Abstract
Endogenous activation of the aryl hydrocarbon receptor (AHR) is required for normal vascular development. This biology led us to investigate the interplay between the AHR and vascular physiology by using an in vitro model of fluid shear stress. Using this system, we show that fluid flow induces a robust AHR-mediated increase in CYP1 expression. Furthermore, we demonstrate that incubation with sheared bovine or human sera is sufficient for AHR activation, indicating that direct cellular exposure to shear stress is not required for this response. Fractionation of sera by size and density revealed the AHR-activating factor to be low-density lipoprotein (LDL). Purified LDL (0.1 mg/ml) from sheared sera induces a 6-fold increase in AHR-mediated signaling as compared with LDL purified from static sera. Similar results were obtained by exposing a purified fraction of LDL to fluid flow, suggesting that shear stress is capable of directly modifying LDL structure and/or function. In addition, we show that LDL can be converted to an AHR-activating species by conventional methods of lipoprotein modification, such as NaOCl oxidation. Finally, we demonstrate that an increased level of AHR-activating LDL is present in the sera of AHR null mice as compared with heterozygous littermates, suggesting a role for the Ahr locus in the physiological response to modified LDL in vivo. Overall, these data demonstrate a previously undescribed relationship between LDL modification and AHR biology and provide a potential explanation for the vascular abnormalities observed in AHR null mice.
Keywords: AHR, cyp1a1, cyp1b1, shear stress
The aryl hydrocarbon receptor (AHR) is a ligand activated basic-helix–loop–helix (bHLH) transcription factor that belongs to the PAS (Per-Arnt-Sim) family of nuclear sensors (1). The AHR binds to numerous environmental contaminants including tetrachlorodibenzo-p-dioxin (TCDD), benzo[a]pyrene, and coplanar polychlorinated biphenyls (2). Upon binding these xenobiotic ligands, the AHR translocates to the nucleus and dimerizes with a second bHLH-PAS protein known as the Ah receptor nuclear translocator (ARNT). The AHR–ARNT complex can then bind to dioxin response elements (DREs) found upstream of target genes, leading to their transcriptional up-regulation. The CYP1 family of cytochrome P450s, CYP1A1, CYP1A2 and CYP1B1, as well as the phase II enzymes GST-A1 and NQO1 are among the genes that are regulated by the AHR (2, 3). The enzymes encoded by these genes display metabolic activity toward many AHR ligands, prompting the idea that this pathway represents an adaptive response to xenobiotic exposure (2, 3).
In addition to its role in the metabolic response to xenobiotics, the AHR also has an important role in vascular development. Mice that harbor a null allele at the Ahr locus display a number of vascular phenotypes. These include a patent ductus venosus (DV), persistent hyaloid arteries in the eye, decreased hepatic perfusion, a confused vascularization in the corneal limbus, and cardiac hypertrophy (4–6). Experiments using Ahr and Arnt hypomorphic mice have demonstrated that, similar to its role in xenobiotic metabolism, the developmental role of AHR requires activation of the receptor and formation of an AHR/ARNT dimer complex (7, 8). Additionally, experiments using tissue-specific AHR null mice have demonstrated that AHR expression within endothelial and/or hematopoietic cells is necessary for closure of the DV (9). Overall, these findings are consistent with the idea that an endogenous activator exists for the AHR and that this signaling pathway is important in vascular physiology.
We were initially compelled to explore the role of shear stress in AHR biology following exploratory studies designed to elucidate AHR-regulated pathways in hemangioblast-lineage cells. In these preliminary gene expression studies, we identified a set of TCDD-inducible genes, which had been characterized as shear stress-inducible (B.J.M., unpublished observation). A subsequent survey of the literature revealed that classical AHR-regulated genes, including CYP1A1 and CYP1B1, have also been documented to be highly induced by fluid flow (10–13). Because of the key role of shear stress in vascular biology, we further explored the link between fluid flow and AHR signaling. These studies have led us to identify modified low-density lipoprotein (LDL) as an endogenous activator of the AHR. In this report, we show that LDL can be converted to an AHR-activating species via multiple routes, including fluid shear stress and NaOCl-induced modification.
Results
AHR Activation by Direct Cellular Exposure to Shear Stress.
In preliminary experiments, several cell lines were used to demonstrate the induction of CYP1 gene expression after fluid shear stress, which was applied using a parallel-plate flow system. The shear-induced levels of CYP1B1 were examined in primary human aorta endothelial cells as well as the murine EOMA endothelioma cell line; levels of CYP1A1 were measured in the human Hep3b and rat 5L hepatoma cell lines. We observed a >3.5-fold induction of CYP1 mRNA in all tested cell lines in response to 12 dyne/cm2 shear stress or 1 nM of the potent AHR agonist TCDD [supporting information (SI) Table 1].
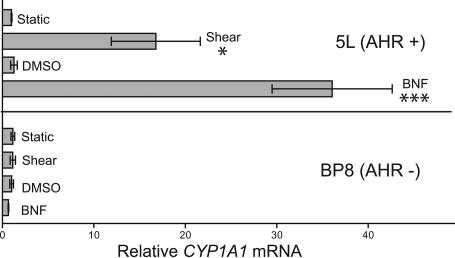
We next tested whether the CYP1A1 response to fluid shear stress depended on the AHR. Toward this end, we used the rat hepatoma cell line BP8, which is deficient in AHR expression, and its parental line 5L, which maintains a high level of AHR expression (14). Each cell line was exposed to 12 dyne/cm2 shear stress or static control conditions for 16 h. As a positive control, static cells were also treated with a 10 μM concentration of the AHR agonist β-napthoflavone (βNF) or DMSO control. The expression of CYP1A1 mRNA was then determined by quantitative RT-PCR (qRT-PCR). In the AHR-expressing 5L cells, induction of CYP1A1 mRNA was observed in response to both shear (17-fold) and βNF (36-fold) treatments (Fig. 1). Conversely, the AHR-deficient BP8 cell line exhibited no induction of CYP1A1 mRNA after either βNF treatment or shear stress (Fig. 1).
Fig. 1.
Shear stress induction of CYP1A1 depends on the AHR. The rat hepatoma 5L cell line and its derived AHR null line, BP8, were exposed in duplicate to 12 dyne/cm2 shear stress, 10 μM βNF, or control conditions for 16 h. CYP1A1 mRNA was quantitated by qRT-PCR. The relative change in mRNA expression was calculated as 2̂(ΔCT). Error bars indicate the minimum and maximum data points. This experiment was repeated, with virtually identical results. ∗, P < 0.05; ∗∗∗, P < 0.001.
AHR Activation by Sheared Serum.
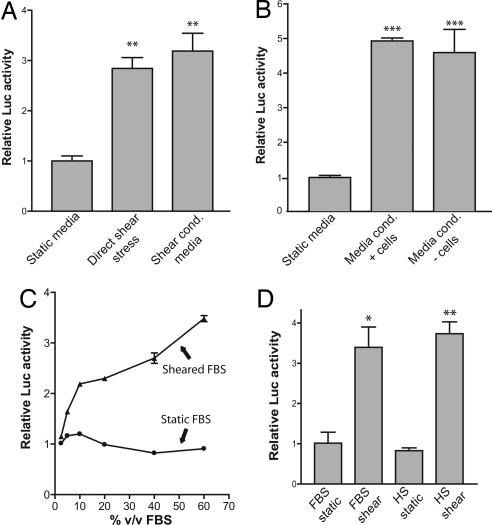
We next tested whether cellular exposure to shear stress resulted in the formation of an AHR activator that might be purified from the “conditioned” culture media. Toward this end, we used the H1L6.1c3 hepatoma cell line, which contains a stably integrated luciferase reporter that is driven by four upstream DREs (15). In the first part of the experiment, slides with confluent cells were placed in perfusion chambers and directly exposed to fluid shear stress. The growth medium “conditioned” by these sheared cells was then used to treat naïve, static cells. We observed that direct shear stress significantly increased DRE-driven luciferase activity (Fig. 2A, direct shear stress). Importantly, we also found that the shear-conditioned media induced a 3-fold increase in luciferase activity, similar to that observed in directly sheared cells. (Fig. 2A, shear-conditioned media).
Fig. 2.
Sheared serum induces DRE-mediated transcription. (A) The H1L6.1c3 cell line, which harbors a stably integrated DRE-driven luciferase reporter, was grown to confluency on fibronectin-coated slides. In the first part of the experiment, cells (n = 3) were directly exposed to fluid shear stress (direct shear stress) or static conditions (static media) for 24 h. The growth medium “conditioned” by this experiment was then used to treat confluent slides of H1L6.1c3 cells for 24 h. (B) Media were conditioned by shear stress by using perfusion chambers that contained slides with confluent Hepa1c1c7 cells (media conditioned + cells) or slides without cells (media conditioned − cells) for 8 h. Each medium was then used to treat H1L6.1c3 cells (n = 3) for 20 h. (C) A DMEM solution supplemented with 60% vol/vol FBS (DMEM+FBS) and a nonsupplemented DMEM solution were sheared for 2 h. After exposure to shear stress, static FBS was added to the sheared DMEM solution to 60% of total volume. Dilutions of the sheared DMEM+FBS (triangles) and sheared DMEM + static FBS (circles) were used to treat H1L6.1c3 cells (n = 3) for 20 h. (D) HS and FBS were exposed to shear stress for 2 h. H1L6.1c3 cells (n = 3) were treated with a 20% vol/vol (v/v) dilution of each serum for 20 h and then assayed for luciferase activity. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P 0.001.
We then tested whether the conditioning of the culture medium depended on cellular exposure to shear stress. Toward this end, we set up two independent perfusion chamber systems. Setup “A” had a perfusion chamber complete with a slide of confluent Hepa1c1c7 mouse hepatoma cells. Setup “B” used a perfusion chamber constructed with a slide that did not have cells. After exposure to shear stress, the medium from each setup was removed and used to treat static H1L6.1c3 cells. We found both the media from setup A (media conditioned + cells) and from setup B (media conditioned − cells) induced luciferase activity (Fig. 2B). The absence of cells within the perfusion chamber did not alter the capacity of sheared media to activate AHR signaling.
To determine the source of the shear-activated factor, unsupplemented DMEM and DMEM containing FBS were exposed to shear stress. Static FBS was then added to the sheared, unsupplemented DMEM. Dilutions were made of each solution with untreated DMEM and used to treat H1L6.1c3 cells. The sheared DMEM+FBS solutions significantly induced DRE-driven luciferase activity at all concentrations ≥5% vol/vol serum as compared with matched controls that were treated with sheared DMEM containing static FBS (P < 0.001) (Fig. 2C). Furthermore, we observed a dose-dependent relationship between the concentration of sheared FBS and the induction of DRE-mediated luciferase activity. To determine the time frame of serum activation, aliquots of culture media were removed at different time points after exposure to shear stress. Each media aliquot was then used to treat H1L6.1c3 cells to assess its ability to activate the AHR. A significant change (P < 0.05) in sera activity was observed in as little as 5 min (SI Fig. 6). Maximal activation was observed after ≥2 h of shear stress (SI Fig. 6).
We next wanted to confirm that sheared serum could induce the transcription of endogenous AHR target genes. To facilitate the treatment of cells with high concentrations of sera, static and sheared sera were concentrated to 1/10 their original volume by using a centrifugal dialysis unit (Amicon) with a 10-kDa molecular mass (MW) limit. The concentrated sera were then diluted in DMEM to the equivalent of 20% or 100% vol/vol serum and used to treat static Hepa1c1c7 cells. The relative levels of Cyp1a1 mRNA were determined by qRT-PCR. The expression of Cyp1a1 was significantly induced by sheared serum as compared with static serum controls at both 20% and 100% vol/vol concentrations (2.6- and 5.8-fold, respectively) (SI Fig. 7).
To determine whether the response to sheared serum was specific to FBS, human serum (HS) was acquired and exposed to fluid flow. DMEM was then supplemented with static or sheared HS and used to treat H1L6.1c3 cells. Static and sheared FBS were used as controls. We found that treatment with sheared HS induced DRE-driven luciferase activity >3-fold when compared with static HS controls (Fig. 2D). The induction of luciferase activity by sheared HS was nearly identical to that seen with sheared FBS.
AHR Activation by Sheared LDL.
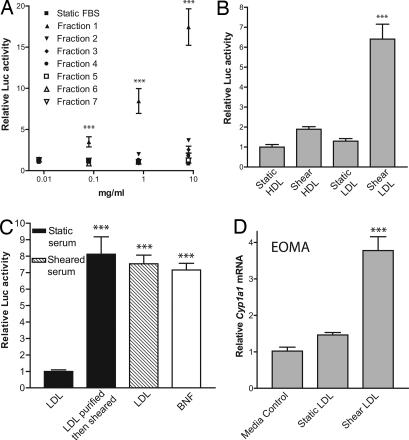
To identify the serum component(s) responsible for AHR activation, FBS was sheared and then separated into fractions by MW using a Superose 6 gel-filtration HPLC column. Seven different eluent fractions were collected and concentrated, with fraction 1 being the highest-MW components and fraction 7 being the lowest-MW components. Dilutions of each fraction were made in DMEM and used to treat H1L6.1c3 cells. We found that fraction 1 activated AHR signaling to a much greater degree than any other fraction (Fig. 3A). At the highest concentration tested, 8 mg/ml, we observed an 18-fold increase in DRE-mediated signaling. Similar results were observed with human serum (data not shown).
Fig. 3.
The AHR is activated by sheared LDL. (A) Sheared FBS was separated by MW using a Superose 6 gel-filtration resin. Seven total fractions were collected and used to treat H1L6.1c3 cells (n = 3) at the indicated concentrations for 18 h. (B) Static and sheared sera were fractionated by sequential density ultracentrifugation. H1L6.1c3 cells (n = 6) were treated for 20 h with 0.1 mg/ml HDL or LDL from each serum and then assayed for luciferase activity. (C) LDL was purified from static serum (black bars) or from serum exposed to shear stress for 18 h at 4C (hatched bar). An aliquot of LDL purified from the static serum was then diluted in PBS and exposed to the same shear stress conditions. Each LDL treatment group was then used to treat H1L6.1c3 cells (n = 5) for 20 h. Cells were treated with 2 μM βNF as a positive control. (D) The EOMA cell line (n = 4) was treated with 0.1 mg/ml static or sheared LDL or an equal volume of PBS for 20 h. The relative level of Cyp1a1 mRNA in each treatment group was determined by using qRT-PCR. ∗∗∗, P < 0.001.
To test the hypothesis that the active serum factor was a lipoprotein complex, LDL (1.006 < d < 1.063) and HDL (1.063 < d < 1.21) fractions were isolated from sheared and static FBS by sequential density ultracentrifugation. We did not detect VLDL (very low-density lipoprotein) (d < 1.006) in the commercially prepared FBS as measured by BCA assay. Each lipoprotein fraction was used to treat H1L6.1c3 cells in serum-free DMEM. The LDL fraction derived from sheared serum induced a significant, 6-fold increase in luciferase activity as compared with the LDL fraction from static serum (Fig. 3B).
We next tested whether purified LDL could be modified to an AHR-activating species by exposure to shear stress. First, we isolated LDL from sheared and static sera. An aliquot of LDL purified from the static serum was then diluted in PBS and exposed to shear stress. We then treated the H1L6.1c3 cell line with LDL that had been exposed to shear stress in whole serum or LDL that had been exposed to shear stress as a purified complex in PBS. Both sheared LDL (shLDL) samples induced a significant increase in luciferase activity as compared with static LDL (Fig. 3C). The luciferase activity induced by 0.1 mg/ml shLDL was similar to that induced by 2 μM βNF.
The effect of fluid flow on LDL activity was further confirmed by using a cone-plate viscometer (16) and human LDL, which was purchased commercially. Purified LDL was diluted to a 0.1 mg/ml concentration in DMEM and exposed to ≈18 dyne/cm2 fluid shear stress for 1 h in the cone-plate system. H1L6.1c3 cells were then incubated in the static or sheared DMEM/LDL solutions for 20 h. The LDL samples exposed to fluid shear stress were found to induce a significant, 3.3-fold increase in DRE-mediated signaling as compared with static controls (SI Fig. 8). Similar results were obtained by exposing human LDL to fluid flow in our standard parallel-plate system (data not shown).
The capacity of shLDL to activate AHR signaling was then tested in additional cell lines. The endothelial-lineage EOMA cell line was treated with 0.1 mg/ml static LDL or shLDL for 8 h and then assayed for Cyp1a1 expression by using qRT-PCR. Incubation with shLDL was found to induce a significant, 3-fold increase in Cyp1a1 mRNA as compared with static controls (Fig. 3D). shLDL was also found to induce a significant increase in DRE-mediated signaling in the human hepatoma 101L cell line (data not shown).
AHR Activation by NaOCl-Modified LDL.
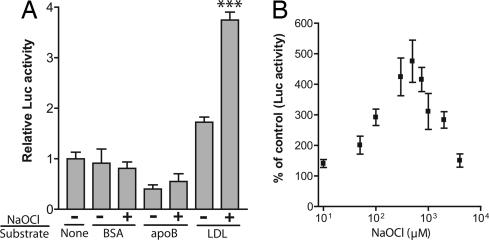
We next asked whether the AHR-mediated response to LDL was specific to flow-induced modification. To test whether conventional forms of modified LDL also induce AHR signaling, static LDL was preincubated with 100 μM NaOCl, dialyzed, and then used to treat H1L6.1c3 cells. As controls, BSA and apoB were also incubated with NaOCl to determine whether the response was specific to LDL. We observed that NaOCl-modified LDL, but not apoB or BSA, induced a significant increase in DRE-mediated signaling (Fig. 4A). Next, we treated H1L6.1c3 cells with LDL, which was preincubated with a range of NaOCl concentrations from 10–4,000 μM. The maximal DRE-mediated response to NaOCl-modified LDL (≈5-fold over untreated LDL) was observed at a concentration of 500 μM NaOCl (Fig. 4B). A significant response (P < 0.01) was observed from 50–2,000 μM NaOCl. No change in cell viability was observed over this concentration range, as measured by trypan blue exclusion (data not shown).
Fig. 4.
The AHR is activated by NaOCl-modified LDL. (A) LDL, BSA, and apoB (all 0.2 mg/ml in PBS) were preincubated in the presence or absence of 100 μM NaOCl for 1 h at 37°C. After extensive dialysis of each sample, H1L6.1c3 cells (n = 4) were treated with 0.1 mg/ml of each sample in serum-free DMEM for 18 h. Statistical comparisons were performed between each NaOCl-modified substrate and its untreated control. (B) LDL was preincubated with the indicated concentration of NaOCl for 1 h at 37°C; a control sample of LDL was incubated without NaOCl. After extensive dialysis, each LDL sample (0.1 mg/ml) was used to treat H1L6.1c3 cells (n = 4) for 18 h. ∗∗∗, P < 0.001.
Sera from AHR Null Mice Contain AHR-Activating LDL.
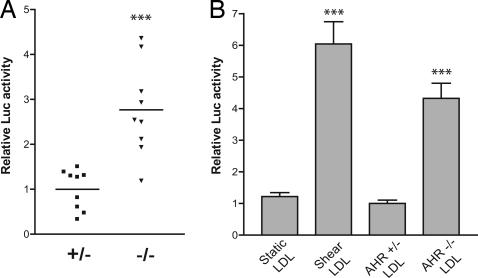
The observation that modified LDL activates the AHR signaling pathway prompted us to examine whether in vivo AHR deficiency results in increased levels of AHR-activating LDL. Toward this end, sera were isolated from nine Ahr+/− or Ahr−/− mice and used to treat H1L6.1c3 cells. We found that the sera derived from Ahr−/− mice induced a significant 2.8-fold increase in mean luciferase activity as compared with Ahr+/− sera (Fig. 5A). To determine whether the activity of null sera was mediated by LDL, we purified LDL from a pool of Ahr−/− and Ahr+/− littermates. The LDL fraction derived from each AHR genotype was then used to treat H1L6.1c3 cells. As a control, cells were treated with LDL purified from static or sheared FBS. The LDL purified from Ahr−/− mice was found to induce a 4-fold increase in DRE-mediated signaling as compared with LDL purified from Ahr+/− sera or static FBS (Fig. 5B).
Fig. 5.
AHR null sera contains AHR-activating LDL. (A) Sera were isolated from nine age-matched AHR null (triangles) or heterozygous mice (squares) and used to treat H1L6.1c3 cells (n = 3) at a 30% vol/vol concentration for 20 h. The horizontal line represents the mean luciferase activity of each group. (B) LDL was purified from the sera of Ahr−/− and Ahr+/− littermates. H1L6.1c3 cells (n = 6) were treated with 0.1 mg/ml LDL from each mouse genotype or with LDL purified from static or sheared FBS for 20 h and then assayed for luciferase activity. ∗∗∗, P < 0.01.
Discussion
Although AHR null mice display a number of vascular defects, the role of AHR signaling in vascular biology is largely unknown. Recent experiments have demonstrated that activation of endothelial and/or hematopoietic AHR is required for normal vascular development (7), but neither the mechanism of receptor activation nor the role of AHR-mediated transcription in vascular biology is understood. We were initially compelled to explore the role of the AHR in shear stress biology after a screen for novel AHR target genes. In this preliminary experiment, we identified previously characterized shear-responsive genes as TCDD-inducible (B.J.M., unpublished observation). A further survey of the literature revealed that the classical AHR-regulated genes CYP1A1 and CYP1B1 are also highly inducible by shear stress (11–13, 17). The overlap between shear and TCDD-induced gene expression changes led us to hypothesize that the AHR could act as a sensor for a shear-induced signal and mediate a transcriptional response to altered fluid flow in the vasculature.
To study the mechanism behind these transcriptional responses to fluid flow, we first used a parallel-plate model of fluid shear stress. The fluid shear stress produced by this flow system is comparable with the levels of shear in the human arterial system (18) and within the range found to induce CYP1 gene expression (12, 13, 17). We first verified that our shear system could replicate the CYP1 response to fluid flow by using endothelial and hepatoma cell lines derived from several mammalian species (SI Table 1). All cell lines tested were found to be responsive to shear stress, indicating that hepatoma cells, in addition to primary vascular endothelial cells, contain all necessary signaling components for the CYP1 response to shear stress. Using an AHR-deficient rat hepatoma cell line and its AHR-expressing parental line, we found that CYP1A1 induction by shear stress completely depended on the AHR (Fig. 1). These findings are consistent with the results of Mufti et al. (19) using an AHR-deficient murine cell line in a microcarrier-attached suspension culture system. Moreover, we demonstrated that a promoter fragment containing four canonical DREs was sufficient to induce reporter activity after cellular exposure to shear stress (Fig. 2A). Together, these data demonstrate that shear stress activates AHR-regulated transcription in a classical DRE-dependent manner.
In contrast to the conventional paradigm of shear-induced cell signaling, we found that activation of the AHR did not depend on direct exposure of cells to fluid flow (Fig. 2). We initially hypothesized that exposure to shear stress could result in the intracellular production of an AHR agonist that might be purified from the conditioned culture media. Although shear-conditioned media did indeed activate AHR signaling, we found that this conditioning process was completely independent of the presence of directly sheared cells (Fig. 2B). Concurrent with these studies, Eskin et al. (17) noted a similar response to shear-conditioned culture media in primary endothelial cells. These independent findings strongly suggested that AHR activation by sheared media was a real, reproducible phenomenon. To determine the source of the activator, we exposed culture media ±FBS to fluid flow and found that AHR activation strictly depended on the concentration of sera present during shearing (Fig. 2C). In a similar manner, we found that shear stress also produces an AHR activator in adult human sera (Fig. 2D). These findings demonstrate that a flow-induced modification occurs in human and bovine as well as fetal and adult sera.
We next set out to identify the factor(s) that mediate AHR activation by sheared serum. As an initial purification step, we fractionated serum by MW using a Superose 6 gel-filtration column. We found that serum activity was contained exclusively in the fraction of highest MW in both human and bovine serum (Fig. 3A and B.J.M., unpublished observations). A search of the literature revealed that this gel resin is routinely used to purify the high-MW lipoprotein complexes present within the early eluent fractions (20). A series of HPLC purifications additionally revealed that the active serum fraction was extremely hydrophobic and exhibited a similar SDS/PAGE pattern to apolipoprotein isoforms (B.J.M., unpublished observations). These observations led us to test the hypothesis that AHR activation by sheared serum was mediated by a modification to lipoproteins. Toward this end, we used a conventional method of lipoprotein purification and tested each fraction for the ability to induce AHR signaling. These experiments revealed that LDL purified from sheared, but not static, serum induced robust AHR signaling in multiple cell lines and species (Fig. 3 B and D and B.J.M., unpublished observations).
We then asked whether the shear-induced change in LDL activity was due to a direct modification of the lipoprotein complex. Toward this end, we used multiple flow systems to expose purified LDL (both bovine and human) to fluid shear stress. We found that LDL was modified to an AHR-activating species independent of other serum factors (Fig. 3C and SI Fig. 8). These data are the first evidence that aspects of the cellular response to shear stress can be mediated through a direct effect on LDL. Moreover, these data strongly suggest that physiological levels of fluid shear are capable of directly modifying LDL structure and/or function. We hypothesize that these effects are directly related to the ability of fluid shear stress to induce misfolding and aggregation of large, hydrophobic sera factors, such as von Willebrand Factor and amyloid precursors (21–23).
The modification of LDL structure/function has been extensively studied due to considerable evidence linking modified LDL, most notably oxidized LDL, to atherosclerosis (24). Numerous treatments have been demonstrated to misfold LDL structure and lead to aggregation, including oxidation, glycation, acetylation, proteolysis, lipolysis, and vortexing (25, 26). Notably, these independent modifications often induce overlapping cellular responses (25). Therefore, we asked whether conventional methods of LDL modification could result in activation of the AHR, similar to that observed with shLDL. To test this hypothesis, we treated cells with LDL that was preincubated with NaOCl, which is commonly used to model LDL oxidation by the monocyte myeloperoxidase pathway (27). We found that NaOCl-modified LDL produced robust activation of the AHR (Fig. 4). In addition, preliminary results indicate that acetylation of LDL is also sufficient for the induction of AHR activity (unpublished observation). It is worth noting, however, that AHR activation appears to depend highly on the degree of LDL modification. Extensive levels of oxidation or fluid agitation, such as vortexing, do not result in an AHR-activating species (Fig. 4B and B.J.M., unpublished observations).
Overall, these results suggest that the AHR pathway can be activated by multiple routes of LDL modification. The mechanism through which multiple “types” of modified LDL result in AHR activation is unclear. We speculate that activation could occur through either a direct or indirect mechanism. In the former scenario, modification could result in increased intracellular delivery of an LDL-derived [pro]-agonist that directly binds to the AHR. Conversely, AHR activation could be the result of an indirect cell response to the modification of LDL structure, e.g., intracellular release of cAMP (28) or arachidonic acid (AA) that results in the downstream production of an AHR agonist. Notably, the flow-induced release of AA has been implicated in the CYP1 response to fluid flow via inhibitors of AA metabolism (29). However, we have not been able to replicate these results and, in some cases, have observed an opposite response (B.J.M., unpublished observation).
In this report, we have demonstrated that LDL can be modified to an AHR-activating species using in vitro models of endogenous vascular physiology. To further demonstrate the relationship between the AHR and LDL biology, we set out to establish that AHR-activating LDL is endogenously produced. Ex vivo detection of AHR-activating LDL, however, is complicated by the presence of numerous physiological pathways designed to prevent the systemic accumulation of toxic, modified LDL (30, 31). We hypothesized, however, that defects in the adaptive, cellular response to AHR-activating LDL would result in increased circulating levels of this factor. Moreover, we speculated that AHR deficiency could constitute such a defect. Both paraoxonase-1, an HDL-associated enzyme with a well established role in preventing LDL oxidation and cardiovascular disease, and NQO1, a shear-inducible gene that maintains the endogenous lipid-soluble antioxidants α-tocopherol-hydroquinone and ubiquinol in their reduced and active forms, are directly regulated by AHR activation (12, 32–34). In addition, exogenous activation of the AHR is sufficient for the induction of the macrophage “foam cell” phenotype (35), a lipid-laden form generated by uptake of modified LDL from the sera (36).
These data, as well as other information, led us to hypothesize that AHR signaling could constitute a nonredundant pathway, which serves to attenuate the accumulation of AHR-activating LDL. According to this hypothesis, exposure to chronic in vivo conditions would result in the increased accumulation of AHR-activating LDL in the sera of AHR null mice, which could then be detected ex vivo. To test this hypothesis, we isolated sera and LDL from AHR null and heterozygous mice and tested their capacity to activate AHR signaling in vitro. We found that both whole sera and purified LDL derived from AHR null mice induced robust AHR-mediated signaling. Sera and LDL obtained from mice with a functional AHR signaling pathway, however, had minimal effect (Fig. 5). These data indicate that AHR-activating LDL can be produced endogenously and support a role for the Ahr locus in mediating an adaptive response to this physiological activator.
Conclusions
Prior studies from our laboratory and others have indicated that an endogenous activator exists for the AHR and that this pathway is important in vascular physiology. In this report, we have identified modified LDL as a previously uncharacterized activator of AHR signaling. LDL can be modified to an AHR-activating species via multiple routes, including fluid shear stress or NaOCl. Moreover, we have shown that AHR deficiency is linked to increased levels of AHR-activating LDL in vivo. Overall, these data support a nonredundant role for the AHR in mediating an adaptive, cellular response to modified LDL, a critical factor in vascular disease.
Materials and Methods
Fluid Shear Stress Experiments.
Cells were exposed to fluid shear stress in a parallel-plate perfusion chamber purchased from Glycotech (Gaithersburg, MD) using a silicon gasket of 0.025-cm height. A Cellmax Quad system (Spectrum Laboratories, Rancho Dominguez, CA) was used to supply pulsatile, laminar fluid flow at setting “4” with a dual “long-pin” setup. These settings corresponded to a 10 ml/min flow rate through the provided length of tubing coupled in series to two perfusion chambers. Fluid shear stress was calculated by using the following equation: τw = 6 μQ/a2b, where τw is wall shear stress in dynes/cm2, μ is fluid viscosity (H2O at 37°C = 0.0076), Q is flow rate in ml/s, a is channel height, and b is channel width. The void volume of the apparatus was ≈40 ml.
Sheared Sera Experiments.
Sera were exposed to fluid flow by using the conditions described above. Unless otherwise noted, serum was exposed to fluid flow overnight (16–20 h).
Lipoprotein Isolation Experiments.
Lipoproteins were purified from sera by sequential density ultracentrifugation, as described (37). Briefly, sera were sequentially adjusted to 1.006, 1.063, and 1.21 mg/ml and ultracentrifuged to obtain very low-density lipoprotein, LDL, and high-density lipoprotein fractions, respectively. The lipoprotein fractions were concentrated and buffer exchanged with PBS by using Amicon Ultra-15 units with a 100-kDa MW limit (Millipore, Billerica, MA). Lipoprotein purity was assayed by nondenaturing acrylamide electrophoresis.
Cone-Plate Viscometer.
Human LDL (Intracel, Frederick, MD) was diluted to a 0.1 mg/ml concentration in DMEM. A 5-ml volume of DMEM/LDL was exposed to 18 dyne/cm2 shear stress for 1 h by using a cone-plate viscometer provided by Andrew Greene (Medical College of Wisconsin), as described (16). A static, control solution of LDL was incubated in an identical setup without rotation of the cone.
NaOCl Modification.
The concentration of reagent-grade NaOCl (Fisher, Hampton, NH) was determined spectrophotometrically by using a molar absorption coefficient for OCl− of 350 cm−1 at 292 nm; NaOCl was then diluted to a working concentration in 1× PBS and adjusted to pH 7.4. LDL, apoB (Biodesign, Saco, ME), or BSA (all 0.2 mg/ml) were incubated in PBS, pH 7.4, for 1 h at 37°C in the presence or absence of NaOCl. Each sample was then extensively dialyzed against PBS plus 0.01% wt/vol EDTA by using Amicon filter units.
Mouse Serum.
Sera were obtained by retroorbital bleed from 12- to 16-week-old female Ahr+/− or Ahr−/− mice, which have been described (38). All mice were fasted for 24 h before serum isolation. The purified LDL experiments were performed by using sera obtained from a pool of female 16-week-old Ahr−/− or Ahr+/− (both n = 5) mice derived from Ahr+/−(female) × Ahr−/− (male) breeding pairs.
Figures.
All data are presented as the mean ± SEM, unless otherwise noted. Statistical significance is indicated as follows: ∗, P < 0.05; ∗∗, P < 0.01 ∗∗∗, P < 0.001.
Statistical Analysis.
The activity of Ahr+/− and Ahr−/− sera was analyzed with a two-tailed t test. All other experiments were analyzed by using a one-way ANOVA with a Neuman–Keuls multiple-comparison test.
Online SI.
Cell lines and tissue culture, qRT-PCR, luciferase assay, size fractionation of sheared sera, and determination of protein concentration are described in SI Methods.
Supplementary Material
Acknowledgments
We thank Susanne McMillan for help in manuscript preparation, Ed Glover for performing retroorbital bleeds, Drs. Naomi Chesler and Ron Magness for valuable conversations relating to fluid shear stress, and the laboratory of Dr. Andrew Greene for use of the cone-plate viscometer. This work was supported by National Institutes of Health Grants R37-ES05703, T32-CA009135, and P30-CA014520.
Abbreviations
- AHR
aryl hydrocarbon receptor
- βNF
β-napthoflavone
- DRE
dioxin response element
- HS
human serum
- LDL
low-density lipoprotein
- MW
molecular mass
- qRT-PCR
quantitative RT-PCR
- shLDL
sheared LDL
- TCDD
tetrachlorodibenzo-p-dioxin.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607296104/DC1.
References
- 1.Gu Y-Z, Hogenesch J, Bradfield C. Annu Rev Pharmacol Toxicol. 2000;40:519–561. doi: 10.1146/annurev.pharmtox.40.1.519. [DOI] [PubMed] [Google Scholar]
- 2.Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. J Biol Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- 3.Schrenk D. Biochem Pharmacol. 1998;55:1155–1162. doi: 10.1016/s0006-2952(97)00591-1. [DOI] [PubMed] [Google Scholar]
- 4.Lahvis G, Pyzalski RW, Glover E, Pitot HC, McElwee MK, Bradfield CA. Mol Pharmacol. 2005;67:714–720. doi: 10.1124/mol.104.008888. [DOI] [PubMed] [Google Scholar]
- 5.Lahvis G, Lindell S, Thomas R, McCuskey R, Murphy C, Glover E, Bentz M, Southard J, Bradfield C. Proc Natl Acad Sci USA. 2000;97:10442–10447. doi: 10.1073/pnas.190256997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thackaberry EA, Gabaldon DM, Walker MK, Smith SM. Cardiovasc Toxicol. 2002;2:263–274. doi: 10.1385/ct:2:4:263. [DOI] [PubMed] [Google Scholar]
- 7.Walisser JA, Bunger MK, Glover E, Harstad EB, Bradfield CA. J Biol Chem. 2004;279:16326–16331. doi: 10.1074/jbc.M400784200. [DOI] [PubMed] [Google Scholar]
- 8.Walisser JA, Bunger MK, Glover E, Bradfield CA. Proc Natl Acad Sci USA. 2004;101:16677–16682. doi: 10.1073/pnas.0404379101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walisser JA, Glover E, Pande K, Liss AL, Bradfield CA. Proc Natl Acad Sci USA. 2005;102:17858–17863. doi: 10.1073/pnas.0504757102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Zhu Y, Rannou F, Lee TS, Formentin K, Zeng L, Yuan X, Wang N, Chien S, Forman BM, Shyy JY. Circulation. 2004;110:1128–1133. doi: 10.1161/01.CIR.0000139850.08365.EC. [DOI] [PubMed] [Google Scholar]
- 11.McCormick SM, Eskin SG, McIntire LV, Teng CL, Lu CM, Russell CG, Chittur KK. Proc Natl Acad Sci USA. 2001;98:8955–8960. doi: 10.1073/pnas.171259298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dekker RJ, van Soest S, Fontijn RD, Salamanca S, de Groot PG, VanBavel E, Pannekoek H, Horrevoets AJ. Blood. 2002;100:1689–1698. doi: 10.1182/blood-2002-01-0046. [DOI] [PubMed] [Google Scholar]
- 13.Mufti NA, Bleckwenn NA, Babish JG, Shuler ML. Biochem Biophys Res Commun. 1995;208:144–152. doi: 10.1006/bbrc.1995.1316. [DOI] [PubMed] [Google Scholar]
- 14.Gottlicher M, Cikryt P, Wiebel FJ. Carcinogenesis. 1990;11:2205–2210. doi: 10.1093/carcin/11.12.2205. [DOI] [PubMed] [Google Scholar]
- 15.Rushing SR, Denison MS. Arch Biochem Biophys. 2002;403:189–201. doi: 10.1016/s0003-9861(02)00233-3. [DOI] [PubMed] [Google Scholar]
- 16.Rieder MJ, Carmona R, Krieger JE, Pritchard KA, Jr, Greene AS. Circ Res. 1997;80:312–319. doi: 10.1161/01.res.80.3.312. [DOI] [PubMed] [Google Scholar]
- 17.Eskin SG, Turner NA, McIntire LV. Endothelium. 2004;11:1–10. doi: 10.1080/10623320490432434. [DOI] [PubMed] [Google Scholar]
- 18.Cunningham KS, Gotlieb AI. Lab Invest. 2005;85:9–23. doi: 10.1038/labinvest.3700215. [DOI] [PubMed] [Google Scholar]
- 19.Mufti NA, Bleckwenn NA, Babish JG, Shuler ML. Biochem Biophys Res Commun. 1995;208:144–152. doi: 10.1006/bbrc.1995.1316. [DOI] [PubMed] [Google Scholar]
- 20.de Silva HV, Mas-Oliva J, Taylor JM, Mahley RW. J Lipid Res. 1994;35:1297–1310. [PubMed] [Google Scholar]
- 21.Hill EK, Krebs B, Goodall DG, Howlett GJ, Dunstan DE. Biomacromolecules. 2006;7:10–13. doi: 10.1021/bm0505078. [DOI] [PubMed] [Google Scholar]
- 22.Siedlecki CA, Lestini BJ, Kottke-Marchant KK, Eppell SJ, Wilson DL, Marchant RE. Blood. 1996;88:2939–2950. [PubMed] [Google Scholar]
- 23.Shankaran H, Alexandridis P, Neelamegham S. Blood. 2003;101:2637–2645. doi: 10.1182/blood-2002-05-1550. [DOI] [PubMed] [Google Scholar]
- 24.Chisolm GM, Steinberg D. Free Radical Biol Med. 2000;28:1815–1826. doi: 10.1016/s0891-5849(00)00344-0. [DOI] [PubMed] [Google Scholar]
- 25.Aviram M. Atherosclerosis. 1993;98:1–9. doi: 10.1016/0021-9150(93)90217-i. [DOI] [PubMed] [Google Scholar]
- 26.Khoo JC, Miller E, McLoughlin P, Steinberg D. Arteriosclerosis. 1988;8:348–358. doi: 10.1161/01.atv.8.4.348. [DOI] [PubMed] [Google Scholar]
- 27.Hazell LJ, Stocker R. Biochem J. 1993;290:165–172. doi: 10.1042/bj2900165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oesch-Bartlomowicz B, Huelster A, Wiss O, Antoniou-Lipfert P, Dietrich C, Arand M, Weiss C, Bockamp E, Oesch F. Proc Natl Acad Sci USA. 2005;102:9218–9223. doi: 10.1073/pnas.0503488102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mufti NA, Shuler ML. Biotechnol Prog. 1996;12:847–854. doi: 10.1021/bp960067j. [DOI] [PubMed] [Google Scholar]
- 30.Niki E, Noguchi N. Mol Cell Biochem. 2002;234–235:19–25. [PubMed] [Google Scholar]
- 31.Murphy JE, Tedbury PR, Homer-Vanniasinkam S, Walker JH, Ponnambalam S. Atherosclerosis. 2005;182:1–15. doi: 10.1016/j.atherosclerosis.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 32.Nioi P, Hayes JD. Mutat Res. 2004;555:149–171. doi: 10.1016/j.mrfmmm.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 33.Mackness B, Hine D, Liu Y, Mastorikou M, Mackness M. Biochem Biophys Res Commun. 2004;318:680–683. doi: 10.1016/j.bbrc.2004.04.056. [DOI] [PubMed] [Google Scholar]
- 34.Gouedard C, Barouki R, Morel Y. Mol Cell Biol. 2004;24:5209–5222. doi: 10.1128/MCB.24.12.5209-5222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogel CF, Sciullo E, Matsumura F. Cardiovasc Toxicol. 2004;4:363–373. doi: 10.1385/ct:4:4:363. [DOI] [PubMed] [Google Scholar]
- 36.Shashkin P, Dragulev B, Ley K. Curr Pharm Des. 2005;11:3061–3072. doi: 10.2174/1381612054865064. [DOI] [PubMed] [Google Scholar]
- 37.Bronzert TJ, Brewer HB., Jr Clin Chem. 1977;23:2089–2098. [PubMed] [Google Scholar]
- 38.Schmidt JV, Su GH, Reddy JK, Simon MC, Bradfield CA. Proc Natl Acad Sci USA. 1996;93:6731–6736. doi: 10.1073/pnas.93.13.6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.