binding regions via these interactions.
Concluding Remarks
The hinge-like opening and closing of the N-lobe/C-lobe interface upon ATP binding or release and the repositioning of the C-helix are two critical events in the protein kinase catalytic cycle. AGC kinases appear to modulate these events through interaction of the C-helix, the interlobe linker, and the αC–β4 loop with three regions of the C-tail: the NLT, AST, and CLT. The residues within the catalytic domain that mediate these interactions both are highly conserved within AGC kinases from diverse organisms and some of these diverge from the canonical EPK residues conserved at corresponding positions within non-AGC kinases. This finding suggests that AGC kinases lack residue interactions that are functionally critical for non-AGC protein kinases and, instead, conserve alternative interactions that are functionally critical for AGC kinases. Hence the apparent functional deficiency of these divergent AGC residues seems to be repaired by their interaction with the C-tail, which thus compensates for these deficiencies. This arrangement also provides a cis-acting regulatory handle upon which other cellular components may act to modulate AGC kinase activity (Fig. 3). Known sites of interaction include the HF motif within the NLT, which binds to PDK1 (46), and the CLT PxxP motif, which acts as an SH3 binding site in PKB (36).
Notably, PDK1 lacks key regions of the C-tail and thus appears to be doubly deficient, presumably because of the requirement for PDK1 activation of a transinteracting C-tail donated by another AGC kinase. The ubiquitous conservation of the C-tail across diverse AGC families from diverse organisms suggests that there is something fundamental about this mode of regulation that goes beyond particular signaling pathways. What might this be?
Because many AGC kinases are involved in transmembrane signaling, AGC features may play a fundamental role in membrane localization or some other requirement generally associated with transmembrane signaling. Thus another potential regulatory component is the plasma membrane itself, association with which might also influence the C-tail and thus catalytic activity in some way. The interface of the CLT and interlobe region conserves a preponderance of basic and aromatic residues, which are prevalent in proteins and peptides that bind to membranes (47, 48). Moreover, a myristyl group that anchors PKA to the membrane docks to a hydrophobic groove formed between the CLT and the interlobe region (49). Notably, in one high-resolution structure of PKA (50), the CLT basic residue (R308PKA in Fig. 4 C Inset) exists in two different conformations: one where it packs up against the AGC basic residues in the αC–β4 loop and another where it hydrogen-bonds to the N-terminal glycine that gets myristylated.
The three regions of the C-tail (the NLT, AST, and CLT) and the CLA may function synergistically as the AGC kinases switches between an active and inactive state, or, given that each of these functional segments can vary independently, they may come into play at different stages of AGC functions, thereby providing multiple layers of control.
Finally, the concept of cis and trans regulation of AGC kinase activity introduced in this study can also be helpful in understanding the regulatory mechanisms of other protein kinase families in that we can begin to classify protein kinase regulation based on whether it is cis, trans, or both. For instance, the mechanism of Cdk2 activation by cyclin can be classified as trans, whereas the regulation of Src kinase by SH2 and SH3 domains can be classified as cis. Understanding how these cis and trans regulatory elements coevolve with the catalytic core will be essential to fully appreciate the mechanisms of the kinase machinery.
Materials and Methods
Contrast hierarchical alignment and interaction network (CHAIN) analysis of AGC kinases was performed as described (27, 28) (for a review see ref. 28). Bayesian statistical models and Markov chain Monte Carlo procedures were used to identify and quantify the degree of selective pressure exerted on residues characteristic of various categories of proteins, in this case EPK and AGC kinases. The selective constraints imposed on AGC kinases are shown in Fig. 1. A detailed description of CHAIN analysis is given in SI Appendix 3.
Supplementary Material
Acknowledgments
We thank members of S.S.T.'s laboratory and Alexandra Newton's laboratory (University of California at San Diego) for helpful discussions. This work was funded by National Institutes of Health Grant IP01DK54441 (to S.S.T.), National Library of Medicine Grant LM06747 (to A.F.N.), and National Institutes of Health Division of General Medicine Grant GM078541 (to A.F.N.).
Abbreviations
- EPK
eukaryotic protein kinase
- C-tail
C-terminal tail
- NLT
N-lobe tether
- AST
active-site tether
- CLT
C-lobe tether
- HF motif
hydrophobic motif
- CLA
C-lobe anchor
- SH3
Src homology 3.
Footnotes
Author contributions: N.K. designed research; N.K. and N.H. performed research; A.F.N. contributed new reagents/analytic tools; N.K. analyzed data; and N.K., S.S.T., and A.F.N. wrote the paper.
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610251104/DC1.
References
- 1.Hanks SK, Hunter T. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- 2.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 3.Johnson LN, Noble ME, Owen DJ. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 4.Huse M, Kuriyan J. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 5.Nolen B, Taylor S, Ghosh G. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 6.Alessi DR, James SR, Downes CP, Holmes AB, Gaffney PR, Reese CB, Cohen P. Curr Biol. 1997;7:261–269. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 7.Pullen N, Dennis PB, Andjelkovic M, Dufner A, Kozma SC, Hemmings BA, Thomas G. Science. 1998;279:707–710. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- 8.Dutil EM, Toker A, Newton AC. Curr Biol. 1998;8:1366–1375. doi: 10.1016/s0960-9822(98)00017-7. [DOI] [PubMed] [Google Scholar]
- 9.Cheng X, Ma Y, Moore M, Hemmings BA, Taylor SS. Proc Natl Acad Sci USA. 1998;95:9849–9854. doi: 10.1073/pnas.95.17.9849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Belham C, Wu S, Avruch J. Curr Biol. 1999;9:R93–R96. doi: 10.1016/s0960-9822(99)80058-x. [DOI] [PubMed] [Google Scholar]
- 11.Peterson RT, Schreiber SL. Curr Biol. 1999;9:R521–R524. doi: 10.1016/s0960-9822(99)80326-1. [DOI] [PubMed] [Google Scholar]
- 12.Currie RA, Walker KS, Gray A, Deak M, Casamayor A, Downes CP, Cohen P, Alessi DR, Lucocq J. Biochem J. 1999;337:575–583. [PMC free article] [PubMed] [Google Scholar]
- 13.Milburn CC, Deak M, Kelly SM, Price NC, Alessi DR, Van Aalten DM. Biochem J. 2003;375:531–538. doi: 10.1042/BJ20031229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thomas CC, Deak M, Alessi DR, van Aalten DM. Curr Biol. 2002;12:1256–1262. doi: 10.1016/s0960-9822(02)00972-7. [DOI] [PubMed] [Google Scholar]
- 15.Nalefski EA, Newton AC. Biochemistry. 2001;40:13216–13229. doi: 10.1021/bi010761u. [DOI] [PubMed] [Google Scholar]
- 16.Fraser ID, Tavalin SJ, Lester LB, Langeberg LK, Westphal AM, Dean RA, Marrion NV, Scott JD. EMBO J. 1998;17:2261–2272. doi: 10.1093/emboj/17.8.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Struppe J, Komives EA, Taylor SS, Vold RR. Biochemistry. 1998;37:15523–15527. doi: 10.1021/bi981326b. [DOI] [PubMed] [Google Scholar]
- 18.Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massague J, Pavletich NP. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 19.Yang J, Cron P, Thompson V, Good VM, Hess D, Hemmings BA, Barford D. Mol Cell. 2002;9:1227–1240. doi: 10.1016/s1097-2765(02)00550-6. [DOI] [PubMed] [Google Scholar]
- 20.Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 21.Balendran A, Biondi RM, Cheung PC, Casamayor A, Deak M, Alessi DR. J Biol Chem. 2000;275:20806–20813. doi: 10.1074/jbc.M000421200. [DOI] [PubMed] [Google Scholar]
- 22.Biondi RM, Cheung PC, Casamayor A, Deak M, Currie RA, Alessi DR. EMBO J. 2000;19:979–988. doi: 10.1093/emboj/19.5.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao T, Toker A, Newton AC. J Biol Chem. 2001;276:19588–19596. doi: 10.1074/jbc.M101357200. [DOI] [PubMed] [Google Scholar]
- 24.Hresko RC, Mueckler M. J Biol Chem. 2005;280:40406–40416. doi: 10.1074/jbc.M508361200. [DOI] [PubMed] [Google Scholar]
- 25.Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Science. 2005;307:1098–1101. doi: 10.1126/science.1106148. [DOI] [PubMed] [Google Scholar]
- 26.Frodin M, Jensen CJ, Merienne K, Gammeltoft S. EMBO J. 2000;19:2924–2934. doi: 10.1093/emboj/19.12.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neuwald AF, Kannan N, Poleksic A, Hata N, Liu JS. Genome Res. 2003;13:673–692. doi: 10.1101/gr.862303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neuwald AF. Trends Biochem Sci. 2006;31:374–382. doi: 10.1016/j.tibs.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 29.Kannan N, Neuwald AF. J Mol Biol. 2005;351:956–972. doi: 10.1016/j.jmb.2005.06.057. [DOI] [PubMed] [Google Scholar]
- 30.Narayana N, Cox S, Nguyen-huu X, Ten Eyck LF, Taylor SS. Structure (London) 1997;5:921–935. doi: 10.1016/s0969-2126(97)00246-3. [DOI] [PubMed] [Google Scholar]
- 31.Chestukhin A, Litovchick L, Schourov D, Cox S, Taylor SS, Shaltiel S. J Biol Chem. 1996;271:10175–10182. doi: 10.1074/jbc.271.17.10175. [DOI] [PubMed] [Google Scholar]
- 32.Lamers MB, Antson AA, Hubbard RE, Scott RK, Williams DH. J Mol Biol. 1999;285:713–725. doi: 10.1006/jmbi.1998.2369. [DOI] [PubMed] [Google Scholar]
- 33.Tsigelny I, Grant BD, Taylor SS, Ten Eyck LF. Biopolymers. 1996;39:353–365. doi: 10.1002/(SICI)1097-0282(199609)39:3%3C353::AID-BIP7%3E3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 34.Hayward S. J Mol Biol. 2004;339:1001–1021. doi: 10.1016/j.jmb.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Lodowski DT, Pitcher JA, Capel WD, Lefkowitz RJ, Tesmer JJ. Science. 2003;300:1256–1262. doi: 10.1126/science.1082348. [DOI] [PubMed] [Google Scholar]
- 36.Jiang T, Qiu Y. J Biol Chem. 2003;278:15789–15793. doi: 10.1074/jbc.M212525200. [DOI] [PubMed] [Google Scholar]
- 37.Bhattacharyya RP, Remenyi A, Good MC, Bashor CJ, Falick AM, Lim WA. Science. 2006;311:822–826. doi: 10.1126/science.1120941. [DOI] [PubMed] [Google Scholar]
- 38.Zhou T, Sun L, Humphreys J, Goldsmith EJ. Structure (London) 2006;14:1011–1019. doi: 10.1016/j.str.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 39.Nolen B, Ngo J, Chakrabarti S, Vu D, Adams JA, Ghosh G. Biochemistry. 2003;42:9575–9585. doi: 10.1021/bi0344331. [DOI] [PubMed] [Google Scholar]
- 40.Padyana AK, Qiu H, Roll-Mecak A, Hinnebusch AG, Burley SK. J Biol Chem. 2005;280:29289–29299. doi: 10.1074/jbc.M504096200. [DOI] [PubMed] [Google Scholar]
- 41.Nguyen JT, Turck CW, Cohen FE, Zuckermann RN, Lim WA. Science. 1998;282:2088–2092. doi: 10.1126/science.282.5396.2088. [DOI] [PubMed] [Google Scholar]
- 42.Lim WA, Richards FM, Fox RO. Nature. 1994;372:375–379. doi: 10.1038/372375a0. [DOI] [PubMed] [Google Scholar]
- 43.Conery AR, Cao Y, Thompson EA, Townsend CM, Jr, Ko TC, Luo K. Nat Cell Biol. 2004;6:366–372. doi: 10.1038/ncb1117. [DOI] [PubMed] [Google Scholar]
- 44.Akamine P, Madhusudan, Wu J, Xuong NH, Ten Eyck LF, Taylor SS. J Mol Biol. 2003;327:159–171. doi: 10.1016/s0022-2836(02)01446-8. [DOI] [PubMed] [Google Scholar]
- 45.Deminoff SJ, Howard SC, Hester A, Warner S, Herman PK. Genetics. 2006;173:1909–1917. doi: 10.1534/genetics.106.059238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balendran A, Casamayor A, Deak M, Paterson A, Gaffney P, Currie R, Downes CP, Alessi DR. Curr Biol. 1999;9:393–404. doi: 10.1016/s0960-9822(99)80186-9. [DOI] [PubMed] [Google Scholar]
- 47.Zhang W, Crocker E, McLaughlin S, Smith SO. J Biol Chem. 2003;278:21459–21466. doi: 10.1074/jbc.M301652200. [DOI] [PubMed] [Google Scholar]
- 48.Strandberg E, Morein S, Rijkers DT, Liskamp RM, van der Wel PC, Killian JA. Biochemistry. 2002;41:7190–7198. doi: 10.1021/bi012047i. [DOI] [PubMed] [Google Scholar]
- 49.Zheng J, Knighton DR, Xuong NH, Taylor SS, Sowadski JM, Ten Eyck LF. Protein Sci. 1993;2:1559–1573. doi: 10.1002/pro.5560021003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang J, Ten Eyck LF, Xuong NH, Taylor SS. J Mol Biol. 2004;336:473–487. doi: 10.1016/j.jmb.2003.11.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.