Abstract
Increased seed production has been a common goal during the domestication of cereal crops, and early cultivators of barley (Hordeum vulgare ssp. vulgare) selected a phenotype with a six-rowed spike that stably produced three times the usual grain number. This improved yield established barley as a founder crop for the Near Eastern Neolithic civilization. The barley spike has one central and two lateral spikelets at each rachis node. The wild-type progenitor (H. vulgare ssp. spontaneum) has a two-rowed phenotype, with additional, strictly rudimentary, lateral rows; this natural adaptation is advantageous for seed dispersal after shattering. Until recently, the origin of the six-rowed phenotype remained unknown. In the present study, we isolated vrs1 (six-rowed spike 1), the gene responsible for the six-rowed spike in barley, by means of positional cloning. The wild-type Vrs1 allele (for two-rowed barley) encodes a transcription factor that includes a homeodomain with a closely linked leucine zipper motif. Expression of Vrs1 was strictly localized in the lateral-spikelet primordia of immature spikes, suggesting that the VRS1 protein suppresses development of the lateral rows. Loss of function of Vrs1 resulted in complete conversion of the rudimentary lateral spikelets in two-rowed barley into fully developed fertile spikelets in the six-rowed phenotype. Phylogenetic analysis demonstrated that the six-rowed phenotype originated repeatedly, at different times and in different regions, through independent mutations of Vrs1.
Keywords: domestication, evolution, grass, transcription factor, vrs1
Domestication of the major modern cereal crops started ≈10,000 years before the present (yBP) (1–4). Throughout the process of cereal domestication, humans have deliberately selected individuals of wild species to emphasize seed recovery (5–7) and improved seed yield (1, 8). One of the most conspicuous instances of this process that occurred in the Near East was the appearance of six-rowed spikes during the domestication of barley (Hordeum vulgare ssp. vulgare; Fig. 1A and B). The barley spike is composed of triplets (each with one central and two lateral spikelets) arranged alternately at rachis nodes. All three spikelets of the modern six-rowed barley cultivars are fully fertile and able to develop into grains, but the lateral spikelets of two-rowed barley are reduced in size and are sterile (Fig. 1 C–G). The reduced lateral spikelets of this phenotype have greatly reduced stamens and a rudimentary ovary and stigma (Fig. 1H) compared with those of the central spikelets (Fig. 1I). Wild barley (H. vulgare ssp. spontaneum), the progenitor of cultivated barley (1, 9), is two-rowed, and its arrow-like triple spikelets, a product of disarticulation of the mature inflorescence due to brittleness of the rachis (main axis), are an adaptive specialization that ensures that the seeds will bypass stones and pebbles and reach soil when they fall to the ground (10). This feature is an evolutionary advantage offered by the two-rowed spikes in nature, and spontaneous six-rowed mutants are eliminated naturally and rapidly from wild barley populations because they lack this adaptation (10). Thus, six-rowed barley occurs primarily as cultivars or weeds (9).
Fig. 1.
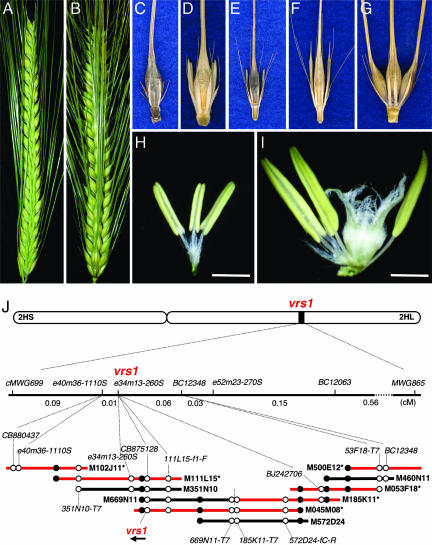
Map-based cloning of barley six-rowed spike gene vrs1. (A) Two-rowed spike. (B) Six-rowed spike. (C–G) One central and two lateral spikelets at a rachis node. (C) Ethiopian landrace var. deficiens; rudimentary lateral spikelets (Vrs1.t). (D) Two-rowed cultivar var. distichon; sterile lateral spikelets (Vrs1.b). (E) Wild barley var. spontaneum; sterile lateral spikelets (Vrs1.b). (F) Wild barley var. proskowetzii; short-awned or tip-pointed lateral spikelets (Vrs1.p). (G) Six-rowed cultivar convar. vulgare; fully fertile and awned lateral spikelets (vrs1.a). (H and I) Staminate floret of lateral spikelet (H) and hermaphroditic floret (I) in central spikelet in Vrs1.b two-rowed cultivar (D). (Scale bars: 2 mm.) (J) High-resolution linkage map and physical map. Six BAC clones (red) were fully sequenced. Open circles indicate markers uniquely assigned to chromosome 2H, of which genetically mapped markers are connected with the high-resolution map by dotted lines. Filled circles indicate repeated markers used for BAC connection. M669N11 and M185K11 are shown head-to-tail, separated by a vertical broken line.
The earliest archaeological specimens of barley seeds were uncovered from preagricultural sites in the Near East dating from 19,000 to 9,000 yBP (1). The specimens showed kernels of a two-rowed barley with a brittle rachis that are essentially identical to present-day wild barley. The earliest domesticated barley (9,500–8,400 yBP) also had two-rowed spikes; cultivation of six-rowed barley started later, with estimated dates ranging from 8,800 to 8,000 yBP (1, 11). Around 7,000–6,000 yBP, when barley was cultivated in the alluvial soils of Mesopotamia and, later, in the soils of Lower Egypt, six-rowed barley soon became dominant, replaced two-rowed barley, and established itself as the most important crop for Near Eastern Neolithic civilizations (1, 11, 12).
The development of a six-rowed spike is controlled by a single allele, vrs1 (formerly v for vulgare), that is recessive to the dominant allele responsible for the two-rowed spike (Vrs1) (13, 14). Although Intermedium spike-c.h (Int-c.h), which occurs in six-rowed barley, is involved in enlarging the size of lateral spikelets, the presence of the recessive gene vrs1 is by itself sufficient to cause two-rowed barley to become six-rowed barley (14). Three additional genes have been identified on different chromosomes through artificially induced mutations; however, none of these genes has been found in known six-rowed cultivars, probably due to the phenotypic disadvantages that occur in these mutants, such as reduced size and reduced fertility of the lateral spikelets on the upper and lower portions of the spikes (14). These observations indicate that Vrs1 has been the primary target of mutation during the evolution of six-rowed barley. It has been assumed that six-rowed barley developed from domesticated two-rowed barley by means of spontaneous mutation (1, 12), but the origin of six-rowed barley has not been confirmed. In the present study, we isolated the vrs1 gene and used studies of this gene to reveal the origin of six-rowed barley.
Results
Map-Based Cloning of Vrs1.
We previously mapped the vrs1 locus to a 0.90-cM interval between cMWG699 and MWG865 (15, 16). In the present study, we narrowed the locus to a 0.07-cM interval between e40m36-1110S and BC12348 by using segregating progeny equivalent to 9,831 gametes (Fig. 1J). The candidate genomic region was covered completely with six bacterial artificial chromosome (BAC) clones by means of chromosome walking. The contig containing vrs1 is composed of 518,343 bp. Annotation showed three predicted genes [see supporting information (SI) Table 1]: HvHox1 and HvEP2 appeared to be intact genes, whereas HvEP1 is highly degenerated and interrupted by several insertions of transposable elements. HvHox1 is the only gene that lies between the flanking markers e40m36-1110S and 53F18-T7 and is thus a likely candidate for Vrs1 (Fig. 1J). The ORF of two-rowed barley encoded a polypeptide composed of 222 amino acid residues, including a homeodomain-leucine zipper motif (HD-ZIP) (see SI Fig. 5).
Expression of Vrs1 in Lateral Spikelet Primordia.
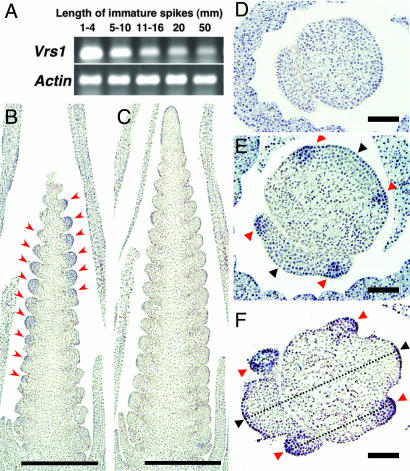
The expression pattern of the HD-ZIP I gene (Vrs1) was highly tissue- and stage-specific in two-rowed barley. Transcription of Vrs1 was abundant during the early developmental stages of the immature spike (1–4 mm long; Fig. 2A), when the central and lateral spikelet primordia became differentiated. (For a description of the stages of spike development, see ref. 17.) Transcript levels were lower, but still moderately abundant, at a length of 5 to 10 mm, but decreased greatly at later stages. In situ hybridization revealed that Vrs1 was expressed only in the lateral spikelet primordia (Fig. 2 B–F). Vrs1 was not detectable at the double-ridge stage, when the central and lateral spikelets remain undifferentiated (Fig. 2D), but Vrs1 expression was clearly detected at the triple-mound stage (Fig. 2E) and at the glume primordium stage (Fig. 2F). During these Vrs1-expressing stages (when the immature spikelets are 1.5 to 2.0 mm long), the primordium of the central spikelet became larger than the primordia of the lateral spikelets. Only the lateral spikelet primordia showed expression of the gene (Fig. 2B); the central spikelet primordia showed no expression (Fig. 2C).
Fig. 2.
Expression pattern of Vrs1 in two-rowed barley. (A) Expression of Vrs1 in immature inflorescences of different developmental stages (1–50 mm). Single-stranded cDNA synthesized from total RNA by using reverse transcriptase was subjected to RT-PCR analysis using primers specific to the 3′-UTR sequence. The barley actin gene was used as a control. (B–F) RNA in situ hybridization analysis of Vrs1. (B and C) Longitudinal serial sections along the row of lateral (B) and central (C) spikelet primordia at glume primordium stage. Red arrowheads in (B) indicate Vrs1 expression. (D–F) Transverse sections at double-ridge stage (D), triple-mound stage (E), and glume primordium stage (F). Red arrowheads indicate lateral spikelet primordia, and black arrowheads indicate central spikelet primordia. Broken lines in F correspond to longitudinal sectioning shown in B and C. (Scale bars: B and C, 500 μm; D–F, 100 μm.)
Mutant Vrs1 Gene.
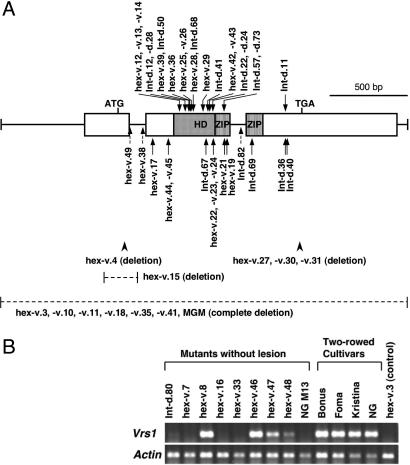
To demonstrate the biological function of Vrs1, we analyzed mutant lines. Among 91 mutant lines reported (14), we included 57 mutant lines (Fig. 3), which were derived from five two-rowed cultivars, of which both original and mutant seeds were available. The hexastichon (hex-v) mutants have six-rowed spikes with fully fertile, well developed, and long-awned lateral spikelets (18) and thus resemble normal six-rowed barley. The Intermedium spike-d (Int-d) mutants produced sterile or partially fertile lateral spikelets with variable awn length, which appeared intermediate to those of two-rowed and six-rowed barley (18). Because allelism of these mutations with vrs1 was confirmed in a previous study (19), morphological changes in the mutants can be attributed to changes in the Vrs1 gene. Lesions in Vrs1 were correlated with morphological changes in 48 mutant lines (Fig. 3A). Of the 48 mutants, 21 revealed a single amino acid substitution, 12 revealed truncation of the protein by a new stop codon, 3 showed a single nucleotide substitution in the conserved splicing sites of introns (change of splicing was confirmed by sequencing their transcripts), 5 had a frameshift mutation caused by a deletion, and 7 revealed complete deletion of the Vrs1 region (based on analysis using many flanking markers; see SI Table 2). These deletions (>182 kb), which were generated by means of irradiation, always resulted in hex-v-type six-rowed spikes under a range of growing conditions (see SI Table 3). The phenotypes observed in mutants that consistently exhibited six-rowed spikes support our hypothesis that complete deletion of Vrs1 occurred. Because the 7 deletion mutants did not show any developmental lesions, Vrs1 appears to be dispensable in barley. We found 9 mutants without any DNA changes throughout the coding region, and RT-PCR analysis showed that 7 of the 9 exhibited reduced or no expression of Vrs1 (Fig. 3B). This result suggests the occurrence of mutational events in the regulatory regions for Vrs1. The remaining 2 mutants showed Vrs1 expression at almost the same level as in the two-rowed cultivars, suggesting that posttranscriptional regulation is involved in these mutant phenotypes. The two mutation lines (hex-v.08 and hex-v.46) were derived from cv. ‘Bonus’ by ethyleneimine exposure and neutron bombardment, respectively, and were allelic to vrs1, as confirmed by crosses with hex-v.3 and hex-v.4 (19).
Fig. 3.
Analysis of mutants allelic to vrs1. (A) Lesions at Vrs1 detected in 48 mutants. Arrows pointing down indicate amino acid substitutions, arrows pointing up with a solid line indicate new stop codons, and the three arrows pointing up with a broken line indicate single-nucleotide substitutions in the introns with a changed splicing. The arrowheads and horizontal broken lines indicate deletions, in which five mutants have a partial deletion and seven mutants have a complete deletion of Vrs1. (B) RT-PCR analysis of nine mutant lines (including a New Golden mutant, NG M13) that did not show any lesions on the Vrs1. Four two-rowed cultivars and a deletion mutant (hex-v.3) were included as positive and negative controls, respectively. NG, New Golden. Total RNA was extracted from immature inflorescences 2–3 mm long.
The Origin of Six-Rowed Barley.
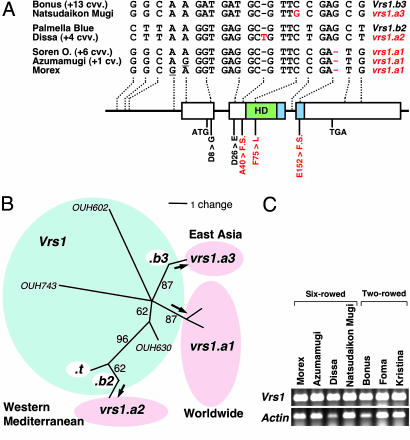
To infer the origin of six-rowed barley, we analyzed the sequence of the Vrs1 region from 15 two-rowed and 16 six-rowed cultivars, one variety (deficiens), and three wild barley lines chosen from different geographical areas. Two alleles in two-rowed barley (Vrs1.b2 in 1 cultivar and Vrs1.b3 in 14 cultivars) and three alleles in six-rowed barley (vrs1.a1 in 10 cultivars, vrs1.a2 in 5 cultivars, and vrs1.a3 in 1 cultivar) were identified by means of haplotype analysis (Fig. 4A). The sequences of each allele were identical except that vrs1.a1 had three subhaplotypes. For vrs1.a2 and vrs1.a3, we could deduce a direct descent from Vrs1.b2 and Vrs1.b3, respectively, as a result of point mutation. Phylogenetic analysis (Fig. 4B) supported the hypothesis that the six-rowed alleles were derived from two-rowed alleles, rather than vice versa, because the wild barley lines (OUH602, OUH630, and OUH743) were outgroups. The vrs1.a2 allele has an insertion of one nucleotide in exon 2, which results in a frame shift of the deduced amino acid sequence (Fig. 4A). The vrs1.a3 allele has a substitution of one nucleotide in which the phenylalanine (F) at amino acid position 75 becomes leucine (L)(Fig. 4A). The F is highly conserved in the DNA-binding domains of plants, animals, and yeasts, as shown by BLASTP analysis against the database of conserved domains (20). Three hex-v mutants (hex-v.12, hex-v.13, and hex-v.14) also revealed the replacement of F by another nucleotide at position 75, supporting the importance of this amino acid in the DNA-binding domain. The vrs1.a3 allele occurred in one six-rowed cultivar (“Natsudaikon Mugi”) in East Asia. The most common allele, vrs1.a1, which is distributed around the world (10 cultivars), has a deletion of one nucleotide in exon 3, which results in a different frame shift (Fig. 4A). The progenitor of the vrs1.a1 allele remains to be identified (Fig. 4B). It was noteworthy that the vrs1.a1 allele has been differentiated into three subhaplotypes by point mutations in the 5′ noncoding region (Fig. 4A). Two- and six-rowed barleys showed equal levels of transcription of Vrs1 (Fig. 4C), thus single-base-pair mutations in vrs1 appear to be responsible for the functional changes observed in all three natural six-rowed alleles.
Fig. 4.
Molecular events occurred in six-rowed barley. (A) Haplotype analysis of the Vrs1 region in two- and six-rowed cultivars. (B) Phylogenetic tree of Vrs1 alleles illustrating the three independent origins of six-rowed barley. Three wild barley lines (“OUH” identifiers) are outgroups. (C) RT-PCR analysis of Vrs1 expression in immature inflorescences of six-rowed and two-rowed barley.
Discussion
The evolution of barley was not a single event but has been an ongoing process of continuous specialization, as suggested by the concept of diffuse centers of differentiation (12). All of the present barley forms have resulted from specialization as a result of cumulative mutations, and tracing the origin of six-rowed barley would require us to find the least-specialized form of barley either conceptually (based on genetic analysis) or archaeologically. One of the main arguments in the debate over the evolution of barley relates to genetic changes between the brittle and nonbrittle rachis and between the two- and six-rowed spikes. We previously inferred the phylogeny of rachis brittleness (21) and of the number of rows of spikelets (22) based on molecular markers that are tightly linked to the btr1/btr2 and vrs1 genes. The genealogy based on each domestication gene (here, a domestication gene is one that motivates humans to domesticate a plant) allows adequate inferences about the origins of cultivated barley. In the present study, cloning of the Vrs1 gene substantiated our proposed phylogeny for the number of rows of spikelets. Wild and cultivated barley are interfertile (9, 23), and mutual introgression of genes appears to be substantial between the two forms (24). Therefore, random markers do not appear to represent the two- and six-rowed types of barley, and evolutionary studies based on DNA markers that are inherited independently of the domestication genes might provide different insights into the specialization of barley (25, 26). In the present paper, we focus exclusively on the origin of six-rowed barley. In a more strict sense, we focus on the Vrs1 gene rather than the whole genome of barley to elucidate the origin of six-rowed barley, even though our data might have some implications for evolution of the species.
An HD-ZIP I Homeobox Gene Determines the Number of Rows of Spikelets in Barley.
Our study revealed that Vrs1 encodes a member of the HD-ZIP class of transcription factors. Analysis of induced mutants clearly confirmed the identity of Vrs1 and demonstrated its biological function. Although complementation analysis by transformation is another option (27), our data, based on as many as 57 independent mutant lines, together with the specific gene expression patterns, confirm that Vrs1 encodes an HD-ZIP I protein. Although HD is universal, the HD-ZIP is unique to the plant kingdom. HD-ZIP protein forms dimers via ZIP and correctly binds by means of HD to dyad-symmetrical recognition sequences of DNA based on the strict spatial relationship between HD and ZIP (28, 29), and the DNA-binding activity of HD-ZIP was demonstrated (30). HD-ZIP proteins have been grouped into four families (I–IV) (31), and the barley VRS1 protein belongs to family I (data not shown). The HD-ZIP III and IV genes are functionally well characterized, being involved in (i) development of the apical meristem, (ii) vascular development, and (iii) establishment of cell fates in epidermal (32). The biological function of the HD-ZIP I and II genes has not yet been clarified, although involvement in plant growth and development was suggested (32). Our study shows a conspicuous association between an HD-ZIP I gene and a plant developmental process. The spatial and temporal specificity of Vrs1 gene expression suggests that VRS1 is a transcription factor involved in the development (suppression) of lateral spikelets in two-rowed barley. Most domestication genes have been found to encode transcription factors (33), and our results agree with these observations.
During the domestication of barley and wheat, rudimentary spikelets or flowers of wild species have been restored to their full functionality to increase seed number (1, 8). In the present study, we demonstrated that loss of function of Vrs1 is sufficient to convert the rudimentary spikelets of two-rowed barley into the fertile spikelets of six-rowed barley. The mutations found in six-rowed cultivars were commonly represented by single-nucleotide polymorphisms, but even more drastic changes such as complete deletion of Vrs1 were also found in several induced mutants (Figs. 3 and 4). This loss of function agrees with the facts that six-rowed spikes are genetically recessive and that two-rowed spikes are controlled by the wild-type allele, which occurs naturally in wild barley. The creation of six-rowed spikes is similar to the gigantism that occurs during domestication because both types of change can be caused by loss of function of genes (34). The dominant nature of Vrs1 and the potential DNA-binding activity of HD-ZIP I proteins suggest that VRS1 is a repressor protein that binds to the DNA of genes that regulate the development of lateral spikelets. Further investigation of the subcellular localization of VRS1 proteins will be necessary to test this hypothesis.
Multiple Origins for Six-Rowed Barley Implied by the Mutant Vrs1/HD-ZIP I Gene.
The number of rows of spikelets is a key characteristic in inferring the origin of cultivated barley, and it has been debated whether the progenitor of cultivated barley was six-rowed (35), two-rowed (12, 36), or both (37, 38). Recently, the two-rowed progenitor hypothesis was supported by (i) archaeological specimens showing preagricultural utilization of wild barley and the existence of domesticated two-rowed remains that were older than six-rowed barley remains, (ii) the dominance of the two-rowed phenotype, and (iii) the rather weedy nature of brittle six-rowed barley in Israel and Tibet (9–12).
Our phylogenetic tree (Fig. 4B) shows two lineages of domesticated two-rowed barley that diverged considerably from each other: Vrs1.b3 stood alone, and the other lineage further differentiated into Vrs1.b2 and Vrs1.t (Fig. 4B). Owing to its provenance in the western Mediterranean, the Vrs1.b2 allele is likely to be an immediate progenitor of the vrs1.a2 six-rowed allele, which predominates in the western Mediterranean. This is an indication that one of the six-rowed alleles originated in this region. Similarly, it is clear that the vrs1.a3 six-rowed allele derived directly from the Vrs1.b3 two-rowed allele by means of a point mutation (Fig. 4A). Indeed, the Korean vrs1.a3 cultivar has great similarities to old European two-rowed cultivars, such as a spring growth habit, nonbrittle rachis due to the recessive btr1 gene (38), lax spikes, a long stem, and the K haplotype of cMWG699, which is specific to two-rowed barley (22). It is interesting to speculate that the vrs1.a3 six-rowed cultivar might have originated from European two-rowed barley introduced into Eastern Asia. Our results suggest that, at least for the vrs1.a2 and vrs1.a3 alleles, six-rowed barley was derived from cultivated two-rowed barley. Because the number of lines of wild barley was limited in the present study, further collection of wild barley will be necessary to find a potential wild ancestor for each allele.
We were unable to resolve the origin of the most common vrs1.a1 six-rowed allele in two-rowed barley (Fig. 4B). A notable feature of the vrs1.a1 allele is that it has three subhaplotypes (Fig. 4A). If we assume the minimum number of changes, the vrs1.a1 allele must have originated from a hypothetical Vrs1.b1 allele in a single, unknown, two-rowed ancestor, and the first vrs1.a1 allele (which today exists in cv. ‘Soren Oomugi 19329’; Fig. 4A) has differentiated into two additional haplotypes by means of background point mutations in the 5′ noncoding region. The vrs1.a1 cultivars correspond to the major Type I cultivars classified by use of a marker tightly linked to vrs1, and the dominant Type I cultivars are distributed throughout the world (22). Because of its predominance and higher haplotype diversity, we hypothesize that the vrs1.a1 allele may represent the most ancient six-rowed allele (dating to 7,000–6,000 yBP), which was widely present in Neolithic agriculture in the Near East.
Our study demonstrates three independent processes for the origins of six-rowed barley (Fig. 4B). (This result does not necessarily mean that barley has a polyphyletic origin as a species, a distinction that is often misunderstood.) We previously hypothesized two independent origins of six-rowed barley, one of which gave rise to the six-rowed barley that has spread around the world and the other of which gave rise to the six-rowed barley endemic to the western Mediterranean (22). Our present study supports the concept of multiple origins of six-rowed barley. We believe these origins are straightforward because we used the DNA sequence encoding the Vrs1 gene itself. Creation of the three alleles (vrs1.a1, vrs1.a2, and vrs1.a3) cannot be deduced through intragenic recombination by mutual hybridization of six-rowed barleys possessing different alleles (Fig. 4A), which suggests that their origins were independent. Divergence of these alleles cannot be explained by simple mutation. Thus, six-rowed barley must have originated repeatedly from cultivated two-rowed or wild barley by means of a loss-of-function mutation of an HD-ZIP I-class homeobox gene. Six-rowed barley originated independently at different times and in different regions (Fig. 4B), probably as a result of conscious selection by delighted farmers who greatly appreciated the improved seed yield.
Implications for the Evolution of Barley.
The DNA sequence of Vrs1 may trace the origin and migration of two-rowed barley. Both two- and six-rowed barleys emerged in Greece between 8,000 and 6,000 yBP, but six-rowed barley dominated in the Balkans and Central Europe (5,000–2,000 yBP) and in Southern Europe and North Africa (7,000–4,000 yBP) (1). After its disappearance from ancient Mesopotamia and Egypt, two-rowed barley does not reappear in the archaeological record in these regions until 1,100 yBP (11). Cultivation of two-rowed barley is virtually unknown in Central and Northern Europe until 1,000 yBP, and it has been assumed that two-rowed barley was introduced into Europe at 900–800 yBP by Crusaders from the Near East, as suggested by Fischbeck (39). In the present study, 14 two-rowed cultivars carrying the Vrs1.b3 allele did not show any polymorphism in the DNA sequence (Fig. 4A). This indicates that the origin of the Vrs1.b3 allele is relatively recent and that the allele has expanded rapidly throughout the world and become dominant, which partially supports Fischbeck's hypothesis (39). The interpretation that Vrs1.b3 is a recent allele agrees with the fact that only one vrs1.a3 six-rowed cultivar (derived from Vrs1.b3) has been detected thus far (Fig. 4A). This hypothesis does not necessarily contradict the hypothesis of Helbaek (11) that two-rowed barley was cultivated earlier than six-rowed barley, because the DNA sequences of archaeological specimens remain unknown. Moreover, the type of lateral spikelet in the ancient domesticated two-rowed cultivars (deficiens, Vrs1.t; distichon, Vrs1.b) is not clear. We favor the hypothesis that the common progenitor of Vrs1.t and Vrs1.b2 occurred earlier than that of Vrs1.b3, based on the genetic distances seen in the allele coalescence and polymorphism results for the Ethiopian and western Mediterranean cultivars (Fig. 4A). Detailed genetic analysis of archaeological barley specimens would help to answer these questions.
The inflorescence architecture in the Poaceae is a continuous story of reduction from a more original “panicle” of spikelets (as seen in rice and oats) to a “spike” of spikelets (40), resulting in three sessile spikelets per node in barley and a single sessile spikelet per node in wheat and rye. In wild Hordeum species, the three spikelets and their slender awns form a light dispersal unit that permits both anemochory and zoochory (9). In two-rowed barley, strict temporal and spatial regulation of Vrs1 expression leads to reduction and sterility of the lateral spikelets. We speculate that either strong alleles or differential expression or regulation of Vrs1 orthologs could lead to complete repression of lateral spikelet formation at inflorescence nodes of other species in the Poaceae, as is found in wheat and rye. A Poaceae-wide assessment of the variability and regulation of Vrs1 orthologs would be an exciting and productive way to improve our understanding of plant development and the evolution of grass species.
Materials and Methods
Plant Materials.
Barley cultivars representing geographical distribution and three wild lines used in this study are maintained in the laboratory of T.K. (see SI Table 4). Mutants consisting of 39 hex-v and 16 Int-d lines were obtained from the Nordic GenBank (Alnarp, Sweden). Two more hex-v-type mutants derived from Japanese two-rowed cultivars New Golden (41) and Misato Golden (42) by gamma irradiation were obtained from T. Makino, National Institute of Crop Science (Tsukuba, Japan).
Genetic Mapping.
Three fine-mapping populations of 6,269 gametes were analyzed previously (15, 16). An additional 1,781 F2 plants of vrs1.a1 × Vrs1.b3 were screened with cMWG699 and BC12063, and recombinants were analyzed further (see SI Table 5). ESTs contigs BC12348 (AJ468022 and CB881790) and BC12063 (BU973565 and BU987455) were converted to sequence-tagged site markers. Other markers were generated from BAC DNA sequences.
BAC Contig and Sequencing.
A BAC library of cv. Morex (43) was obtained from Clemson University Genomic Institute (Clemson, SC) and has been screened by PCR (27). BAC contig was made by chromosome walking (see SI Methods). Shotgun sequencing of the BACs was carried out by the standard method (44). Assembled sequence contigs were correctly ordered, oriented with information of the cloning sites of the vector and the end-sequences of the bridge subclones, and annotated (see SI Methods). Contig gaps were filled by full-sequencing the bridge subclones either by primer walking or by GPS-1 transposon sequencing (New England Biolabs, Ipswich, MA).
RNA Analysis.
Immature inflorescences were developmentally staged by measuring under a microscope. RNAs were extracted from leaves or immature inflorescences excised from 2-month-old plants by using TRIzol (Invitrogen, Carlsbad, CA). First-strand cDNA was generated by using SuperScript II (Invitrogen). RT-PCR and full-length cDNA sequencing were carried out with gene-specific primers (see SI Methods).
In Situ Hybridization Analysis.
A 315-bp RT-PCR fragment including only the 3′-UTR of Vrs1 was amplified by using primers M111L15F84380 (5′-CAT ACT TAA CGC ACG CCT AGA GATC-3′) and M111L15R84670 (5′-TAG CTG CTG CCG CCG CCA AAT CCTC-3′) and cloned into pCR4-TOPO vector (Invitrogen); after digestion by EcoRI, the insert was subcloned into pBluescript II KS (+) vector (Stratagene, La Jolla, CA). Two clones with different insert orientations were linearized by using NotI and were used as templates to generate antisense and sense probes by using T3 RNA polymerase. Hybridization with a digoxigenin-labeled RNA probe, and immunological detection, were conducted. In situ hybridization was conducted according to the methods of Kouchi and Hata (45), with thorough modifications for improved reactions (see SI Methods).
Haplotype Analysis.
Genomic DNAs (2.1-kb) including Vrs1 were amplified from wild barley, cultivars, and mutants by PCR using the gene-specific primer pair M111L15F84204 (5′-GAA AGA TGA TTG CCA ACT ACC-3′) and M111L15R86329 (5′-GTC ATA ACT CGG CAA ACA TAG-3′), and their DNA sequences were determined by using internal primers designed by taking advantage of the BAC sequence (see SI Methods).
Phylogenetic Analysis.
A phylogenetic tree was constructed by the neighbor-joining method (46) using PAUP 4.0b10 (47). The DNA sequences of single haplotypes or subhaplotypes represented the cultivars sharing identical sequences. Phylogenetically informative insertions/deletions (indels) were included for analysis. All substitutions and indels were weighted equally. Bootstrap analysis with 1,000 replicates was performed.
Supplementary Material
Acknowledgments
We thank S. Okamura, M. Sameri, T. Ueda, T. Akihiro, N. Hara, S. Toki, and Y. Nagamura for help and advice and H. Kitano, M. Ashikari, D. Saisho, N. Minaka, D. Vaughan, and H. Knüpffer for discussions. This work was funded by research grants MP1113b (Rice Genome Project) and GD3006 (Green Techno Project) from the Ministry of Agriculture, Forestry, and Fisheries of Japan.
Abbreviations
- HD-ZIP
homeodomain-leucine zipper
- yBP
years before present.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The sequences reported in this paper have been deposited in the EMBL/GenBank/DDBJ database (accession nos. AB259782, AB259783, and EF067844).
This article contains supporting information online at www.pnas.org/cgi/content/full/0608580104/DC1.
References
- 1.Zohary D, Hopf M. Domestication of Plants in the Old World. New York: Oxford Univ Press; 2000. [Google Scholar]
- 2.Khush G. Plant Mol Biol. 1997;35:25–34. [PubMed] [Google Scholar]
- 3.Salamini F, Özkan H, Brandolini A, Schäfer-Pregl R, Martin W. Nat Rev Genet. 2002;3:429–441. doi: 10.1038/nrg817. [DOI] [PubMed] [Google Scholar]
- 4.Doebley J. Annu Rev Genet. 2004;38:37–59. doi: 10.1146/annurev.genet.38.072902.092425. [DOI] [PubMed] [Google Scholar]
- 5.Li C, Zhou A, Sang T. Science. 2006;311:1936–1938. doi: 10.1126/science.1123604. [DOI] [PubMed] [Google Scholar]
- 6.Konishi S, Izawa T, Lin S, Ebana K, Fukuta Y, Sasaki T, Yano M. Science. 2006;213:1392–1996. doi: 10.1126/science.1126410. [DOI] [PubMed] [Google Scholar]
- 7.Simons K, Fellers J, Trick H, Zhang Z, Tai Y-S, Gill B, Faris J. Genetics. 2006;172:547–555. doi: 10.1534/genetics.105.044727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harlan JR, De Wet JMJ, Price EG. Evolution (Lawrence, Kans) 1973;27:311–325. doi: 10.1111/j.1558-5646.1973.tb00676.x. [DOI] [PubMed] [Google Scholar]
- 9.von Bothmer R, Jacobsen N, Baden C, Jørgensen RB, Linde-Laursen I. An Ecogeographical Study of the Genus Hordeum. 2nd Ed. Vol 7. Rome: International Plant Genetic Resources Institute; 1995. Systematic and Ecogeographic Studies on Crop Genepools. [Google Scholar]
- 10.Zohary D. Proceedings of the First International Barley Genetics Symposium, Wageningen: Barley Genetics I; Wageningen, The Netherlands: Pudoc Centre for Agricultural Publications and Documentations; 1963. pp. 27–31. [Google Scholar]
- 11.Helbaek H. Science. 1959;130:365–372. doi: 10.1126/science.130.3372.365. [DOI] [PubMed] [Google Scholar]
- 12.Harlan JR. Barley: Origin, Botany, Culture, Winterhardiness, Genetics, Utilization, Pests. Washington, DC: U.S. Department of Agriculture; 1968. pp. 9–31. Agriculture Handbook No 338. [Google Scholar]
- 13.Ubisch G. Z Ind Abs Ver. 1916;17:120–152. [Google Scholar]
- 14.Lundqvist U, Franckowiak JD, Konishi T. Barley Genet Newsletter. 1997;26:22–516. [Google Scholar]
- 15.Komatsuda T, Tanno K. Hereditas. 2004;141:68–73. doi: 10.1111/j.1601-5223.2004.01820.x. [DOI] [PubMed] [Google Scholar]
- 16.He C, Sayed-Tabatabaei BE, Komatsuda T. Genome. 2004;47:1122–1129. doi: 10.1139/g04-073. [DOI] [PubMed] [Google Scholar]
- 17.Kirby EJM, Appleyard M. Cereal Development Guide. Warwickshire, UK: Cereal Unit, National Agricultural Centre, Stoneleigh, Kenilworth; 1981. [Google Scholar]
- 18.Gustafsson A, Lundqvist U. Hereditas. 1980;92:229–236. [Google Scholar]
- 19.Lundqvist U, Lundqvist A. Hereditas. 1988;108:13–26. [Google Scholar]
- 20.Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, et al. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komatsuda T, Maxim P, Senthil N, Mano Y. Theor Appl Genet. 2004;109:986–995. doi: 10.1007/s00122-004-1710-0. [DOI] [PubMed] [Google Scholar]
- 22.Tanno K, Taketa S, Takeda K, Komatsuda T. Theor Appl Genet. 2002;104:54–60. doi: 10.1007/s001220200006. [DOI] [PubMed] [Google Scholar]
- 23.Linde-Laursen I, Heslop-Harrison J, Shepherd K, Taketa S. Hereditas. 1997;126:1–16. [Google Scholar]
- 24.Abdel-Ghani A, Parzies H, Omary A, Geiger H. Theor Appl Genet. 2004;109:588–595. doi: 10.1007/s00122-004-1657-1. [DOI] [PubMed] [Google Scholar]
- 25.Kilian B, Özkan H, Kohl J, von Haeseler A, Barale F, Deusch O, Brandolini A, Yucel C, Martin W, Salamini F. Mol Genet Genomics. 2006;276:230–241. doi: 10.1007/s00438-006-0136-6. [DOI] [PubMed] [Google Scholar]
- 26.Badr A, Müller K, Schäfer-Pregl R, El Rabey H, Effgen S, Ibrahim H, Pozzi C, Rohde W, Salamini F. Mol Biol Evol. 2000;17:499–510. doi: 10.1093/oxfordjournals.molbev.a026330. [DOI] [PubMed] [Google Scholar]
- 27.Stein N, Perovic D, Kumlehn J, Pellio B, Stracke S, Streng S, Ordon F, Graner A. Plant J. 2005;42:912–922. doi: 10.1111/j.1365-313X.2005.02424.x. [DOI] [PubMed] [Google Scholar]
- 28.Ruberti I, Sessa G, Lucchetti S, Morelli G. EMBO J. 1991;10:1787–1791. doi: 10.1002/j.1460-2075.1991.tb07703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sessa G, Morelli G, Ruberti I. EMBO J. 1993;12:3507–3517. doi: 10.1002/j.1460-2075.1993.tb06025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meijer AH, de Kam RJ, d'Erfurth I, Shen W, Hoge JHC. Mol Genet Genomics. 2000;263:12–21. doi: 10.1007/pl00008671. [DOI] [PubMed] [Google Scholar]
- 31.Sessa G, Carabelli M, Ruberti I, Lucchetti S, Baima S, Morelli G. In: Plant Molecular Biology. Coruzzi G, Puigdomenech P, editors. Vol H81. Berlin: Springer; 1994. pp. 412–426. NATO ASI Series. [Google Scholar]
- 32.Henriksson E, Olsson A, Johannesson H, Johansson H, Hanson J, Engström P, Söderman E. Plant Physiol. 2005;139:509–518. doi: 10.1104/pp.105.063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doebley J. Science. 2006;312:1318–1319. doi: 10.1126/science.1128836. [DOI] [PubMed] [Google Scholar]
- 34.Lester RN, Daunay M-C. Schriften Genet Ressourcen. In: Knüpffer H, Ochsmann J, editors. Rudolf Mansfeld and Plant Genetic Resources. Vol. 22. 2003. pp. 137–152. [Google Scholar]
- 35.Åberg E. Symbolae Botanicae Upsalienses. 1940;4:1–156. [Google Scholar]
- 36.Bakhteyev F. Proceedings of the First International Barley Genetics Symposium, Wageningen: Barley Genetics I; Wageningen, The Netherlands: Pudoc Centre for Agricultural Publications and Documentations; 1963. pp. 1–18. [Google Scholar]
- 37.Freisleben R. Der Züchter. 1940;11:257–272. [Google Scholar]
- 38.Takahashi R. Demerec M. Advances in Genetics. Vol 7. New York: Academic; 1955. pp. 227–266. [Google Scholar]
- 39.Fischbeck G. In: Barley Science: Recent Advance from Molecular Biology to Agronomy of Yield and Quality. Slafer GA, Molina-Cano JL, Savin R, Araus JL, Romagosa I, editors. New York: Food Products Press; 2002. pp. 1–14. [Google Scholar]
- 40.Vegetti AC, Anton AM. Flora. 1995;190:225–228. [Google Scholar]
- 41.Makino T, Furusho M, Fukuoka T. Barley Genet Newsletter. 1995;24:122. [Google Scholar]
- 42.Matsui K, Makino T. Sogo Nogyo Shiken Kenkyu Seiseki Keikaku Gaiyoshu 5, Winter Crops. Tsukuba, Japan: National Agricultural Research Center; 1997. No I8-053-3. [Google Scholar]
- 43.Yu Y, Tomkins JP, Waugh R, Frisch DA, Kudrna D, Kleinhofs A, Brueggeman RS, Muehlbauer GJ, Wise RP, Wing RA. Theor Appl Genet. 2000;101:1093–1099. [Google Scholar]
- 44.International Rice Genome Sequencing Project. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- 45.Kouchi H, Hata S. Mol Genet Genomics. 1993;238:106–119. doi: 10.1007/BF00279537. [DOI] [PubMed] [Google Scholar]
- 46.Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford Univ Press; 2000. [Google Scholar]
- 47.Swofford D. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 1998. Version 4. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.