Abstract
Successful infection by fungal pathogens depends on subversion of host immune mechanisms that detect conserved cell wall components such as β-glucans. A less common polysaccharide, α-(1,3)-glucan, is a cell wall constituent of most fungal respiratory pathogens and has been correlated with pathogenicity or linked directly to virulence. However, the precise mechanism by which α-(1,3)-glucan promotes fungal virulence is unknown. Here, we show that α-(1,3)-glucan is present in the outermost layer of the Histoplasma capsulatum yeast cell wall and contributes to pathogenesis by concealing immunostimulatory β-glucans from detection by host phagocytic cells. Production of proinflammatory TNFα by phagocytes was suppressed either by the presence of the α-(1,3)-glucan layer on yeast cells or by RNA interference based depletion of the host β-glucan receptor dectin-1. Thus, we have functionally defined key molecular components influencing the initial host–pathogen interaction in histoplasmosis and have revealed an important mechanism by which H. capsulatum thwarts the host immune system. Furthermore, we propose that the degree of this evasion contributes to the difference in pathogenic potential between dimorphic fungal pathogens and opportunistic fungi.
Keywords: cell wall, fungal pathogenesis, virulence factor, dectin-1, macrophage
The mammalian innate immune system detects invading microbial pathogens in part by recognition of pathogen-associated molecular patterns (PAMPs) and utilizes this information to tailor an appropriate immune response. The corresponding host molecules used, termed pattern-recognition receptors (PRRs), detect molecules that characterize broad classes of microbes. Many of these PRRs are highly expressed on front-line immune cells, particularly macrophages and dendritic cells. For fungi, the polysaccharide-rich cell wall is a major source of PAMPs, and it comprises the initial structure sampled by cells of the immune system. Identified PRRs important for the detection of fungal surface components include the Toll-like receptors TLR2 and TLR4, the collectins SP-A and SP-D, pentraxin-3, the CR3 integrin, and C-type lectins, all of which appear to detect fungal-associated carbohydrates (1). Recognition of fungal-surface polysaccharides initiates immediate responses such as phagocytosis, production of antimicrobial compounds, and induction of proinflammatory cytokines that activate and recruit other immune effector cells. Recent work on the nonclassical C-type lectin, dectin-1, has defined its substrate to be oligomers of β-(1,3)-glucan (2), a constituent of the cell wall of all fungi and a potent immunostimulatory molecule that induces TNFα production by macrophages.
Successful pathogens must therefore have mechanisms to avoid or counteract detection by host PRRs. Such mechanisms are likely to distinguish primary pathogens that cause disease in normal hosts from opportunistic pathogens that require suppression of the host immune system. The primary pathogenic fungi include Histoplasma capsulatum, Paracoccidioides brasiliensis, Blastomyces dermatitidis, Coccidioides spp., and Cryptococcus neoformans. With the exception of Cryptococcus, each of these species is “dimorphic,” growing as a saprophytic mold form at ambient temperatures (i.e., the soil environment) and a parasitic yeast (or spherule) form at mammalian body temperatures. Mold-produced conidia (or spores) that are inhaled into the lung germinate into yeast, and this conversion is absolutely required for pathogenicity (3, 4), suggesting that yeast phase-specific characteristics include determinants important for virulence.
One such yeast phase-specific component is α-(1,3)-glucan, a homopolymer of glucose with α-glycosidic linkages, which has been linked to fungal virulence. The cell walls of most medically important fungi contain α-(1,3)-glucan, and reduction in α-(1,3)-glucan, through either laboratory passage of Histoplasma, Blastomyces, and Paracoccidioides (5–7) or genetic loss of α-(1,3)-glucan synthase (AGS1) in Histoplasma (8), has no effect on in vitro growth but severely attenuates virulence in murine respiratory infection models. However, the mechanism by which α-(1,3)-glucan facilitates the pathogenesis of dimorphic fungi has not been determined. We present evidence showing that Histoplasma cell wall α-(1,3)-glucan blocks host PRR recognition of the fungal PAMP β-glucan, enabling Histoplasma yeast to avoid detection as a fungal invader.
Results
Histoplasma α-(1,3)-Glucan Production Alters the Exposed Yeast Cell Surface.
As one of the first fungal structures sampled by the host immune surveillance system, the Histoplasma yeast cell wall is likely organized to influence the initial host–pathogen interaction. Respiratory infection by Histoplasma occurs by inhalation of mold-produced conidia that germinate into the parasitic yeast form on exposure to mammalian body temperatures. Although mold-form Histoplasma entirely lacks α-(1,3)-glucan, germination of Histoplasma conidia at 37°C into yeast cells initiates production of this polysaccharide, which we detected only on the emergent yeast cell wall and not the conidial structure (Fig. 1A–C). This finding suggests that the immediate synthesis of α-(1,3)-glucan is required specifically for pulmonary infection and yeast survival.
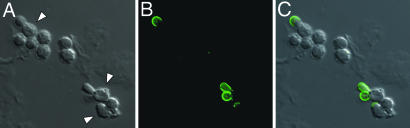
Fig. 1.
α-(1,3)-Glucan is produced during the transition from conidia to yeast. Histoplasma conidia were incubated for 18 h at 37°C in F-12 medium containing 10% FBS and visualized by direct interference contrast microscopy (DIC) (A) and immunofluorescence microscopy (B) after staining with an antibody specific for α-(1,3)-glucan. Arrowheads indicate germinating conidia with emergent yeast buds that have a smoother cell surface. (C) Merged DIC and immunofluorescence images show that α-(1,3)-glucan is present only around the yeast buds and not the conidial structure.
The cell walls of wild-type Histoplasma yeast contain primarily three polysaccharides: chitin, β-glucans [β-(1,3)- and β-(1, 6)-linked], and α-(1,3)-glucan (9–12). Immunofluorescence localization in cross-sections of wild-type yeast cells with α-(1,3)- and β-(1,3)-glucan-specific antibodies showed a nonhomogeneous spatial distribution (Fig. 2A–C). Although some overlap exists between α-(1,3)-glucan and β-(1,3)-glucan, merging the images suggests that the cell wall is somewhat layered, with α-(1,3)-glucan being more external (Fig. 2 C and D). This layered organization supports earlier biochemical analyses that used serial α-glucanase and β-glucanase treatments to quantify cell wall polysaccharides as well as ultrastructural studies in which alkali treatment of isolated cell walls removed an outer fibrillar pattern (9, 11). To confirm this spatial organization, α-(1,3)-glucan and β-(1,3)-glucan were localized by immunoelectron microscopy of Histoplasma yeast (Fig. 2E). Quantitation of the gold particle labeling corroborates the immunofluorescence localization patterns, with ≈50% of α-(1,3)-glucan being located more distal to the yeast cell membrane than β-(1,3)-glucan (Fig. 2 F and G). This organization suggests that the α-(1,3)-glucan layer may conceal the underlying β-glucans from detection by the host.
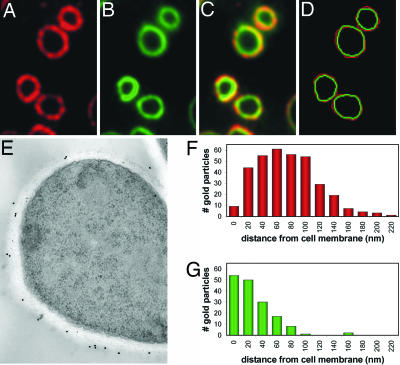
Fig. 2.
α-(1,3)-Glucan comprises the outer cell wall layer of yeast. Wild-type Histoplasma yeasts were fixed, and antibodies specific for α-(1,3)-glucan (A) and β-(1,3)-glucan (B) were used to localize the respective polysaccharides. (C) Merged α-(1,3)-glucan and β-(1,3)-glucan localizations show a somewhat layered spatial organization. (D) Tracings of the α-(1,3)- and β-(1,3)-glucan outer perimeters (red and green, respectively) were overlaid, confirming that α-(1,3)-glucan forms the outermost surface. (E) Representative immunoelectron micrograph (n = 25) in which α-(1,3)-glucans and β-glucans were labeled in wild-type yeast cells with 18-nm and 12-nm gold particles, respectively. (F and G) Histograms showing the distribution of individual α-(1,3)-glucan (F; n = 342) and β-(1,3)-glucan (G; n = 162) labeling in electron micrographs, calculated as the perpendicular distance from the cell membrane.
α-(1,3)-Glucan Prevents Recognition of Histoplasma Yeast by dectin-1.
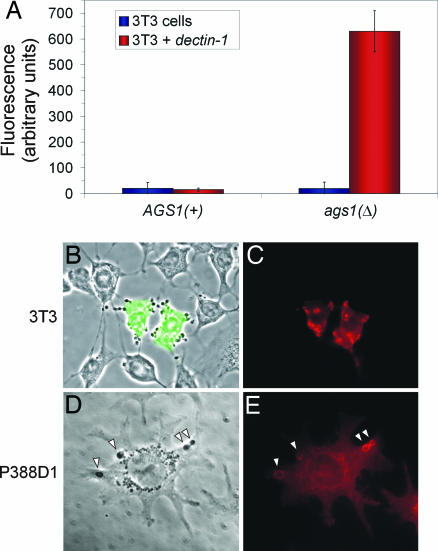
Because dectin-1 functions as the primary β-glucan receptor on host cells, we tested whether α-(1,3)-glucan could block dectin-1 recognition of Histoplasma cell wall β-glucans. Histoplasma is an intracellular pathogen of macrophages and potentially interacts with a number of components on the phagocyte surface, some of which are used by phagocytes in sampling foreign particles. Thus, to test dectin-1 recognition of Histoplasma yeast, we examined binding of Histoplasma cells to 3T3 fibroblasts expressing a murine dectin-1 transgene (3T3 cells do not normally express endogenous dectin-1). Histoplasma yeasts that lack α-(1,3)-glucan [ags1(Δ)] readily bound to 3T3 cells expressing dectin-1, whereas yeast cells containing α-(1,3)-glucan [AGS1(+)] were not recognized (Fig. 3A). This recognition was completely dependent on dectin-1 because 3T3 cells without the dectin-1 transgene failed to bind either Histoplasma strain. Furthermore, dectin-1 localized to sites of contact with ags1(Δ) yeast cells, as seen in 3T3 cells transiently transfected with a dectin-1 transgene (Fig. 3 B and C). When Histoplasma was cocultured with P388D1 murine macrophage-like cells, dectin-1 localization was enriched on phagolysosomal compartments containing ags1(Δ) yeasts (Fig. 3 D and E). Thus, murine dectin-1 can recognize Histoplasma yeasts, but only in the absence of the α-(1,3)-glucan cell wall layer.
Fig. 3.
The α-(1,3)-glucan layer blocks recognition of Histoplasma yeast by dectin-1. (A) Live Histoplasma yeasts lacking α-(1,3)-glucan [ags1(Δ)] readily bind to dectin-1-expressing 3T3 cells, whereas yeasts possessing α-(1,3)-glucan [AGS1(+)] do not bind. Binding was measured by coincubation of Histoplasma yeast with confluent monolayers of 3T3 cells or dectin-1-expressing 3T3 cells followed by quantitation of bound yeasts with the fungal fluorescent stain Uvitex 3BSA. (B and C) Binding of ags1(Δ) yeast cells depends on dectin-1 expression. (B) Merged phase-contrast and GFP fluorescence images show that only 3T3 cells expressing dectin-1 (marked by cotransfection of a gfp expression plasmid) bind ags1(Δ) yeast. (C) Immunofluorescence labeling of cells with an antibody to dectin-1 shows enrichment of dectin-1 protein at sites of contact with Histoplasma ags1(Δ) yeast. (D and E) P388D1 macrophage-like cells were incubated with ags1(Δ) yeast (arrowheads) for 30 min, and dectin-1 localization was determined. Phase-contrast image (D) and immunofluorescent labeling with an antibody to dectin-1 (E) show enrichment of dectin-1 around Histoplasma-containing phagosomal compartments.
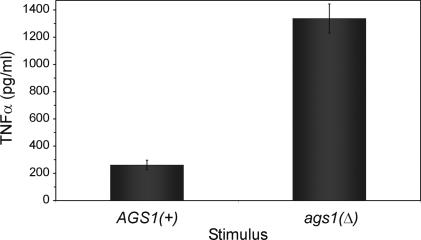
To determine whether the masking of Histoplasma β-(1,3)-glucan by α-(1,3)-glucan has functional significance, we examined whether α-(1,3)-glucan can prevent production of the proinflammatory cytokine TNFα by mammalian phagocytic cells. Alveolar macrophages are the principal innate immune cells that encounter Histoplasma yeasts; however, unactivated macrophages are unable to prevent the intracellular proliferation of Histoplasma unless activated by cytokines (13, 14). TNFα has been shown to be a critical component of the host response to Histoplasma because loss of TNFα or TNFα receptors 1 or 2 exacerbates experimental histoplasmosis (15–17). When we infected P388D1 macrophage-like cells with live Histoplasma yeasts, TNFα was released; however, ags1(Δ) yeasts stimulated 5-fold greater amounts of TNFα as isogenic yeasts possessing the α-(1,3)-glucan outer layer [AGS1(+); Fig. 4].
Fig. 4.
α-(1,3)-Glucan reduces TNFα production by phagocytic cells in response to Histoplasma yeasts. ELISA-based quantitation of TNFα produced by P388D1 phagocytic cells in response to 3-h infection by live Histoplasma yeast shows increased TNFα stimulation by yeasts that lack the α-(1,3)-glucan polysaccharide compared with yeasts surrounded by α-(1,3)-glucan. All results represent data obtained from triplicate infections, with error bars corresponding to the SD.
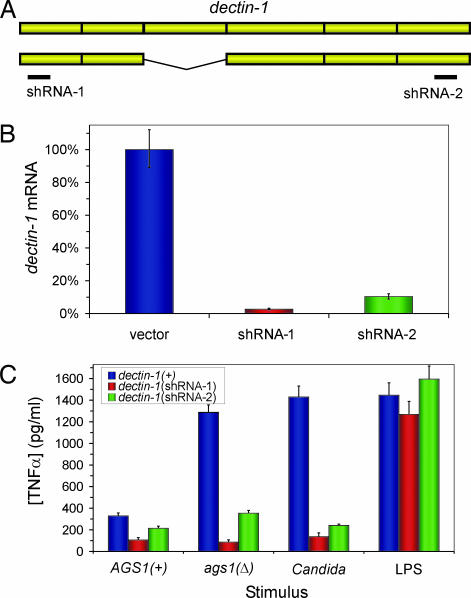
To assess whether dectin-1 was the primary PRR mediating the stimulation of TNFα production by exposed Histoplasma β-glucans, we depleted the dectin-1 receptor from phagocytes. Because no dectin-1 knockout mouse was available, we used RNAi to generate stably transfected phagocytic cell lines with greatly reduced dectin-1 function. Because two primary isoforms of dectin-1 are produced by murine monocytic lineage cells (18, 19), short-hairpin RNAs (shRNAs) were designed that simultaneously targeted both isoforms (Fig. 5A). Expression of either of the two shRNAs from the human U6 promoter in P388D1 cells depleted dectin-1 to <10% of vector-transformed cells as determined by dectin-1 immunofluorescence (data not shown) and by quantitative RT-PCR (Fig. 5B). RNAi-based depletion of host cell dectin-1 abrogated TNFα production normally stimulated by ags1(Δ) yeasts (Fig. 5C). In fact, TNFα production was reduced to similar levels either by deficiency of the host PRR dectin-1 or by the presence of α-(1,3)-glucan around Histoplasma yeast. Reduction in murine dectin-1 also decreased the production of TNFα in response to live Candida albicans yeasts, providing genetic evidence of the contribution of dectin-1 to host detection of this fungal pathogen as well (Fig. 5C). The loss of fungal-stimulated TNFα production was specific because identical phenotypes were observed with the two independent dectin-1 (RNAi) lines that target nonoverlapping regions of the dectin-1 transcript. Furthermore, TNFα production was specifically correlated with dectin-1 because RNAi of dectin-1 did not impair the stimulation of TNFα by LPS, which is detected through the unrelated TLR4 PRR (Fig. 5C).
Fig. 5.
dectin-1 mediates the functional response to Histoplasma yeasts that lack α-(1,3)-glucan. (A) Schematic representation depicts the two major splice isoforms of dectin-1 and the location targeted for RNAi by two dectin-1 shRNAs. (B) Depletion of dectin-1 was confirmed by quantitative RT-PCR of dectin-1 mRNA in P388D1 clonal lines harboring an empty RNAi vector or shRNAs targeting dectin-1. (C) TNFα produced by P388D1 lines in response to live Histoplasma yeasts with and without the α-(1,3)-glucan cell wall layer [AGS1(+) and ags1(Δ), respectively], live C. albicans cap1/cap1 yeast (the cap1 mutation impairs filamentous growth of Candida), or 39 endotoxin units/ml of endotoxin (LPS) was quantified by ELISA. The dectin-1(+) cell line used was a stable transfectant line expressing a gfp-targeting shRNA. Results represent data obtained from three infections, and error bars indicate SD.
Discussion
We present here mechanistic insight as to how cell wall α-(1,3)-glucan promotes fungal virulence: interference with β-glucan recognition and with the subsequent activation of immune responses. This conclusion was made possible by the simultaneous manipulation of both sides of the interaction through the use of isogenic Histoplasma strains and alteration of the levels of dectin-1 in host cells. Given the spatial arrangement of the cell wall, the simplest model for interference is that α-(1,3)-glucan overlays and obscures presentation of β-glucan polymers. However, the participation of other yeast surface molecules dependent on α-(1,3)-glucan localization cannot be ruled out. Notably, the parasitic forms of the other dimorphic fungal pathogens also possess α-(1,3)-glucan, suggesting that β-glucan masking by α-(1,3)-glucan may be a conserved pathogenic mechanism among this group of fungi that cause respiratory and systemic disease.
In macrophages infected by Histoplasma, TNFα production in response to yeasts lacking the α-(1,3)-glucan shield was not completely eliminated by RNAi of dectin-1. Either residual dectin-1 protein is present in the RNAi lines, or additional, as yet undetermined, pathogen-associated molecules on the surface of Histoplasma are recognized at a low level. Consistent with the latter hypothesis, Histoplasma yeasts that possess the α-(1,3)-glucan cell wall layer do stimulate low levels of TNFα. Nevertheless, the β-glucan–dectin-1 PAMP–PRR pairing constitutes a major means of identification of Histoplasma as a fungal intruder, and recognition is largely abolished by the presence of the α-(1,3)-glucan cell wall layer, which provides one explanation for the greatly attenuated virulence of the ags1(Δ) Histoplasma strain.
Limitation of β-glucan exposure is one mechanism shaping the overall pathogenic potential of different medically important fungi. No α-(1,3)-glucan receptor PRR has been identified to date. Organisms that present α-(1,3)-glucan as the most external cell wall layer may thus effectively mask their β-glucan signature and avoid alerting the host immune system. Consistent with this hypothesis, the parasitic forms of the dimorphic fungal pathogens each possess α-(1,3)-glucan and can cause disease even in the face of normal host immune function. The fungal pathogen Aspergillus fumigatus also appears to limit β-glucan exposure by as yet unknown mechanisms because no β-glucan is detected on resting conidia until the conidia germinate (20–22). The opportunistic fungal pathogen C. albicans lacks α-(1,3)-glucan and does not typically cause disease in healthy hosts. Wild-type C. albicans yeasts stimulate substantial TNFα production by macrophages and are recognized by dectin-1, indicating significant β-glucan exposure (23–25). Pharmacologic or genetic perturbation of the Candida cell wall can increase β-glucan display, explaining the further virulence attenuation under such conditions (26). Interestingly, Histoplasma ags1(Δ) yeasts, which also lack α-(1,3)-glucan, stimulated TNF production to a level comparable with that elicited by C. albicans (Fig. 4C). In normal mammalian hosts, these two types of yeast have reduced capacity to cause significant disease. Thus, concealment of β-glucans from the host by α-(1,3)-glucan is not only an important virulence mechanism for Histoplasma, but it also helps explain mechanistically the higher native pathogenicity of dimorphic fungi.
Materials and Methods
Fungal Cell Culture.
Isogenic AGS1(+) and ags1(Δ) strains of H. capsulatum were derived from the clinical isolate G186A (ATCC 26029) and have been described in ref. 8. Histoplasma yeasts were grown at 37°C with 95% air/5% CO2 in HMM supplemented with 100 μg/ml uracil (27). For solid medium, 0.6% agarose and 25 μM FeSO4 were added. Dispersed Histoplasma yeasts were obtained by growth of liquid cultures to late exponential phase, removal of large yeast clumps by low-speed centrifugation (60 s at 100 × g), three washes with Ham's F-12 (Invitrogen, Carlsbad, CA), and counting with a hemacytometer. Conidia used in germination studies were obtained from Histoplasma strain G184A (ATCC 26027) grown for 4 weeks at room temperature on 0.9% yeast extract/2% agar plates. Conidia were harvested by flooding plates with 0.9% yeast extract, washing twice with 5-min centrifugations at 8,000 × g, and lysing contaminating hyphal fragments by dispersing through a needle. Conidia were resuspended in F12 medium containing 10% FBS and incubated with 95% air/5% CO2 at 37°C for 18 h. C. albicans cap1/cap1, a strain defective for germ tube formation (28), was used in TNFα assays to maintain yeast morphology during the assay time course. C. albicans was grown overnight in YPD medium to exponential phase growth (A600 between 5 and 8), washed three times with F-12, and counted with a hemacytometer.
Mammalian Cell Culture, Transfection, and Retrovirus Production.
3T3 fibroblasts, 3T3 + dectin-1 stable transformants (24), and 293T cells for dectin-1 RNAi retrovirus production were grown at 37°C with 95% air/5% CO2 in DMEM with 10% serum; P388D1 cells were similarly maintained in F-12 medium with 10% serum. For infections, yeasts and cells were coincubated in HMM-M (29) with 100 μg/ml uracil and 10% serum. Stable RNAi lines were generated by selection and limiting dilution of transformed or transfected cells. The gfp shRNA and dectin-1 shRNA-2 lines were generated by Lipofectamine2000 (Invitrogen)-mediated transfection of P388D1 cells with shRNA-containing pSIREN-based vectors (Clontech, Mountain View, CA). Vector-only and dectin-1 shRNA-1 lines were generated by retroviral transformation of P388D1 cells with pSIREN or dectin-1 shRNA-1-retroviral particles pseudotyped with vesicular stomatitis virus envelope protein G (VSVg). Retrovirus was produced through transient three-plasmid transfection of 293T cells with the appropriate pSIREN vector, MMLV-gag-pol, and VSVg expression plasmids (30). Targeted sequences for RNAi of dectin-1 were GACAGCTTCCTATCAAGAA (shRNA-1) and GATGGATATACTCAATTAG (shRNA-2), and for RNAi of gfp, GCTGACCCTGAAGTTCATCT.
Microscopy.
Visualization of α-(1,3)-glucan was performed on germinating conidia by using the α-(1,3)-glucan antibody MOPC-104E (Sigma, St. Louis, MO) after fixation of organisms in 4% paraformaldehyde in PBS for 30 min at room temperature. The MOPC-104E antibody specifically recognizes α-(1,3)-glucan because only α-(1,3)-linked and not β-linked glycosyl polysaccharides could block the antibody (31, 32). Furthermore, loss of MOPC-104E immunofluorescence staining in Histoplasma cell wall mutants correlates perfectly with loss of α-(1,3)-glucan by biochemical assays (33). Localization of α- and β-glucans in yeast cells was determined by fixation of AGS1(+) yeast in 2% glutaraldehyde in 100 mM phosphate buffer (pH 7.2) for 2 h at 4°C and postfixation in 1% osmium tetroxide (Polysciences, Inc., Warrington, PA) for 1 h at 4°C. Samples were dehydrated in a graded series of ethanol and propylene oxide before embedding in Eponate 12 resin (Ted Pella, Inc., Redding, CA). For analysis by fluorescence microscopy, 300-nm sections were immunolabeled with antibodies MOPC-104E and a monoclonal β-(1,3)-glucan-specific antibody (Biosupplies Australia, Parkville, Australia), probed with fluorophore-conjugated anti-IgM and anti-IgG secondary antibodies, respectively, then visualized with an Olympus BX-60 microscope (Center Valley, PA) with a ×100 objective. The β-(1,3)-glucan monoclonal antibody has previously been shown to be specific for β-(1,3)-linked glucans (34) and has been used in biochemical and immunofluorescence studies of fungal cell walls (26, 35). Cell wall glucan perimeters were produced by manually tracing the outermost boundary of fluorescence after importing fluorophore-specific micrographs into ImageJ (National Institutes of Health, Bethesda, MD). This procedure was done in blinded fashion to eliminate bias in perimeter placement. Fluorophore-specific tracings were pseudocolored and overlaid to generate the combined image. For ultrastructural images, 70- to 90-nm sections were labeled with the same α-(1,3)- and β-(1,3)-glucan-specific antibodies and probed with 18-nm and 12-nm colloidal gold-conjugated secondary antibodies (Jackson ImmunoResearch, West Grove, PA). Sections were subsequently stained with uranyl acetate and lead citrate and viewed with on a JEOL 1200EX transmission electron microscope (JEOL USA, Inc., Peabody, MA). Immunofluorescent localization of dectin-1 in 3T3 cells and P388D1 cells used a dectin-1 antibody (AF1756; R&D Systems, Minneapolis, MN) and a Cy3-conjugated anti-goat IgG secondary antibody after fixation of cells in 4% formaldehyde in PBS for 60 min.
Histoplasma–dectin-1 Binding.
Recognition of Histoplasma by dectin-1 was determined by infecting 2 × 104 3T3 or 3T3+dectin-1 cells with Histoplasma yeast (multiplicity of infection, 100:1) for 60 min, washing twice with PBS to remove unbound yeast, staining remaining yeast with the chitin-binding dye Uvitex3BSA (Ciba, Tarrytown, NY) for 10 min, washing an additional four times with PBS, and releasing bound yeast from cells with PBS plus 1% Triton X-100. Uvitex3BSA fluorescence, which is linearly proportional to the number of yeast (between 5 × 103 and 6 × 105; data not shown), was quantified by a fluorometer (Beacon 2000; PanVera, Madison, WI) with a 388-nm excitation filter and a 480-nm emission filter.
TNFα Assay.
P388D1 cells were seeded into 96-well plates at 2 × 104 cells per well. Histoplasma and Candida cap1/cap1 yeast cells were resuspended in HMM-M (29) with 100 μg/ml uracil and 10% serum at multiplicities of infection of 5:1 and 2:1, respectively, to account for the smaller size of Histoplasma yeast cells. Yeasts and murine cells were cocultured for 3 h at 37°C with 95% air/5% CO2, and then supernatants were removed and assayed for TNFα by ELISA (Assay Designs, Ann Arbor, MI). Lactate dehydrogenase (LDH) activity was used to normalize the results for variations in P388D1 cell densities. Relative LDH activity was determined by using a commercial LDH assay (Sigma) on lysates of cells in uninfected wells.
Acknowledgments
We thank P. Sundstrom for the C. albicans cap1 mutant, G. Brown for the stable dectin-1 3T3 transformants, S. Stewart for retroviral system reagents, and the Washington University Microbiology Microscopy Core for ultrastructural imaging services.This work was supported by Public Health Service Research Grants AI25584 and AI61298 (to W.E.G.) and by Damon Runyon Cancer Research Postdoctoral Fellowship DRG-1769-03 (to C.A.R.).
Abbreviations
- PAMP
pathogen-associated molecular pattern
- PRR
pattern-recognition receptor
- shRNA
short-hairpin RNA
- TLR
Toll-like receptor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Brown GD. Nat Rev Immunol. 2006;6:33–43. doi: 10.1038/nri1745. [DOI] [PubMed] [Google Scholar]
- 2.Palma AS, Feizi T, Zhang Y, Stoll MS, Lawson AM, Diaz-Rodriguez E, Campanero-Rhodes MA, Costa J, Gordon S, Brown GD, et al. J Biol Chem. 2006;281:5771–5779. doi: 10.1074/jbc.M511461200. [DOI] [PubMed] [Google Scholar]
- 3.Nemecek JC, Wuthrich M, Klein BS. Science. 2006;312:583–588. doi: 10.1126/science.1124105. [DOI] [PubMed] [Google Scholar]
- 4.Medoff G, Sacco M, Maresca B, Schlessinger D, Painter A, Kobayashi GS, Carratu L. Science. 1986;231:476–479. doi: 10.1126/science.3001938. [DOI] [PubMed] [Google Scholar]
- 5.Klimpel KR, Goldman WE. Infect Immun. 1988;56:2997–3000. doi: 10.1128/iai.56.11.2997-3000.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.San-Blas G, San-Blas F, Serrano LE. Infect Immun. 1977;15:343–346. doi: 10.1128/iai.15.2.343-346.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hogan LH, Klein BS. Infect Immun. 1994;62:3543–3546. doi: 10.1128/iai.62.8.3543-3546.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rappleye CA, Engle JT, Goldman WE. Mol Microbiol. 2004;53:153–165. doi: 10.1111/j.1365-2958.2004.04131.x. [DOI] [PubMed] [Google Scholar]
- 9.Reiss E. Infect Immun. 1977;16:181–188. doi: 10.1128/iai.16.1.181-188.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Domer JE. J Bacteriol. 1971;107:870–877. doi: 10.1128/jb.107.3.870-877.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanetsuna F, Carbonell LM, Gil F, Azuma I. Mycopathol Mycol Appl. 1974;54:1–13. doi: 10.1007/BF02055967. [DOI] [PubMed] [Google Scholar]
- 12.Pine L, Boone CJ. J Bacteriol. 1968;96:789–798. doi: 10.1128/jb.96.3.789-798.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brummer E, Stevens DA. Clin Exp Immunol. 1995;102:65–70. doi: 10.1111/j.1365-2249.1995.tb06637.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newman SL, Gootee L, Bucher C, Bullock WE. Infect Immun. 1991;59:737–741. doi: 10.1128/iai.59.2.737-741.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deepe GS, Jr, Gibbons RS. J Infect Dis. 2006;193:322–330. doi: 10.1086/498981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allendoerfer R, Deepe GS., Jr J Immunol. 2000;165:2657–2664. doi: 10.4049/jimmunol.165.5.2657. [DOI] [PubMed] [Google Scholar]
- 17.Allendoerfer R, Deepe GS., Jr J Immunol. 1998;160:6072–6082. [PubMed] [Google Scholar]
- 18.Yokota K, Takashima A, Bergstresser PR, Ariizumi K. Gene. 2001;272:51–60. doi: 10.1016/s0378-1119(01)00528-5. [DOI] [PubMed] [Google Scholar]
- 19.Heinsbroek SE, Taylor PR, Rosas M, Willment JA, Williams DL, Gordon S, Brown GD. J Immunol. 2006;176:5513–5518. doi: 10.4049/jimmunol.176.9.5513. [DOI] [PubMed] [Google Scholar]
- 20.Steele C, Rapaka RR, Metz A, Pop SM, Williams DL, Gordon S, Kolls JK, Brown GD. PLoS Pathog. 2005;1:e42. doi: 10.1371/journal.ppat.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gersuk GM, Underhill DM, Zhu L, Marr KA. J Immunol. 2006;176:3717–3724. doi: 10.4049/jimmunol.176.6.3717. [DOI] [PubMed] [Google Scholar]
- 22.Hohl TM, Van Epps HL, Rivera A, Morgan LA, Chen PL, Feldmesser M, Pamer EG. PLoS Pathog. 2005;1:e30. doi: 10.1371/journal.ppat.0010030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gantner BN, Simmons RM, Underhill DM. EMBO J. 2005;24:1277–1286. doi: 10.1038/sj.emboj.7600594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown GD, Gordon S. Nature. 2001;413:36–37. doi: 10.1038/35092620. [DOI] [PubMed] [Google Scholar]
- 25.Brown GD, Herre J, Williams DL, Willment JA, Marshall AS, Gordon S. J Exp Med. 2003;197:1119–1124. doi: 10.1084/jem.20021890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wheeler RT, Fink GR. PLoS Pathog. 2006;2:e35. doi: 10.1371/journal.ppat.0020035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Worsham PL, Goldman WE. J Med Vet Mycol. 1988;26:137–143. [PubMed] [Google Scholar]
- 28.Bahn YS, Sundstrom P. J Bacteriol. 2001;183:3211–3223. doi: 10.1128/JB.183.10.3211-3223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eissenberg LG, Schlesinger PH, Goldman WE. J Leukocyte Biol. 1988;43:483–491. doi: 10.1002/jlb.43.6.483. [DOI] [PubMed] [Google Scholar]
- 30.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen IS, Hahn WC, Sharp PA, et al. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potter M. Physiol Rev. 1972;52:631–719. doi: 10.1152/physrev.1972.52.3.631. [DOI] [PubMed] [Google Scholar]
- 32.Leon MA, Young NM, McIntire KR. Biochemistry. 1970;9:1023–1030. doi: 10.1021/bi00806a043. [DOI] [PubMed] [Google Scholar]
- 33.Marion CL, Rappleye CA, Engle JT, Goldman WE. Mol Microbiol. 2006;62:970–983. doi: 10.1111/j.1365-2958.2006.05436.x. [DOI] [PubMed] [Google Scholar]
- 34.Meikle PJ, Bonig I, Hoogenraad NJ, Clarke AE, Stone BA. Planta. 1991;185:1–8. doi: 10.1007/BF00194507. [DOI] [PubMed] [Google Scholar]
- 35.Zhong Q, Gvozdenovic-Jeremic J, Webster P, Zhou J, Greenberg ML. Mol Biol Cell. 2005;16:665–675. doi: 10.1091/mbc.E04-09-0808. [DOI] [PMC free article] [PubMed] [Google Scholar]