Abstract
p-Hydroxyphenylacetate hydroxylase from Acinetobacter baumannii is a two-component system consisting of a NADH-dependent FMN reductase and a monooxygenase (C2) that uses reduced FMN as substrate. The crystal structures of C2 in the ligand-free and substrate-bound forms reveal a preorganized pocket that binds reduced FMN without large conformational changes. The Phe-266 side chain swings out to provide the space for binding p-hydroxyphenylacetate that is oriented orthogonal to the flavin ring. The geometry of the substrate-binding site of C2 is significantly different from that of p-hydroxybenzoate hydroxylase, a single-component flavoenzyme that catalyzes a similar reaction. The C2 overall structure resembles the folding of medium-chain acyl-CoA dehydrogenase. An outstanding feature in the C2 structure is a cavity located in front of reduced FMN; it has a spherical shape with a 1.9-Å radius and a 29-Å3 volume and is interposed between the flavin C4a atom and the substrate atom to be hydroxylated. The shape and position of this cavity are perfectly fit for housing the oxygen atoms of the flavin C4a-hydroperoxide intermediate that is formed upon reaction of the C2-bound reduced flavin with molecular oxygen. The side chain of His-396 is predicted to act as a hydrogen-bond donor to the oxygen atoms of the intermediate. This architecture promotes the nucleophilic attack of the substrate onto the terminal oxygen of the hydroperoxyflavin. Comparative analysis with the structures of other flavoenzymes indicates that a distinctive feature of monooxygenases is the presence of specific cavities that encapsulate and stabilize the crucial hydroperoxyflavin intermediate.
Keywords: enzyme catalysis, flavin, structural enzymology, oxygen reactivity
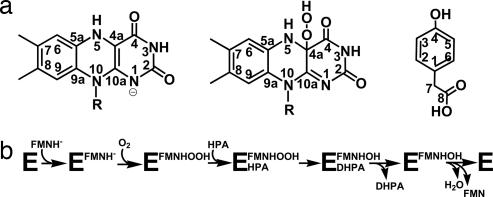
Flavoprotein monooxygenases use dioxygen to insert an oxygen atom into a substrate and have been found to be involved in a wide variety of biological reactions (1–3). The fundamental property of these enzymes is their ability to promote formation and stabilization of the C4a-hydroperoxyflavin (Fig. 1a) resulting from the reaction of the protein-bound reduced flavin with dioxygen. This key intermediate donates an oxygen atom to the substrate, generating the unstable C4a-hydroxyflavin that eliminates one molecule of water to yield oxidized flavin (5). Understanding the structural bases governing functional properties of monooxygenases is crucial to address one of the most fascinating issues in flavoenzymology: the ability of flavoenzymes to differentially react with molecular oxygen.
Fig. 1.
Overview of substrate and reaction properties of C2. (a) Structures of fully reduced flavin, C4a-hydroperoxyflavin and HPA; the face of flavin shown here is the re side. The pKa value of the two-electron reduced flavin is 6.5, and many flavoenzymes are known to preferentially bind the anionic form of the reduced cofactor (4). On these bases, we have assumed that the C2-bound flavin is in the anionic state. (b) Scheme of the catalytic cycle of HPA hydroxylase C2 (7).
In recent years, a new group of flavoprotein monooxygenases has been identified. These enzymes consist of two components: a reductase generating reduced flavin and a hydroxylase using reduced flavin to catalyze substrate monooxygenation (6). p-hydroxyphenylacetate hydroxylase from Acinetobacter baumannii catalyzes hydroxylation of p-hydroxyphenylacetate (HPA) to 3,4-dihydroxyphenylacetate (Fig. 1a). HPA hydroxylase has unusual features in both sequence and catalysis. The smaller reductase component of HPA hydroxylase (C1) performs HPA-stimulated NADH-dependent reduction of free FMN, which is subsequently transferred to the larger monooxygenase component (C2) and used for reaction with dioxygen and HPA monooxygenation (Fig. 1b). Specificity for FMN is conferred by C1, whereas C2 works equally well with both reduced FMN (FMNH−; Fig. 1a) and reduced FAD (7–10). C2 can effectively stabilize the C4a-hydroperoxyflavin intermediate for minutes, and, at high concentration of HPA, a stable dead-end complex between C4a-hydroxyflavin and HPA is observed.
Here, we present crystal structures of C2 in the apoenzyme form and of its complexes with FMNH− (C2:FMNH−) and HPA (C2:FMNH−:HPA). Structural analysis reported here provides a framework for the mechanisms of catalysis and reveals structural features that account for the stability of C4a-hydroperoxyflavin intermediate in monooxygenases.
Results
Overall Structure.
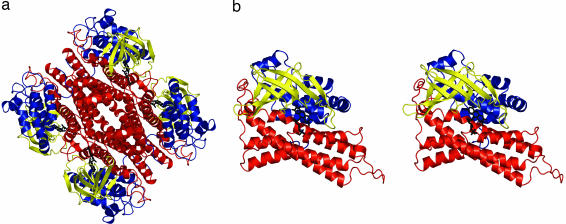
The C2 component of HPA hydroxylase from A. baumannii is a tetramer of four identical subunits, each with 422 aa (Fig. 2a). The tetrameric structure is in accordance with the apparent molecular weight of the native enzyme as observed by size-exclusion chromatography (8). The four crystallographically independent monomers are essentially identical (rms deviations between equivalent Cα atoms <0.2 Å). Each C2 monomer is made of three domains: the N-terminal domain, the β-sheet domain, and the C-terminal domain (Fig. 2b). Extensive interactions between the subunits of the tetramer are mediated by helical bundles of the C-terminal domains.
Fig. 2.
Overall structure of C2. (a) Ribbon diagram of the 222 symmetric C2 tetramer. N-terminal domains (residues 24–143) are in blue, β-sheet domains (144–237) are in yellow, and C-terminal domains (238–422) are in red. FMNH− is in black sticks. (b) Ribbon stereo diagram of the C2 monomer. This view is obtained from rotating the top monomer of a by 45° around the vertical axis. Produced with PyMOL [DeLano, W. L. (2002) www.pymol.org].
A search of the Protein Data Bank (11) with the program DALI (12) identified pig medium-chain acyl-CoA dehydrogenase as the closest structural homolog (rms deviation of 2.8 Å for 356 Cα atoms with 15% sequence identity), followed by Fusarium oxysporum nitroalkane oxidase (rms deviation of 3.1 Å for 362 Cα atoms with 15% sequence identity). The topological similarity extends to the quaternary structure because all of them are homotetrameric. Acyl-CoA dehydrogenases (13) and nitroalkane oxidase (14) belong to the class of dehydrogenases/oxidases and, therefore, do not have a close functional relationship to C2. An accurate comparison between C2 and these structural homologs reveals that the main differences are restricted to the loops lining the flavin- and substrate-binding sites. The folding topology of C2 is unrelated to that exhibited by other flavin-dependent monooxygenases/hydroxylases of known three-dimensional structure, such as p-hydroxybenzoate hydroxylase (15), yeast flavin-containing monooxygenase (16), and phenylacetone monooxygenase (17).
FMNH− Binding.
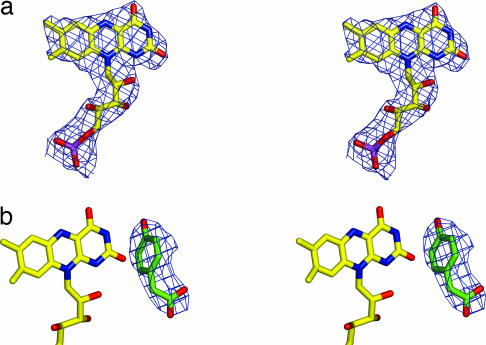
At the position where medium-chain acyl-CoA dehydrogenase binds the catalytic portion of FAD (13), the structure of the apo form of C2 displays a wide pocket filled by ordered water molecules. The structure of the binary C2:FMNH− complex obtained by crystal soaking (Fig. 3a) shows that FMNH− binds in this pocket (Fig. 2b). The binding site lies between the β-sheet and C-terminal domains and is completed by residues from the C-terminal domain of a neighboring subunit, indicating that the tetrameric arrangement is required for enzyme function. The rms deviation between the Cα atoms of the apoenzyme and binary complex structures is 0.23 Å. This finding implies that only minor conformational changes occur upon FMNH− binding with a tilt of the indole moiety of Trp-169 as the most evident one. The isoalloxazine ring is buried in the protein, whereas the phosphate group and ribityl 2′ OH and 3′ OH are solvent-accessible (Fig. 4). With such an arrangement, the additional AMP moiety of FAD would easily hang out in the solvent or rest on the tetramer surface; this observation explains why both FMNH−and FADH− work equally well in the hydroxylation reaction (7, 9).
Fig. 3.
Crystallographic data for the C2:FMNH− (a) and C2:FMNH−:HPA (b) complexes. The stereo pictures show the fitting of the FMNH− and HPA atoms of the refined models in the unbiased 4-fold averaged electron-density maps (1σ contour level). Carbon is shown in yellow (flavin) or green (HPA), nitrogen is shown in blue, oxygen is shown in red, and phosphorus is shown in purple.
Fig. 4.
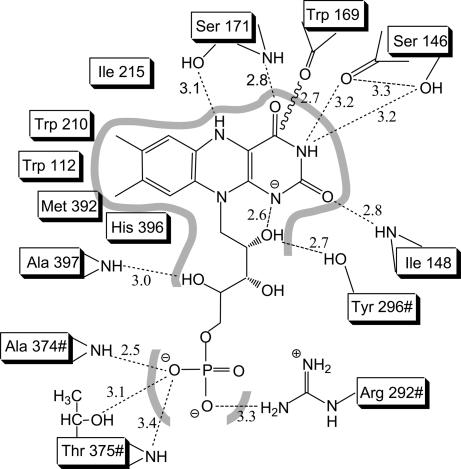
Interactions between FMNH− and C2. Distances are in Å and refer to subunit A of C2:FMNH−:HPA structure; # indicates residues from subunit D.
FMNH− is embedded in a protein scaffold formed by several stretches of residues: 112–116, 146–148, 169–171, 210–220, 292–296#, 374–375#, and 392–397 (where # indicates residues from a different subunit). As generally observed in flavoenzymes, the flavin is kept in position by a network of hydrogen bonds and hydrophobic interactions (Fig. 4). An unusual feature is the backbone carbonyl of Trp-169 that points toward the isoalloxazine plane at 2.7 Å from its C4, making it impossible for a hydroperoxy adduct to protrude on the si side of the flavin. Trp-169 side chain is approximately coplanar with the isoalloxazine and is responsible for making the si side of flavin solvent-inaccessible. A corresponding tryptophan is conserved with similar orientation in medium-chain acyl-CoA dehydrogenase and nitroalkane oxidase (13, 14) and may represent a signature residue for the flavoenzymes sharing this folding topology.
C2 binds FMNH− more tightly than oxidized FMN with a 200-fold difference in Kd (7). Consistently, no FMN bound to crystals has been observed when oxidized FMN was used in cocrystallizations. No clear structural explanation for C2 preference for reduced cofactor arises from structural analysis. It appears that, rather than a single structural element, the overall balance of interactions between protein and flavin is responsible for the differential binding affinity.
Substrate Binding.
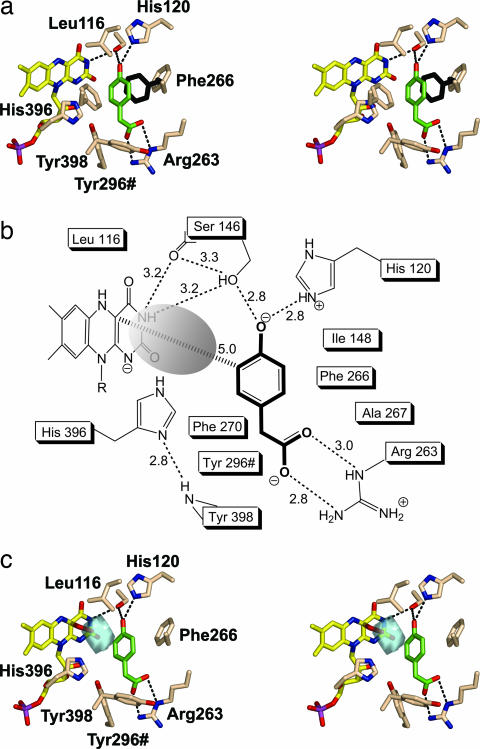
Crystals of the ternary C2:FMNH−:HPA complex were obtained by soaking preformed crystals in a solution containing FMNH− and HPA. The electron-density map reveals the presence of an additional peak, clearly consistent with a bound HPA molecule (Fig. 3b). The presence of substrate rather than product most likely reflects a very low oxygen concentration created by the oxygen-scavenging effect of FMNH− in the soaking solution. HPA binds on the re side of the flavin, and its aromatic ring lies on a plane roughly perpendicular to the plane of the flavin (Fig. 5a). The virtual projection of the aromatic ring of HPA on the flavin ring falls immediately aside the pyrimidine moiety, over the region of O2 atom. The shortest HPA-flavin distance is 3.6 Å between HPA C3 and flavin C2, whereas the distance between HPA C3 and flavin C4a (i.e., between the substrate atom to be hydroxylated and the flavin locus where the C O bond of the adduct forms) is 5.0 Å (Figs. 1a and 5b). The substrate is inaccessible to solvent, surrounded by a cluster of nine hydrophobic residues and kept in position by hydrogen bonds established by its two hydrophilic substituents. Several residues interact with both HPA and FMNH− (Tyr-296#, Ser-146, His-396, Leu-116, and Ile-148), and FMNH− is directly involved in HPA binding (Fig. 5b). These features are consistent with the biochemical observation that HPA can bind to the enzyme only after FMNH− binding (7) (Fig. 1b).
O bond of the adduct forms) is 5.0 Å (Figs. 1a and 5b). The substrate is inaccessible to solvent, surrounded by a cluster of nine hydrophobic residues and kept in position by hydrogen bonds established by its two hydrophilic substituents. Several residues interact with both HPA and FMNH− (Tyr-296#, Ser-146, His-396, Leu-116, and Ile-148), and FMNH− is directly involved in HPA binding (Fig. 5b). These features are consistent with the biochemical observation that HPA can bind to the enzyme only after FMNH− binding (7) (Fig. 1b).
Fig. 5.
The structure of C2:FMNH−:HPA complex. (a) Stereo image of the active site of monomer A with HPA bound. The color code is as in Fig. 3 except for protein carbons (pale pink). The position of Phe-266 before HPA binding is shown in black. For clarity, some residues of the substrate pocket and labels of Ser-146 and Phe-270 are not shown. (b) Interactions of the protein-bound HPA. (c) Modeling of a flavin C4a-hydroperoxide adduct. The figure shows a stereo image of the cavity (cyan) in front of flavin C4a and the surrounding residues in the C2:FMNH−:HPA complex. The cavity in the active site was detected by using the program VOIDOO (18) with a sphere of 1.2-Å radius as probe. The distal oxygen atom of the adduct fits within the cavity at 3.1 Å from the substrate atom to be hydroxylated (C3 of HPA). Substrate HPA is shown in green. Ser-146 label is not shown for the sake of clarity.
Comparison between the structures of C2, C2:FMNH− and C2:FMNH−:HPA illustrates an interesting feature. In the apoenzyme and binary complex structures, Phe-266 side chain occupies the place of the HPA ring; upon substrate binding, Phe-266 swings out changing its χ1 angle from +140° to −140° and creating the HPA pocket (Fig. 5a). This movement is the only clear structural variation induced by substrate binding. There is no evidence for domain motions to allow HPA access to its binding site. Rather, admission of the substrate into the active site probably involves movement of the Arg-263 side chain that directly interacts with the HPA carboxylate group and might act as a gate to the active site (Fig. 5 a and b).
The Cavity in Front of Flavin C4a and Oxygen Reaction.
An outstanding feature of the C2 structure is a well defined cavity identified in the binary and ternary complexes, located just in front of the pyrimidine ring of FMNH−. The cavity has an approximately spherical shape, with a 1.9-Å radius and a 29-Å3 volume (Fig. 5 b and c) and is interposed between flavin C4a and substrate C3 atoms. The cavity is outlined by the ribityl 2′ OH, the isoalloxazine, and the side chains of Trp-112, Leu-116, His-120, Ser-146, Ile-148, Phe-266, Phe-270, His-396, and Tyr-296#; its size is perfectly fit for housing the two additional oxygen atoms of the C4a-hydroperoxyflavin. A modeling experiment in which a model of the C4a-hydroperoxyflavin has been superimposed to FMNH− in the ternary complex, shows that the distal oxygen of the flavin intermediate would be located within the cavity at 3.1 Å from HPA C3 (Fig. 5c).
Discussion
Implications for Catalysis.
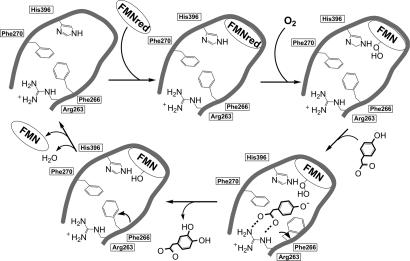
Analysis of the crystal structures provides a framework for the catalytic events in the C2 active site (7). A preorganized binding site (top left in Fig. 6) recruits free reduced FMN (produced by HPA hydroxylase C1 component in vivo). The protein-bound reduced cofactor reacts with molecular oxygen to produce C4a-hydroperoxy-FMN (top right in Fig. 6), which is housed in the C4a cavity. Subsequently, the HPA substrate binds to the enzyme, inducing the movement of the Phe-266 side chain. The HPA hydrophilic groups are contacted through Ser-146, His-120, and Arg-263, whereas the substrate C3 atom is positioned at ≈ 3 Å from the distal oxygen of C4a-hydroperoxy-FMN (bottom right in Fig. 6). These stereochemical features in the interactions between the hydroperoxyflavin and the aromatic substrate promote substrate monooxygenation that occurs in a solvent-protected environment. After releasing the 3,4-dihydroxyphenylacetate product, C4a-hydroxy-FMN (bottom left in Fig. 6) eliminates H2O, and the resulting FMN can dissociate from the enzyme, which is ready to undergo a new cycle.
Fig. 6.
Scheme for the catalytic events in the C2 active site.
The position of His-396 in the active site is intriguing. The Nδ1 atom of its side chain is H-bonded to the backbone nitrogen of Tyr-398 (Fig. 5b), whereas its Nε2 atom is perfectly positioned to act as a hydrogen-bond donor to either the proximal and/or the distal oxygen atoms of the C4a-hydroperoxyflavin (Fig. 5c). Moreover, the Nε2 atom can communicate with the solvent through the 2′ OH group of the FMN ribityl chain (Fig. 5a). These observations suggest that His-396 can have an active role in catalysis, possibly acting as proton donor in the reaction between reduced flavin and oxygen that leads to the formation of the C4a-hydroperoxyflavin intermediate (Fig. 6). Likewise, His-396 might be involved in the protonation of the C4a-hydroxyflavin that forms after substrate monooxygenation.
In flavoprotein monooxygenases, the first step in the reaction of reduced flavin with oxygen is the formation of a caged radical pair, leading to oxidized flavin through the formation of either flavin C4a-peroxide or C4a-hydroperoxide (2, 3, 5). The former case is observed in Baeyer–Villiger monooxygenases (such as cyclohexanone monooxygenase) (19) or bacterial luciferase (20) in which the flavin C4a-peroxide acts as a nucleophile attacking the substrate. On the contrary, a flavin C4a-hydroperoxide has been well documented in flavin-containing monooxygenases (21) and aromatic hydroxylases (22–24), in which the terminal oxygen of hydroperoxyflavin acts as an electrophile in an aromatic substitution reaction. The three-dimensional structure suggests that an electrophilic aromatic substitution mechanism is likely to occur also in the C2 reaction. The hydrogen bonds between HPA O4 and two side chains (Ser-146 and His-120) could favor preferential binding of the deprotonated form of the substrate (Fig. 5b), which would be more reactive than the protonated form in promoting the electrophilic attack by the C4a-hydroperoxyflavin (25) (Fig. 6).
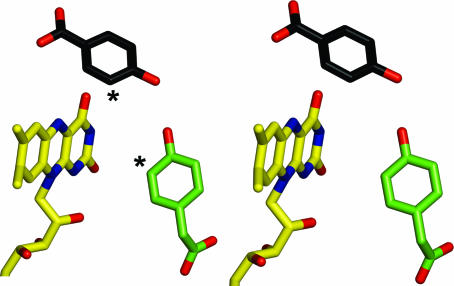
An electrophilic aromatic substitution mechanism has been shown to occur in p-hydroxybenzoate hydroxylase, a single-component flavin-dependent monooxygenase that catalyses a reaction similar to that carried out by C2 (the hydroxylation of 4-hydroxybenzoate) (22). Inspection of the three-dimensional structures shows that the geometry of substrate-binding site is very different in the two enzymes. In C2, the substrate faces the re side of the flavin (Fig. 5a) and is in direct contact with the C2, O2, and N3 atoms of FMNH−. Conversely, in p-hydroxybenzoate hydroxylase (15) the substrate binds on the edge of the flavin ring and interacts with the O4, N5, and C6 atoms of the cofactor (Fig. 7; see Fig. 1 for atomic numbering). However, in both binding modes, the C3 atom of the substrate (HPA in C2 and p-hydroxybenzoate in p-hydroxybenzoate hydroxylase) is properly positioned to be hydroxylated by the C4a-hydroperoxyflavin intermediate. It is remarkable that enzymes that catalyze the same type of reaction by using a similar mechanism are so different in the geometry of substrate–flavin interactions and active-site architecture.
Fig. 7.
The different substrate-binding modes in p-hydroxybenzoate hydroxylase and C2. To compare the two enzymes, the flavin ring atoms of p-hydroxybenzoate hydroxylase (PDB ID code 1PBE) were superposed on the equivalent FMN atoms of the C2:FMNH−:HPA complex. The resulting rotation and translation operators were applied to p-hydroxybenzoate hydroxylase coordinates. The carbon atoms of p-hydroxybenzoate of p-hydroxybenzoate hydroxylase are in black, whereas the carbons of the flavin ring and HPA of C2 are shown in yellow and green, respectively. The asterisks indicate the sites of hydroxylation of the two substrates.
Structural Bases of Oxygen Reactivity.
C2 is a very effective hydroxylase with a second-order rate constant of ≈106 M−1·s−1 for the reaction with dioxygen. Binding of HPA decreases the rate constant to ≈5 × 104 M−1·s−1 (7). A possible explanation for this effect is that protein-bound HPA, precisely fitting among nine hydrophobic side chains (Fig. 5b), hampers access of dioxygen to the cavity in front of C4a. In the absence of HPA, molecular oxygen may have easier access to the cavity by exploiting the inherent flexibility of Phe-266 side chain. This sort of protection exerted by HPA may also account for the stability of the dead-end complex that forms when HPA binds to the C2:C4a-hydroxyflavin (7).
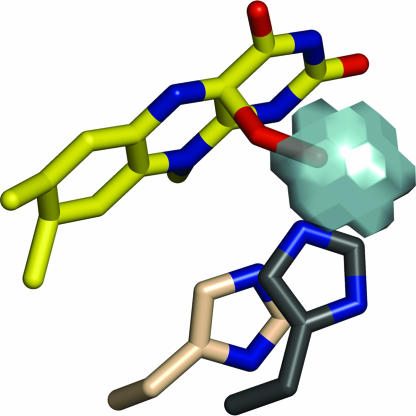
The presence of the cavity in front of C4a is a key feature providing a basis to understand stabilization of C4a-hydroperoxyflavin. The cavity is a precisely packed hydrophobic cage that has the dual role of providing a hydrogen-bond donor (His-396) and creating a solvent-free environment to prevent rapid breakdown of the unstable flavin intermediate (Fig. 5c). Cavities in proximity to flavin have been observed in other monooxygenases. In phenylacetone monooxygenase (17), a Baeyer–Villiger monooxygenase, a cavity is located on the re side of the flavin; a flexible arginine side chain is proposed to stabilize the peroxyflavin intermediate. In p-hydroxybenzoate hydroxylase (15, 26), 3-hydroxybenzoate hydroxylase (27) and tryptophan 7-halogenase (28) a cavity on the re side of flavin houses the oxygen atoms of the hydroperoxyflavin. Conversely, a survey of known structures of flavoprotein oxidases, in which occurrence of a hydroperoxyflavin intermediate is typically not detected, reveals that these enzymes do not exhibit similar C4a cavities (29). In this regard, inspection of the structure of glucose oxidase (30) can be particularly insightful in that this oxidase shares with C2 a considerably high reaction rate constant (≈106 M−1·s−1) and the presence of a histidine residue close to the flavin (31). However, the His side chain in glucose oxidase is shifted with respect to the flavin, leaving no specific cavity for housing a flavin C4a-adduct (Fig. 8). On the basis of these considerations, a specific feature of monooxygenases appears to be the presence of a cavity with optimum geometry for encapsulating and stabilizing the C4-hydroperoxyflavin. Molecular oxygen is forced to approach flavin C4a inside this cavity, where the flavin intermediate can be effectively stabilized by properly positioned hydrogen-bonding groups.
Fig. 8.
Comparison between the active sites of C2 and glucose oxidase, a fast oxygen-reacting oxidase that does not stabilize flavin C4a-hydroperoxide. A histidine side chain located in front of the flavin is present in both enzymes (His-516 in glucose oxidase and His-396 in C2), although with shifted position with respect to the flavin C4 locus. The different positioning of the active site His residues is visualized by superimposing the flavin ring atoms of glucose oxidase (PDB ID code 1GAL) on the equivalent atoms of C2. The picture shows His-516 of glucose oxidase (carbons in dark gray) and the model for the C4a-hydroperoxyflavin and His-396 of C2 (carbons in pale pink). Site-directed mutagenesis studies have shown that the protonated form of His-516 is largely responsible for the high reaction rate of oxygen with the reduced flavin in glucose oxidase (31).
Materials and Methods
Crystallization.
A. baumannii C2 protein (native and selenomethionine-substituted) was expressed and purified by using the protocols published in refs. 8 and 9. with slight modifications [see supporting information (SI) Text]. Crystals of both wild-type and selenomethionine-substituted protein were obtained by using the sitting-drop vapor-diffusion method at 20°C, by mixing equal volumes of protein solution [5 mg/ml in 30 mM Mops (pH 7.0)/1 mM DTT/20 mM FMN/2 mM HPA] and a reservoir solution containing 0.7 M sodium citrate and 0.1 M sodium Hepes (pH 7.5). Despite the high FMN concentration used in crystallization, the initial electron-density maps showed neither FMN nor HPA bound to the protein, reflecting the low affinity of C2 for FMN (7). Considering that the Kd for binding to C2 is 1.2 ± 0.2 μM for FMNH− and 250 ± 50 μM for oxidized FMN (7), a soaking protocol involving the use of the reductase C1 component (10) to generate FMNH− was established. Soaking of C2 crystals in a stabilizing solution containing 4 μM C1, 0.5 mM FMN, 1 mM HPA, and 2 mM NADH allowed us to solve the structures of the binary C2:FMNH− complex (soaking time 5 min) and ternary C2:FMNH−:HPA complex (soaking time 30 min). The longer soaking time required to obtain the C2:FMNH−:HPA complex is likely to reflect the fact that binding of the substrate occurs only after binding of FMNH− (7) (Fig. 1b).
Structure Determination.
Data were collected at the European Synchrotron Radiation Facility (Grenoble, France) and processed with the CCP4 package (32, 33). Despite the identical crystallization conditions, the native and selenomethionine-substituted proteins crystallize in different crystal forms (Table 1). The structure was solved by the single-wavelength anomalous diffraction method. Selenium sites were found by using SHELXD (35), and phasing was performed with SHARP (34). The resulting electron-density map was used in molecular replacement calculations to solve the structure of the native enzyme. Phases were further improved by multiple-crystal density averaging with the program DMMULTI (32) that produced electron density maps of excellent quality. All of the subsequent model building and refinement calculations were done by using data from the native crystals. The initial model was traced with ARP/wARP (36), whereas the programs COOT (37) and Refmac5 (38) were used for model rebuilding and refinement. These programs were also used for crystallographic analysis of the structures of the enzyme complexes. Details for data collection and processing are given in Tables 1 and 2.
Table 1.
Data collection and phasing statistics
| Selenomethionine apoenzyme* | Apoenzyme | C2:FMNH− | C2:FMNH−:HPA | |
|---|---|---|---|---|
| Space group | I4122 | I222 | I222 | I222 |
| a, b, c, Å | 182.0, 182.0, 236.4 | 91.4, 182.1, 287.3 | 94.3, 183.4, 284.4 | 92.0, 181.3, 286.2 |
| Asymmetric unit | 3 monomers | 4 monomers | 4 monomers | 4 monomers |
| Resolution, Å | 3.0 | 2.3 | 2.8 | 2.8 |
| Rsym†,‡ | 0.20 (0.53) | 0.09 (0.48) | 0.09 (0.47) | 0.10 (0.45) |
| Completeness,† % | 100 (100) | 99.1 (94.8) | 99.5 (98.8) | 100 (100) |
| Redundancy† | 28.8 (29.6) | 4.6 (3.5) | 4.9 (4.2) | 5.1 (5.1) |
| I/σ(I)† | 23.0 (4.2) | 12.8 (1.9) | 12.3 (1.7) | 13.3 (2.9) |
| Unique reflections | 3,9943 | 105,406 | 60,763 | 59,270 |
| 〈FOM〉 (centric/acentric)§ | 0.2/0.4 | — | — | — |
| Phasing power | 1.8 | — | — | — |
—, Not applicable.
*High Rsym value for the selenomethionine data was due to the considerable redundancy of the data set and a partial decay of the crystal during data collection. Phasing calculations including all data led to maps that were of higher quality compared with those calculated by including more homogenous subsets of reflections with lower Rsym values. The selenomethionine protein exhibits the same quaternary structure (a 222 symmetric tetramer; Fig. 2a) as the native enzyme. The asymmetric unit of selenomethionine crystals contains three protein monomers; one monomer is part of a tetramer whose subunits are related by the crystallographic twofold axes, whereas the other two monomers are part of a tetramer whose subunits are related by noncrystallographic and crystallographic twofold axes that are orthogonal to each other.
†Data for the highest resolution shell are given in parentheses.
‡Rsym = ΣhΣi I(hkl) − 〈I(hkl)〉 /ΣhΣiI(hkl).
§FOM, overall figure of merit calculated by the program SHARP (34).
Table 2.
Refinement statistics
| Apoenzyme* | C2:FMNH−* | C2:FMNH−:HPA* | |
|---|---|---|---|
| Resolution, Å | 2.3 | 2.8 | 2.8 |
| R-factor† | 0.218 (0.323) | 0.250 (0.346) | 0.217 (0.282) |
| Rfree† | 0.235 (0.347) | 0.287 (0.376) | 0.235 (0.297) |
| No. of nonhydrogen protein atoms | 12,472 | 12,516 | 12,516 |
| No. of water molecules | 375 | — | 26 |
| No. of substrate atoms | — | — | 4 × 11 |
| No. of FMNH− atoms | — | 4 × 31 | 4 × 31 |
| Rmsd bond length, Å° | 0.010 | 0.015 | 0.009 |
| Rmsd bond angle,‡ ° | 1.08 | 1.39 | 1.08 |
| Ramachandran plot§ | |||
| Most favorable region, % | 92.3 | 91.6 | 91.8 |
| Disallowed regions, % | 0.0 | 0.1 | 0.1 |
*No electron density has been detected for residues 1–23; solvent-exposed loops corresponding to residues 30–39, 187–191, and 293–302 are unstructured.
†Rfactor = Σ‖Fobs | − | Fcalc‖/Σ | Fobs |. Rfree is the Rfactor value for 5% of the reflections excluded from the refinement. Data for the highest resolution shell are given in parentheses.
‡Rms deviations from ideal values calculated with REFMAC5 (38).
§Figures from PROCHECK (39).
Supplementary Material
Acknowledgments
This work was supported by grants from the Italian Ministry of Science (Relevant National Interest Projects and Italian Fund for Basic Research programs) and by Fondazione Cariplo (A.M.) and Thailand Research Fund Grants RMU4880028 and RTA478006, and Faculty of Science, Mahidol University (P.C.). M.P. is a recipient of Royal Golden Jubilee Ph.D. Program Scholarship PHD/0151/2547 of the Thailand Research Fund.
Abbreviations
- C1
reductase component of HPA hydroxylase
- C2
monooxygenase component of HPA hydroxylase
- FMNH−
two-electron reduced FMN
- HPA
p-hydroxyphenylacetate.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2JBR, 2JBT, and 2JBS).
This article contains supporting information online at www.pnas.org/cgi/content/full/0608381104/DC1.
References
- 1.Ghisla S, Massey V. Eur J Biochem. 1989;181:1–17. doi: 10.1111/j.1432-1033.1989.tb14688.x. [DOI] [PubMed] [Google Scholar]
- 2.Palfey BA, Ballou DP, Massey V. Active Oxygen in Biochemistry. Glasgow, Scotland, UK: Chapman & Hall; 1995. [Google Scholar]
- 3.Palfey BA, Massey V. Comprehensive Biological Catalysis. Vol 3. San Diego: Academic; 1998. pp. 83–154. [Google Scholar]
- 4.Ghisla S, Massey V. Biochem J. 1986;239:1–12. doi: 10.1042/bj2390001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Massey V. J Biol Chem. 1994;269:22459–22462. [PubMed] [Google Scholar]
- 6.Ballou DP, Entsch B, Cole LJ. Biochem Biophys Res Commun. 2005;338:590–598. doi: 10.1016/j.bbrc.2005.09.081. [DOI] [PubMed] [Google Scholar]
- 7.Sucharitakul J, Chaiyen P, Entsch B, Ballou DP. J Biol Chem. 2006;281:17044–17053. doi: 10.1074/jbc.M512385200. [DOI] [PubMed] [Google Scholar]
- 8.Chaiyen P, Suadee C, Wilairat P. Eur J Biochem. 2001;268:5550–5561. doi: 10.1046/j.1432-1033.2001.02490.x. [DOI] [PubMed] [Google Scholar]
- 9.Thotsaporn K, Sucharitakul J, Wongratana J, Suadee C, Chaiyen P. Biochim Biophys Acta. 2004;1680:60–66. doi: 10.1016/j.bbaexp.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Sucharitakul J, Chaiyen P, Entsch B, Ballou DP. Biochemistry. 2005;44:10434–10442. doi: 10.1021/bi050615e. [DOI] [PubMed] [Google Scholar]
- 11.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holm L, Sander C. J Mol Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 13.Kim JJ, Wang M, Paschke R. Proc Natl Acad Sci USA. 1993;90:7523–7527. doi: 10.1073/pnas.90.16.7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagpal A, Valley MP, Fitzpatrick PF, Orville AM. Biochemistry. 2006;45:1138–1150. doi: 10.1021/bi051966w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schreuder HA, Prick PA, Wierenga RK, Vriend G, Wilson KS, Hol WG, Drenth J. J Mol Biol. 1989;208:679–696. doi: 10.1016/0022-2836(89)90158-7. [DOI] [PubMed] [Google Scholar]
- 16.Eswaramoorthy S, Bonanno JB, Burley SK, Swaminathan S. Proc Natl Acad Sci USA. 2006;103:9832–9837. doi: 10.1073/pnas.0602398103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malito E, Alfieri A, Fraaije MW, Mattevi A. Proc Natl Acad Sci USA. 2004;101:13157–13162. doi: 10.1073/pnas.0404538101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleywegt GJ, Jones TA. Acta Crystallogr D. 1994;50:178–185. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 19.Sheng D, Ballou DP, Massey V. Biochemistry. 2001;40:11156–11167. doi: 10.1021/bi011153h. [DOI] [PubMed] [Google Scholar]
- 20.Macheroux P, Ghisla S, Hastings JW. Biochemistry. 1993;32:14183–14186. doi: 10.1021/bi00214a017. [DOI] [PubMed] [Google Scholar]
- 21.Jones KC, Ballou DP. J Biol Chem. 1986;261:2553–2559. [PubMed] [Google Scholar]
- 22.Entsch B, Cole LJ, Ballou DP. Arch Biochem Biophys. 2005;433:297–311. doi: 10.1016/j.abb.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 23.Ortiz-Maldonado M, Ballou DP, Massey V. Biochemistry. 1999;38:8124–8137. doi: 10.1021/bi990560e. [DOI] [PubMed] [Google Scholar]
- 24.Chaiyen P, Sucharitakul J, Svasti J, Entsch B, Massey V, Ballou DP. Biochemistry. 2004;43:3933–3943. doi: 10.1021/bi035734d. [DOI] [PubMed] [Google Scholar]
- 25.Entsch B, van Berkel WJ. FASEB J. 1995;9:476–483. doi: 10.1096/fasebj.9.7.7737455. [DOI] [PubMed] [Google Scholar]
- 26.Gatti DL, Palfey BA, Lah MS, Entsch B, Massey V, Ballou DP, Ludwig ML. Science. 1994;266:110–114. doi: 10.1126/science.7939628. [DOI] [PubMed] [Google Scholar]
- 27.Hiromoto T, Fujiwara S, Hosokawa K, Yamaguchi H. J Mol Biol. 2006;364:878–896. doi: 10.1016/j.jmb.2006.09.031. [DOI] [PubMed] [Google Scholar]
- 28.Dong C, Flecks S, Unversucht S, Haupt C, van Pée KH, Naismith JH. Science. 2005;309:2216–2219. doi: 10.1126/science.1116510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mattevi A. Trends Biochem Sci. 2006;31:276–283. doi: 10.1016/j.tibs.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 30.Hecht HJ, Kalisz HM, Hendle J, Schmid RD, Schomburg D. J Mol Biol. 1993;229:153–172. doi: 10.1006/jmbi.1993.1015. [DOI] [PubMed] [Google Scholar]
- 31.Roth JP, Klinman JP. Proc Natl Acad Sci USA. 2003;100:62–67. doi: 10.1073/pnas.252644599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collaborative Computational Project, Number 4 (CCP4) Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 33.Leslie AG. Acta Crystallogr D. 1999;55:1696–1702. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- 34.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Acta Crystallogr D. 2003;59:2023–2030. doi: 10.1107/s0907444903017694. [DOI] [PubMed] [Google Scholar]
- 35.Schneider TR, Sheldrick GM. Acta Crystallogr D. 2002;58:1772–1779. doi: 10.1107/s0907444902011678. [DOI] [PubMed] [Google Scholar]
- 36.Perrakis A, Morris R, Lamzin VS. Nat Struct Biol. 1999;6:458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Acta Crystallogr D Biol. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Murshudov GN, Vagin AA, Dodson EJ. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.