Abstract
Ongoing climate change has affected the ecological dynamics of many species and is expected to impose natural selection on ecologically important traits. Droughts and other anticipated changes in precipitation may be particularly potent selective factors, especially in arid regions. Here we demonstrate the evolutionary response of an annual plant, Brassica rapa, to a recent climate fluctuation resulting in a multiyear drought. Ancestral (predrought) genotypes were recovered from stored seed and raised under a set of common environments with descendant (postdrought) genotypes and with ancestor×descendant hybrids. As predicted, the abbreviated growing seasons caused by drought led to the evolution of earlier onset of flowering. Descendants bloomed earlier than ancestors, advancing first flowering by 1.9 days in one study population and 8.6 days in another. The intermediate flowering time of ancestor×descendant hybrids supports an additive genetic basis for divergence. Experiments confirmed that summer drought selected for early flowering, that flowering time was heritable, and that selection intensities in the field were more than sufficient to account for the observed evolutionary change. Natural selection for drought escape thus appears to have caused adaptive evolution in just a few generations. A systematic effort to collect and store propagules from suitable species would provide biologists with materials to detect and elucidate the genetic basis of further evolutionary shifts driven by climate change.
Keywords: contemporary evolution, global climate change, life history theory, local adaptation, plant phenology
Many species have shifted phenology (the seasonal timing of reproduction and other life history events) in response to ongoing climate change (1–3). For example, a recent study reviewing flowering times (FT) in 461 plant species showed a trend of earlier flowering with climate warming (1), and another study showed shifts in plant flowering and bird and butterfly arrival dates in Mediterranean habitats (4). These shifts are largely attributed to rising temperatures, but anticipated changes in precipitation (5) may also affect phenology, especially in arid regions. Observed shifts in phenology are due in part to direct effects of climate on physiological and developmental rates (phenotypic plasticity). However, climate change can impose natural selection on phenology and thereby cause genetically based evolutionary shifts. These shifts may occur rapidly, providing important opportunities for the study of adaptive evolution in natural populations.
Abundant evidence has accumulated over the past several decades showing that natural selection can cause evolutionary change in just a few generations (6, 7). Several cases of contemporary evolution implicate climate change as a selective agent, using two general protocols. The first compares contemporary and previous data on natural populations. This approach has shown shifts over the past few decades in the frequencies of climate-associated isozyme alleles and chromosome inversions across latitudinal gradients in Drosophila (8–10). Similarly, pitcher plant mosquitoes from northern latitudes, where growing seasons have lengthened, now enter winter diapause at shorter photoperiods than they did in the 1970s, while more southern populations remain unchanged (11). The second protocol involves monitoring individuals in natural populations and inferring genetically based changes from the phenotypic resemblance between descendants and ancestors. This method has demonstrated a genetic shift toward earlier parturition dates in red squirrels over the 1990s after increased artic spring temperatures (12) and showed shifts in beak morphology of Darwin's finches after drought changed food availability (13). These two general approaches provide convincing evidence for evolution by showing temporal changes in gene frequencies or phenotypes. However, by necessity, these methods evaluate ancestral and descendant generations at different times and under potentially nonidentical conditions, and so some important questions on the adaptive nature and genetic basis of these changes cannot be fully addressed.
We used a third experimental approach applicable to any species that can be stored in a dormant state. This approach compares phenotypic and fitness values of ancestral, descendant, and ancestral×descendant hybrid genotypes grown simultaneously under conditions that mimic the pre- and postchange environments. This method has several advantages. Ancestors and descendants are reared together under the same controlled conditions so that phenotypic differences between ancestors and descendants can be partitioned into components due to genetic change and due to phenotypic plasticity. By simulating pre- and postchange conditions and measuring fitness in both environments, it is possible to determine whether the descendant genotypes are better adapted to novel conditions, or, conversely, that they have lost adaptation to past conditions. The construction of hybrid lines provides information on the genetic basis and architecture of trait changes, allowing phenotypic shifts to be partitioned into additive versus dominant gene effects (14). This approach thus combines the logic of the reciprocal transplant (15) and the line cross (14) experimental protocols, which have explored evolutionary divergence between populations, to investigate evolutionary changes within populations. Previous studies have compared ancestors and descendants (without hybrids) to demonstrate the evolutionary response of Escherichia coli to elevated temperature in the laboratory (16) and in natural populations of Daphnia to study adaptation to water pollution (17). We used this general protocol to examine changes in phenology after drought.
We examined the evolutionary response of FT in field mustard, Brassica rapa L. (Brassicaceae), during a regional climate anomaly. We define FT as the number of days for a plant to go from germination to production of its first flower.
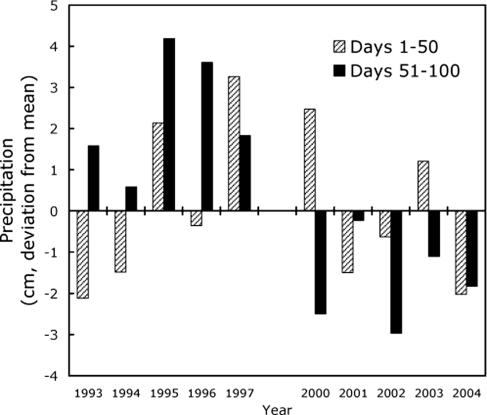
In southern California the growing season for B. rapa is normally terminated in late spring by the end of the rainy season. Winter rainfall data for an 11-year period at our study site indicate a switch from above average to below average precipitation (Fig. 1), leading to abbreviated growing seasons from 2000 to 2004. Although periodic precipitation patterns are common because of the influence of El Niño–Southern Oscillation cycles in this region, the series of dry years from 2000 to 2004 represents an extreme drought event (18, 19). Life history theory predicts that the optimal FT in annual plants will be shorter with shorter growing seasons (20, 21). This led to the prediction that the abbreviated growing seasons imposed by the extended drought would lead to the evolution of earlier FT in B. rapa.
Fig. 1.
Early and late winter precipitation at the Irvine Ranch Water District (<3 km from the study sites) from the National Climate Data Center. We calculated cumulative precipitation during the growing season (100 days after the first 72-h period with rainfall ≥1 cm). Mean cumulative precipitation for days 1–50 and days 51–100 for each of the five growing seasons leading up to 1997 and 2004 were calculated, and values are plotted as deviation from this mean. Shaded bars indicate cumulative precipitation during the first 50 days after rainfall. Black bars show cumulative precipitation in the subsequent 50 days.
Fortuitously, we collected B. rapa seed in 1997, before the drought, and then again in 2004 from two populations. The Newport Harbor (Dry Site) population occupies sandy, fast drying soils, whereas the population at San Joaquin Freshwater Marsh Reserve (Wet Site) occupies soils that retain moisture later into the growing season (22). A previous study (22) using field-collected seeds grown in the greenhouse showed earlier FT of Dry Site than Wet Site genotypes, supporting the evolutionary relationship between season length and FT. The same study also demonstrated significant selection pressure for earlier FT at both sites during drought.
Here we report experiments to determine whether an evolutionary shift in FT occurred during the extended drought, whether this phenological change was adaptive, and whether the rate of evolutionary change observed matched predictions based on estimates of heritability and selection intensity. We first created F1 lines of ancestors, descendants, and hybrids to ameliorate environmental and maternal effects (see Materials and Methods). In one experiment we grew the ancestral, descendant, and hybrid F1 seeds in the greenhouse under three different watering treatments to simulate short, medium, and long growing seasons [supporting information (SI) Fig. 4]. This experiment allowed us to determine whether an evolutionary shift in FT had occurred and whether this change was adaptive. In a second experiment we grew pedigreed ancestral and descendants genotypes to estimate FT heritability. Selection intensity was estimated from previous work on the same populations in the field.
Results
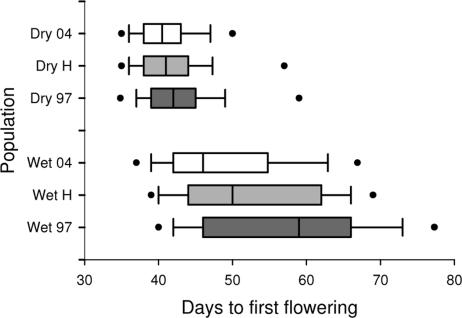
Descendant genotypes had earlier FT than ancestral genotypes, showing a shift to earlier flowering onset after the drought (Fig. 2). Failure-time analysis showed significantly earlier FT for descendants than ancestors across all treatments in both populations (Table 1 and SI Table 2). Under long season treatments (where near-zero mortality allows expression of the full range of FT genes present) mean FT advanced by 1.9 days in the Dry Site population and by 8.5 days in the Wet Site population (Fig. 2). The evolutionary rates, as measured in haldanes (change in mean per generation, in phenotypic standard deviation units), were 0.039 and 0.101 for the Dry and Wet Sites, respectively. The mean FT of the hybrids did not differ significantly from the mid-point between ancestors and descendants (Dry Site: n = 300, t = 1.02, df = 98, P = 0.30; Wet Site: n = 300, t = 0.40, df = 97, P = 0.69), which indicates an additive genetic basis for the observed change. In addition, FT was earlier for Dry Site than for Wet Site genotypes (Table 1 and SI Table 2), confirming earlier observations (22) and suggesting local adaptation.
Fig. 2.
Box plots of the evolution of time to first flowering. Shown are box plots of FT for B. rapa plants from the Wet Site (Wet) and Dry Site (Dry) populations from the ancestral 1997 × 1997 (97), descendant 2004 × 2004 (04), and hybrid (H) crosses in the long season treatment. Days to first flowering is the number of days between germination (first emergence of cotyledons) and first flowering (first full opening of first flower). The center bar is the median, and the boxes, lines, and dots represent the 25th to 75th, 10th to 90th, and 5th to 95th percentiles, respectively. Descendants flowered earlier than ancestors in both populations and across all treatments.
Table 1.
FT means
| Population | Watering treatment | FT |
χ2 | ||
|---|---|---|---|---|---|
| Ancestral | Hybrid | Descendant | |||
| Wet Site | Short | 53.4 ± 1.01 | 50.4 ± 0.93 | 46.2 ± 0.93 | 15.76∗∗∗ |
| Wet Site | Medium | 58.2 ± 0.97 | 53.3 ± 0.94 | 49.1 ± 0.90 | 13.71∗∗ |
| Wet Site | Long | 58.8 ± 0.87 | 54.1 ± 0.87 | 50.1 ± 0.86 | 55.88∗∗∗ |
| Dry Site | Short | 42.6 ± 0.88 | 41.3 ± 0.88 | 41.0 ± 0.87 | 14.54∗∗∗ |
| Dry Site | Medium | 42.2 ± 0.87 | 41.3 ± 0.88 | 41.4 ± 0.87 | 13.47∗∗ |
| Dry Site | Long | 42.8 ± 0.87 | 42.1 ± 0.87 | 41.0 ± 0.87 | 10.38∗∗ |
Shown are the least square means ± SE days to flowering (FT) of B. rapa in the Wet Site and Dry Site populations in the short, medium, and long season treatments for the ancestral (1997) and descendant (2004) collection lines and their hybrids. The χ2 value is from a failure-time analysis testing the effect of collection year for each population by treatment group.
∗P < 0.05;
∗∗P < 0.01;
∗∗∗P < 0.001
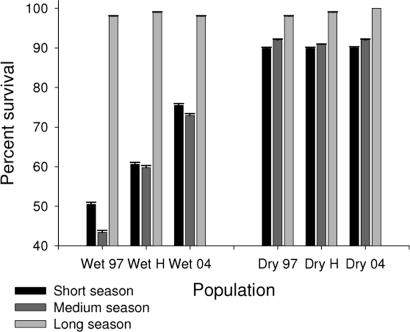
To determine whether descendants were better adapted than ancestors to abbreviated growing seasons, such as drought imposes, we examined survival and fecundity. Survival was nearly 100% in the long season treatment for all lines (Fig. 3). Under the drought-mimicking short season treatment, survival was 90% for the Dry Site population but only 62% for the Wet Site population (n = 600, χ2 = 53.8, df = 1, P < 0.0001), which is consistent with the hypothesis of local adaptation by these populations. Generations did not differ in survival for the Dry Site population but did for the Wet Site population (n = 300, χ2 = 33.5, df = 2, P < 0.0001), with survival under the short season treatment 50% for the ancestral line, 60% for the hybrids, and 75% in the descendant lines. Trends for fecundity were similar but were not statistically significant. Thus, the postdrought genotypes appear better adapted to short seasons than the predrought genotypes.
Fig. 3.
Local adaptation across space and time. Shown is the percentage (mean ± SE) of B. rapa plants surviving from Wet Site (Wet) and Dry Site (Dry) populations from ancestral 1997 (97), descendant 2004 (04), and hybrid (H) crosses in the short (black bars), medium (dark gray bars), and long (light gray bars) season treatments. Higher survival of postdrought (2004) than predrought (1997) genotypes under short season conditions in the Wet Site shows adaptation to recent conditions. Higher survival of Dry Site than Wet Site genotypes under the short season treatment shows adaptation to local conditions. Survival was analyzed with a categorical model using a linear response function.
Does the observed evolutionary change in FT in B. rapa conform to expectations based on the intensity of selection observed in the field (22)? This question can be addressed by comparing observed changes to those predicted from a standard model of quantitative genetics, the breeders equation: R = nSh2 (14) (see Materials and Methods). The variable R is the evolutionary response to selection (in this case the difference between the mean FT of the ancestral and descendant generations). The number of generations between ancestors and descendants is n, and S is the selection differential per generation, calculated as the covariance between relative fitness and FT (assuming that selection intensity is invariant). The last term, h2, is the heritability of FT, which is the proportion of phenotypic variance in FT explained by the additive effects of allelic differences among plants at loci contributing to FT. If all variance in FT is due to environmental factors, heritability is zero, and no evolutionary response is possible. High heritability and strong selection differentials cause rapid evolution (14).
We estimated heritability of FT in the ancestral generation using parent–offspring regression (14) (see Materials and Methods). In the 1997 generation h2 = 0.29 [95% confidence limit (CL) = 0.03–0.55] for the Dry Site population and h2 = 0.46 (95% CL = 0.23–0.68) for the Wet Site. Thus, nearly one-third to one-half of the total phenotypic variance in FT can be attributed to the additive genetic component, suggesting that change over a few generations of selection is possible.
By using data from a previous study (22) with the Dry and Wet site populations in the field, we estimated selection differentials on FT to be S = −4.02 days (95% CL = −6.28, −1.81) for the Dry Site population and S = −7.67 days (95% CL = −10.45, −5.24) for the Wet Site population during the 2003 season (negative values indicate selection for earlier FT).
Combining these greenhouse estimates for heritability and the field estimates for the 2003 selection differential, the expected evolutionary response over the seven generations in the Dry Site is predicted to be −8.2 days (95% CL = −0.03, −16.35) compared with the observed −1.9 days. In the Wet Site population the expected response is −24.7 days (95% CL = −9.61, −39.78) compared with the observed −8.5 days. Thus, on the assumption that selection in 2003 was representative for all seven generations, the observed response to selection is within the error limits of predictions for the Dry Site but less than predicted for the Wet Site.
Discussion
Our results provide evidence for a rapid, adaptive evolutionary shift in flowering phenology after a climatic fluctuation. The shift to earlier FT in B. rapa after only a few generations adds to growing evidence that evolution is not always a slow, gradual process but can occur on contemporary time scales in natural populations (7). The evolutionary rates in our study, although rapid, are within the range reported in a review of microevolution (23). The rate of change in FT we found was less than predicted based on heritabilities in the ancestral generation and on the selection differentials in the field in 2003. Selection intensity in 2003 could have been stronger than average for the 7 years between the 1997 and 2004 generations. The drought did not begin until 2000, so strong selection for accelerated flowering was probably imposed for only 5 of the 7 years. Additionally, heritability was measured under greenhouse conditions, where reduced environmental variability could inflate the estimate of this parameter. Finally, if a fraction of seeds have multiyear dormancy in the wild (i.e., the population has a seed bank) some of the 2004 plants would be fewer than seven generations removed from the 1997 plants, which means that selection would have had fewer opportunities to act. This possibility is under study.
The evolution of shorter FT during the drought agrees with predictions from life history theory (20, 21). Long season conditions should select for delayed flowering because plants that spend more time in exponential growth phase will accumulate disproportionately more resources and thus produce many more offspring once flowering begins. When seasons are short, however, plants that delay flowering are unable to mature seeds before conditions deteriorate lethally. Although plants can also cope with drought through tolerance (increasing water use efficiency), our results indicate that B. rapa adapted to this climate anomaly by an escape strategy of earlier FT.
We show evidence for both local adaptation over space and adaptation to recent conditions over time. Dry Site genotypes had higher fitness (survival times fecundity) than Wet Site genotypes under short season conditions. Thus, Dry Site genotypes are better adapted to faster-drying soils than Wet Site genotypes, which supports the hypothesis of local adaptation. Additionally, Wet Site genotypes from the postdrought generation had higher fitness under short seasons than genotypes from the predrought generation. Descendant genotypes from the wet site are thus better adapted to drought conditions than ancestral genotypes, which is evidence for adaptation to recent conditions. We did not detect the same increase in adaptation to short seasons in the Dry Site population, perhaps because genotypes at this site are already better adapted to drought conditions. This interpretation is supported by lower FT heritability and weaker selection at the Dry Site and by the greater evolutionary response in the Wet Site compared with the Dry Site genotypes.
If we had simply observed that FT was earlier in 2004 than it was in 1997, we would have been unable to determine whether this change represented a plastic response of individuals to new conditions or a genetically based evolutionary shift. Our experimental design “resurrected” dormant ancestors and raised them side-by-side with their descendants. Thus, by holding environmental conditions constant, this design allowed phenotypic differences between the generations to be attributed to evolutionary (i.e., genetic) change. Including hybrids in the design provided some information on the genetic architecture underlying evolutionary change.
Evolutionary adaptation to climate change may be especially important for the persistence of flowering plants because limited dispersal ability and habitat fragmentation may prevent plants from migrating with geographic shifts in favorable climate envelopes (24, 25). Although we found rapid evolution of FT in B. rapa, other species may be less successful in adapting to climate change, and evolution may be considerably slower for other traits. Populations with sufficient genetic variance for an evolutionary response may fail to adapt if abrupt environmental change depresses reproductive rates below replacement levels (26). Sustained and rapid climate change could deplete genetic variance faster than it can be replenished by mutation (27). Finally, antagonistic genetic correlations and trade-offs (28) between two or more selected traits may lead to an evolutionary stalemate between conflicting adaptive responses. In contrast to other traits, the evolution of flowering phenology may be particularly responsive to selection because variation in FT leads to assortative mating (29), which inflates genetic variance (14).
The selective impact of climate change will offer an unprecedented opportunity for exploring the adaptive process. By rearing ancestral, descendant, and hybrid generations of the same population across an array of environments, biologists will be able to measure the rate of evolutionary change, test its adaptive significance, and determine whether genetic constraints limit response. As the costs of sequencing and ancillary technologies continue to drop, it will become feasible to pinpoint the genetic basis for evolutionary change. With this in mind, we propose a coordinated effort to collect and store seeds and other suitable genetic materials from multiple populations of a number of species so that future biologists will have the materials they need to implement the “resurrection protocol” for a better understanding of evolutionary processes.
Materials and Methods
Study System.
B. rapa (syn. campestris) L. (Brassicaceae) is a weedy, self-incompatible winter annual that germinates October through January and flowers January through April in coastal California, where it was introduced ≈300 years ago. The study populations were Newport Harbor (Dry Site) and San Joaquin Marsh (Wet Site), both in Orange County, California, and located ≈3 km apart (22). Seeds were collected at 0.5-m intervals along transects in May 1997 and June 2004 from 400 to 1,800 plants per site.
Phase I: Amelioration of Environmental Effects.
To reduce seed condition, storage, and maternal effects, we produced F1 plants by planting field-collected seeds in the greenhouse at the University of California, Irvine. The resulting plants were assigned to 1997 × 1997, 1997 × 2004, and 2004 × 2004 crossing groups within each population. Germination rates of the F1 plants were >94% for all groups and did not differ by generation for either the Dry Site (df = 2, χ2 = 3.5, P = 0.18) or the Wet Site (df = 2, χ2 = 1.8, P = 0.41). For a subset (n = 36) of the seeds, there was also no difference between the generations in seed mass for the Dry Site (df = 2, F = 0.02, P = 0.88) or the Wet Site (df = 2, F = 0.00, P = 0.97). The creation of the F1 lines thus appears to have reduced differences in seed quality between the collection years.
Phase II, Experiment I: FT Evolution.
F1 seeds were stratified at 4°C on moist filter paper for 5 days and planted into 25-cm pots containing equal volumes of sand and potting medium in the greenhouse at the University of California, Irvine. Six plants were grown in each pot: one each from the ancestor, descendant, and hybrid generations from both populations. Pots were watered daily to saturation for 33 days. Water was withheld starting on day 33 for the short season, day 51 for the medium season, and day 81 for the long season treatments (SI Fig. 4). We applied 1.4 g of fertilizer (20-20-20 NPK; Grow More, Gardena, CA) per liter of tap water at planting and biweekly for 6 weeks. There were 100 replicates of each watering treatment by cross by population combination, for a total of 1,800 plants. Blocks contained four replicates of each treatment. Germination and flowering dates were recorded daily, and seed pods were collected when ripe. All flowers of all plants were pollinated by hand every 3 days. The effects of population, generation, watering treatment, block, and their interactions on fecundity (natural log seed weight) were assessed with a mixed-model ANOVA with block as a random effect. Degrees of freedom for the watering treatment effect were adjusted to account for the fact that the pot was the experimental unit.
Phase II, Experiment II: Heritability.
To obtain the heritability estimates, we planted 12–18 full-sibling progeny seeds from each 1997 × 1997 cross from Phase I into the same type of pots and soil as their parents. Dates of germination and first flowering were recorded. Experiments I and II were conducted simultaneously and in the same greenhouse.
Heritability for the ancestral generation was estimated by regressing family mean FT of the offspring in Phase II over the mid-parent FT in Phase I. The heritabilities are estimated by the regression coefficients (14). All FT values were normalized before analysis to remove the effects of different parent and offspring environments on trait variance.
Statistical Analysis.
We used failure-time analysis, also known as survival analysis (30), to test for genetic differences in FT among the ancestral, descendant, and hybrid generations. Failure-time analysis measures between-group differences in the time to a discrete event in an individual. This method is preferred for analyzing FT in plants because FT is often not normally distributed and because the analysis can accommodate censored values (30). Censoring occurred in this experiment because some of the plants in the short and medium season died from water deprivation late in the experiment before they had flowered. Thus, although the exact FT for these could not be recorded, it was known they had not flowered by the time of death. We used the LIFEREG procedure in SAS (version 9.1; SAS Institute, Cary, NC), testing a model that included the following factors: population (Dry Site and Wet Site), generation (ancestor, descendant, and hybrid), watering treatment (short, medium, and long), and all two- and three-way interactions. This analysis produces a Wald χ2 test statistic. The analysis specified a gamma distribution for the errors, and a Lagrange multiplier confirmed that the gamma fit the data better than the lognormal distribution. After a global analysis we performed separate secondary analyses for each population.
Heritability for the ancestral generation was estimated by regression of family mean FT of the F1 plants over the mid-parent FT (14). The heritabilities are equivalent to the regression coefficients. All offspring in this experiment were thoroughly watered daily, and minimal mortality occurred, so estimates did not have to account for missing values. All FT values were normalized before analysis to remove the effects of different parent and offspring environments on trait variance.
Confidence intervals for heritabilities and selection differentials were estimated by bootstrapping using 1,000 resampling iterations. Monte Carlo simulations were used to estimate confidence intervals for responses to selection. A set of 1,000 pairs of heritability and selection differential values were randomly generated based on the observed means and standard errors of these parameters. The product of the paired heritability and selection differential values were multiplied by seven (generations) to yield a set of simulated response to selection values. The means and standard errors of the responses to selection were calculated from the distribution of the simulated response values.
Supplementary Material
Acknowledgments
We thank the following people for help with data collection and plant maintenance: K. Afshar, V. Chandrasekaran, A. Dick, A. Franks, L. Gonzalez, C. Herman, L. Hua, E. Ko, T. Kossler, P. Le, K. Musser, A. Ng, M. Ngugen, A. Ogura, P. Rath, K. Torosian, P. Tran, W. Yang, A. M. Weis, and E. Weiss. D. Franke made the 1997 seed collections. Thoughtful comments on previous versions of the manuscript were provided by P. Morrel, C. Weinig, and M. T. Weis. M. Franks assisted with editing. This research was supported by National Science Foundation Grants DEB-0345030 and DEB-0440595 (to A.E.W.).
Abbreviations
- FT
flowering time
- CL
confidence limit.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608379104/DC1.
References
- 1.Parmesan C, Yohe GA. Nature. 2003;421:37–42. doi: 10.1038/nature01286. [DOI] [PubMed] [Google Scholar]
- 2.Both C, Bouwhuis S, Lessells CM, Visser ME. Nature. 2006;441:81–83. doi: 10.1038/nature04539. [DOI] [PubMed] [Google Scholar]
- 3.Barbraud C, Weimerskirch H. Proc Natl Acad Sci USA. 2006;103:6248–6251. doi: 10.1073/pnas.0510397103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peñuelas J, Filella I, Comas P. Global Change Biol. 2002;8:531–544. [Google Scholar]
- 5.McCarthy JJ, Canziani OF, Leary NA, Doken DJ, White KS. Climate Change 2001: Impacts, Adaptation, and Vulnerability. Cambridge, UK: Cambridge Univ Press; 2001. [Google Scholar]
- 6.Kinnison MT, Hendry AP. Genetica. 2001;112:145–164. [PubMed] [Google Scholar]
- 7.Reznick DN, Ghalambor CK. Genetica. 2001;112:183–198. [PubMed] [Google Scholar]
- 8.Rodríguez-Trelles F, Rodríguez MA. Evol Ecol. 1998;12:829–838. [Google Scholar]
- 9.Umina PA, Weeks AR, Kearney MR, Hoffmann AA. Science. 2005;308:691–693. doi: 10.1126/science.1109523. [DOI] [PubMed] [Google Scholar]
- 10.Levitan M, Etges WJ. BMC Evol Biol. 2005;5:4–15. doi: 10.1186/1471-2148-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradshaw WE, Holzapfel CM. Proc Natl Acad Sci USA. 2001;98:14509–14511. doi: 10.1073/pnas.241391498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Realé D, McAdam AG, Boutin S, Berteaux D. Proc R Soc London Ser B. 2003;270:591–596. doi: 10.1098/rspb.2002.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant PR, Grant BR. Science. 2002;296:707–711. doi: 10.1126/science.1070315. [DOI] [PubMed] [Google Scholar]
- 14.Lynch M, Walsh JB. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 15.Linhart YB, Grant MC. Annu Rev Ecol Syst. 1996;27:237–277. [Google Scholar]
- 16.Bennett AF, Lenski RE, Mittler JE. Evolution (Lawrence, Kans) 1992;46:16–30. doi: 10.1111/j.1558-5646.1992.tb01981.x. [DOI] [PubMed] [Google Scholar]
- 17.Hairston NG, Lampert W, Caceres CE, Holtmeier CL, Weider LJ, Gaedke U, Fischer JM, Fox JA, Post DM. Nature. 1999;401:446. doi: 10.1111/j.0014-3820.2001.tb00736.x. [DOI] [PubMed] [Google Scholar]
- 18.Hoerling M, Kumar A. Science. 2003;229:691–694. doi: 10.1126/science.1079053. [DOI] [PubMed] [Google Scholar]
- 19.McCabe GJ, Palecki MA, Betancourt JL. Proc Natl Acad Sci USA. 2004;101:4136–4141. doi: 10.1073/pnas.0306738101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohen D. Am Nat. 1976;110:801–807. [Google Scholar]
- 21.Kozlowski J. Trends Ecol Evol. 1992;7:15–19. doi: 10.1016/0169-5347(92)90192-E. [DOI] [PubMed] [Google Scholar]
- 22.Franke DM, Ellis AG, Dharjwa M, Freshwater M, Fujikawa M, Padron A, Weis AE. Int J Plant Sci. 2006;167:83–92. [Google Scholar]
- 23.Bone E, Farres A. Genetica. 2001;112:165–182. [PubMed] [Google Scholar]
- 24.Davis MB, Shaw RG. Science. 2001;292:673–679. doi: 10.1126/science.292.5517.673. [DOI] [PubMed] [Google Scholar]
- 25.Jump AS, Peñuelas J. Ecol Lett. 2005;8:1010–1020. doi: 10.1111/j.1461-0248.2005.00796.x. [DOI] [PubMed] [Google Scholar]
- 26.Gomulkiewicz R, Holt RD. Evolution (Lawrence, Kans) 1995;49:201–207. doi: 10.1111/j.1558-5646.1995.tb05971.x. [DOI] [PubMed] [Google Scholar]
- 27.Burger R, Lynch M. Evolution (Lawrence, Kans) 1995;49:151–163. doi: 10.1111/j.1558-5646.1995.tb05967.x. [DOI] [PubMed] [Google Scholar]
- 28.Etterson JR, Shaw RG. Science. 2001;294:151–154. doi: 10.1126/science.1063656. [DOI] [PubMed] [Google Scholar]
- 29.Weis AE, Kossler TM. Am J Bot. 2004;91:825–836. doi: 10.3732/ajb.91.6.825. [DOI] [PubMed] [Google Scholar]
- 30.Fox GA. In: Design and Analysis of Ecological Experiments. Scheiner SM, Gurevitch J, editors. Oxford: Oxford Univ Press; 2001. pp. 253–289. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.