Abstract
Myelodysplastic syndromes (MDS) are among the most frequent hematologic malignancies. Patients have a short survival and often progress to acute myeloid leukemia. The diagnosis of MDS can be difficult; there is a paucity of molecular markers, and the pathophysiology is largely unknown. Therefore, we conducted a multicenter study investigating whether serum proteome profiling may serve as a noninvasive platform to discover novel molecular markers for MDS. We generated serum proteome profiles from 218 individuals by MS and identified a profile that distinguishes MDS from non-MDS cytopenias in a learning sample set. This profile was validated by testing its ability to predict MDS in a first independent validation set and a second, prospectively collected, independent validation set run 5 months apart. Accuracy was 80.5% in the first and 79.0% in the second validation set. Peptide mass fingerprinting and quadrupole TOF MS identified two differential proteins: CXC chemokine ligands 4 (CXCL4) and 7 (CXCL7), both of which had significantly decreased serum levels in MDS, as confirmed with independent antibody assays. Western blot analyses of platelet lysates for these two platelet-derived molecules revealed a lack of CXCL4 and CXCL7 in MDS. Subtype analyses revealed that these two proteins have decreased serum levels in advanced MDS, suggesting the possibility of a concerted disturbance of transcription or translation of these chemokines in advanced MDS.
Keywords: biomarker, chemokine, proteomics, hematologic malignancy
Myelodysplastic syndromes (MDS) are among of the most frequent hematologic stem cell malignancies in elderly patients (1, 2) and the cause of a reduction of median life expectancy to 5–50 months because of infections and bleeding (3). Up to 45% of the cases transform to acute myeloid leukemia (3). Peripheral blood cytopenias and bone marrow (BM) dysplasia are the hallmarks of MDS and the mainstay of its diagnosis. However, morphologic recognition of BM cell dysplasia requires extensive experience and can be subjective. Thus, interobserver correlation among examiners can be poor, e.g., as shown for dyserythropoiesis (R = 0.27) and dysgranulopoiesis (R = 0.45) (4). Other diseases such as nutritional deficiencies, viral infections, autoimmune disorders, or treatment with cytotoxic drugs may mimic the MDS phenotype and should be excluded before diagnosing MDS (5, 6). Chromosomal abnormalities are present in only 40% of patients (7). Further diagnostic features are sparse and lack sensitivity and specificity.
Despite the discovery of chromosomal abnormalities, gene mutations (8), and aberrant hypermethylation (9), the pathophysiology of MDS has remained elusive (6). Ineffective hematopoiesis has convincingly been attributed to increased apoptosis in the BM, but whether apoptosis is the primary defect or the consequence of other insults to the hematopoietic stem cell or its environment is unknown. Lately, it has been speculated that apoptosis is a reactive phenomenon fueled by cytokines (10). Numerous reports have shown abnormal serum levels of growth factors and cytokines in MDS, e.g., increased levels of IL-1, IL-1 receptor antagonist (IL-1 RA), and TNF-α (11), thrombopoietin (TPO), IL-6, and IL-8 (12), or b-FGF and hepatocyte growth factor (13). It is conceivable that cytogenetic abnormalities and hypermethylation translate into qualitative and quantitative alterations of protein expression. This hypothesis prompted us to perform a comprehensive analysis of the serum proteome to reveal molecular features that may aid in diagnosing MDS and provide insights into the biology of MDS.
Surface-enhanced laser desorption/ionization TOF MS (SELDI-TOF-MS) has been used for protein profiling in clinical studies (14–16). We and others have improved the early methodological approach (17), demonstrating that SELDI-TOF-MS can generate long-term reproducible and reliable proteomic information (18, 19). Hence, we used SELDI-TOF-MS to generate serum proteome profiles from patients with MDS and patients with conditions resembling MDS (non-MDS cytopenia). We found a profile that predicts MDS with an accuracy of 80% and validated this prediction twice, including a prospectively collected independent validation set. Finally, using tandem MS, we identified CXC chemokine ligands (CXCL)4 and CXCL7 as two differential proteins, corroborated their decreased serum levels with antibodies, and showed that they might represent previously uncharacterized markers of advanced MDS.
Results
Patient Characteristics.
The clinical characteristics of our patients are summarized in Table 1. The distribution of MDS types is similar to prior studies (ref. 3; Table 2). Patients with non-MDS cytopenia included 39 cases with autoimmune disorders, 19 with clonal hematologic disorders other than MDS, and 14 with miscellaneous conditions [supporting information (SI) Table 4].
Table 1.
Characteristics of patients with MDS and non-MDS cytopenia in the first blood collection set
| Patient's characteristics | MDS (n = 74) | Non-MDS cytopenia (n = 39) | P |
|---|---|---|---|
| Age, years | 67 (20–84) | 57 (20–88) | 0.001 |
| Male/female (n=) | 44/30 | 11/28 | <0.001 |
| (59.5%/40.5%) | (28.2%/71.8%) | ||
| Leukocytes, ×103 per μl | 3.8 (0.9–22.3) | 4.2 (0.6–133) | 0.179 |
| Hemoglobin, g/dl | 9.1 (4.2–13.3) | 12.2 (6.0–15.6) | <0.001 |
| Platelets, × 103 per μl | 113.5 (8–748) | 157 (5–426) | 0.102 |
| Peripheral blood blasts | 0 (0–29) | 0 (0–0) | 0.004 |
| Serum LDH, units/liters | 180 (122–666) | 170 (113–519) | 0.102 |
| Free serum Hb, μg/ml | 17 (3–93) | 17 (3–142) | 0.779 |
All values are medians (range in parentheses) except for gender, which is presented in absolute frequencies (percentages in parentheses). P was calculated by the Mann–Whitney test, except for gender, where the test of proportions was used.
Table 2.
Representation of MDS FAB types in our study compared with an earlier published large cohort of patients with MDS
| FAB types | Current study cohort, no. of patients, % (n = 122) | Ref. 3, no. of patients, % (n = 1,600) |
|---|---|---|
| RA | 56 (46) | 418 (26) |
| RARS | 17 (14) | 328 (20) |
| RAEB | 36 (29) | 344 (22) |
| RAEB-t | 7 (6) | 273 (17) |
| CMML | 6 (5) | 237 (15) |
RAEB-t, RA with excess of blasts in transformation; CMML, chronic myelomonocytic leukemia.
Supervised Analysis to Develop a Predictive Serum Proteome Profile for MDS.
After randomizing the samples from our first collection set into a learning and a validation set, we generated serum proteome profiles by means of SELDI-TOF-MS. We analyzed the mass spectra of our learning set (n = 72) with a supervised pattern recognition algorithm and discovered 32 multiprotein patterns associated with the distinction between MDS and non-MDS cytopenia (P < 0.001) (SI Fig. 6). We then performed class prediction on the learning set and obtained optimal accuracy with an 81-peak k-nearest-neighbor (k-nn) predictor (SI Table 5). Its accuracy by leave-one-out cross-validation was 81.9%, with a sensitivity of 83.3% and a specificity of 79.2% (Table 3) (P < 0.001 by Fisher's and class-label permutation tests).
Table 3.
Performance of the predictive serum proteome profile
| Performance | Learning, % | First validation, % | Second validation, % |
|---|---|---|---|
| Accuracy | 81.9* | 80.5† | 79.0‡ |
| Sensitivity | 83.3 | 80.8 | 91.6 |
| Specificity | 79.2 | 80.0 | 60.6 |
The predictor was derived from a learning set (n = 72) by means of pattern recognition and k-nn analysis and tested on a first independent validation set (n = 41). A prospectively collected second sample set (n = 81) run 5 months later was used as a second independent validation set.
*Fisher's exact test, P = 3 × 10–7; permutation P = 0.001.
†Fisher's exact test, P = 2 × 10–4.
‡Fisher's exact test, P = 6 × 10–7.
Validation of the MDS Serum Proteome Profile in an Independent Set of Patients.
To validate the association of our profile with MDS, we applied the 81-marker k-nn predictor to the first independent validation set (n = 41). We again observed an accuracy of 80.5% (Fisher's test, P < 0.001; 95% confidence interval, 68–92%), a sensitivity of 80.8%, and a specificity of 80.0% (Table 3). Statistically significant differences in Hb levels, peripheral blood blast counts, age, and gender between MDS and non-MDS cytopenia patients (Table 1) may presumably affect protein profiles; however, in a multivariate logistic regression model, controlling for these variables, the profile remained an independent predictor of MDS in the validation set [P < 0.001, odds ratio, 13.3; 95% confidence interval (CI), 1.5–111.1%], as well as in the learning and validation sets together (P < 0.001, odds ratio 16.7, 95% CI, 4.6–58.8%). Because peripheral blood blasts can be a diagnostic marker for MDS, we further tested the independence of our profile from peripheral blood blast counts by removing those samples that had peripheral blood blasts (n = 5) from the validation set. This removal resulted in predictive accuracy, sensitivity, and specificity of 83.3%, 85.7%, and 80%, respectively.
To reduce the likelihood of a potential bias due to sampling from different hospitals, we repeated the performance test after removing, in the validation set, all (n = 6) samples from the smaller sample source (St. Johannes Hospital, Duisburg, Germany). We found a nearly identical accuracy of 77.1% (Fisher's test, P = 0.003), whereas five of the six excluded samples were correctly predicted.
Finally, because in vitro and in vivo hemolysis may affect proteomic profiles, we examined serum lactate dehydrogenase (LDH) and free serum Hb levels in the two patient groups. There were no statistically significant differences between MDS and non-MDS cytopenia patients (Table 1), excluding the possibility of spurious differences in proteomic profiles due to release of intracellular proteins after accidental in vitro hemolysis during serum preparation or in vivo hemolysis related to the disease. Accordingly, the mass spectral peaks that likely represent Hb-α and -β (15,100 and 15,900 Da, respectively) did not significantly differ between the two patient groups.
Second Validation of the Predictive MDS Serum Proteome Profile in a Prospectively Collected Independent Sample Set.
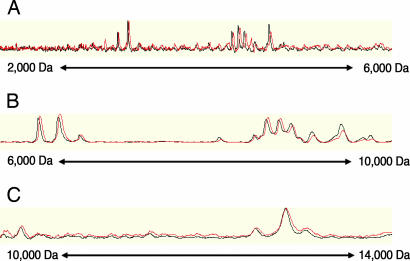
To further validate the over-time reproducibility and predictive robustness of our profile, a second independent validation serum set, prospectively collected from additional 81 patients (48 MDS and 33 non-MDS cytopenia) and six reference samples from our previous analysis were subjected to the identical experimental procedure 5 months after the first collection set had been processed. Over time reproducibility was assessed by comparing the spectra of our reference samples to the spectra of the same reference samples generated 5 months earlier. No significant differences were observed (Fig. 1) when determining the variation of the spectra from our six reference samples. Using peaks with a signal-to-noise ratio ≥3, we obtained median coefficients of variation ranging between 10% and 28%, depending on the combination of serum preparation and protein array type under investigation.
Fig. 1.
Overlay view of a reference serum sample. Protein mass spectra from the same reference serum sample, generated 5 months apart. Black trace, first experiment series; red trace, second experiment series (second independent validation set). Traces are displayed minimally offset to enhance visibility of both traces. Molecular mass is shown from 2 to 6 kDa (A), 6 to 10 kDa (B), and 10 to 14 kDa (C).
Our 81-marker k-nn predictor was applied to the second independent validation set without any modification of the predictive model. Its diagnostic accuracy remained robust (79.0%) and statistically significant (Fisher's test, P < 0.001; 95% confidence interval, 72–88%). Sensitivity was 91.6% and specificity, 60.6%. The relative loss in specificity may be related to the somewhat different composition of the non-MDS cytopenia group in the second blood collection set (see SI Text).
Identification of CXCL4 and CXCL7 as Proteomic Markers for MDS.
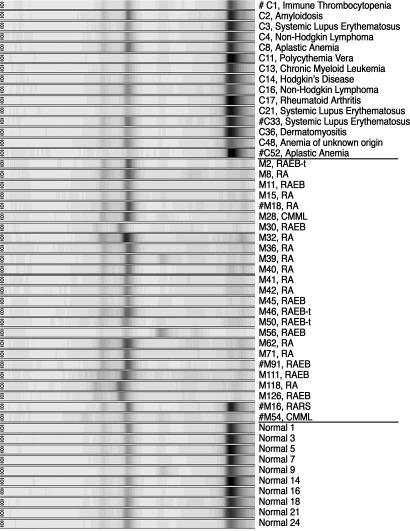
We sought to identify proteomic markers and to validate their differential serum levels in MDS vs. non-MDS cytopenia patients with independent antibody assays. We selected two differential protein peaks from the mass spectra with sufficient molecular-mass distance from other protein peaks and with generally high relative detection signals. A differential protein peak from the pH 5 fraction with a molecular mass of 7,786 Da (Fig. 2) was identified by tandem MS as the CXCL4 after serial fractionation. Likewise, we identified the 9,319-Da protein peak from the pH 9 fraction as CXCL7.
Fig. 2.
Levels of protein band at 7,786 Da in different sample groups. Spectra from serum fraction pH 5 on weak cationic exchange protein arrays displayed in a digitally simulated gel view. The x axis is scaled in daltons. C, non-MDS cytopenia; M, MDS; #, a platelet protein extract was available for CXCL4 Western blot analysis.
The median levels of CXCL4 and CXCL7 were 2.3- and 2.9-fold lower in MDS serum than in non-MDS cytopenia serum (Mann—Whitney test, P = 0.001 and P = 0.02, respectively), as measured by mass spectrometric profiling. Similarly, the median levels of CXCL4 and CXCL7 were 2.2- and 4.0-fold lower in MDS than in normal serum.
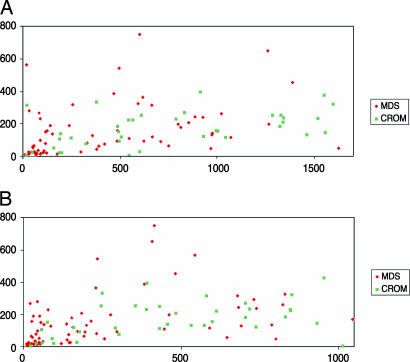
CXCL4 and CXCL7 are CXC chemokines located in the α-granules of platelets, and therefore their serum levels may simply reflect platelet counts. Correlation analyses of these two chemokine serum levels and platelet counts revealed a modest correlation (Spearman test, r = 0.40 for CXCL4 and r = 0.50 for CXCL7) (Fig. 3). Multivariate logistic regression analysis demonstrated that serum levels of CXCL4 and CXCL7 were significantly associated with MDS independent of the platelet count (P < 0.001 and P = 0.05, respectively).
Fig. 3.
Serum CXCL levels as determined by ELISA plotted against platelet counts of patients with MDS or cytopenia for reasons other than MDS (CROM). Both serum CXCL levels were significantly different between MDS and CROM patients (both P < 0.001), whereas platelet counts were not (P = 1.02). CXCL4 (A) and CXCL7 (B) serum levels are on the x axis; platelet counts are on the y axis.
Corroboration of Differential CXCL4 and CXCL7 Serum Levels with Immunoassays.
Immunoassay measurement of CXCL4 and CXCL7 serum levels in all first collection samples (n = 137) revealed that median intensities for CXCL4 and CXCL7 were significantly lower in MDS (344.2 and 146.6 arbitrary units, respectively) than in non-MDS cytopenia (608.8 and 457.4; Mann–Whitney test, P = 0.003, and P < 0.001, respectively). Normal samples displayed slightly higher serum levels (782.5 and 516.5) than non-MDS cytopenia samples. These antibody-assay results correlated positively with our results from mass spectrometric profiling (Spearman test, r = 0.75 for CXCL4, and r = 0.61 for CXCL7).
We tested in a prospectively collected third sample set (n = 32) whether our finding of differential CXCL levels would hold true for patients with cytopenia due to HIV, hepatitis C, or chemotherapy, who were not included in the first two sample collections and again found that median intensities for CXCL4 and CXCL7 were significantly lower in MDS (266.9 and 113.7 arbitrary units, respectively) than in HIV (9,096.2 and 3,614.9; Mann—Whitney test, P = 0.003 and P = 0.002, respectively) or chemotherapy-induced cytopenia (1,528.7 and 376.2; Mann–Whitney test, P = 0.005 and P = 0.0036, respectively).
CXCL4 Protein Expression Levels in Platelets and Plasma.
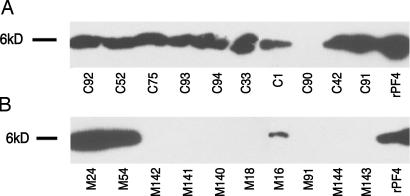
Decreased CXCL4 levels in MDS serum could be due to platelets displaying a lower expression or a deficient release of CXCL4. Western blot analyses on platelet-free plasma and platelets from 10 MDS and 10 non-MDS cytopenia patients did not detect any CXCL4 protein in plasma from both groups. In contrast, in protein extracts from platelets, we detected CXCL4 in 9 of 10 samples from non-MDS cytopenia patients but in only 3 of 10 patients with MDS (Fisher's test, P = 0.005) (Fig. 4).
Fig. 4.
Western blotting of platelet proteins. (A and B) Analysis of extracts from 10 patients with non-MDS cytopenia (A) and from 10 MDS patients (B). rPF4, recombinant platelet factor 4 (= CXCL4, positive control).
CXCL4 and CXCL7 Serum Levels Are Proteomic Markers for Advanced MDS.
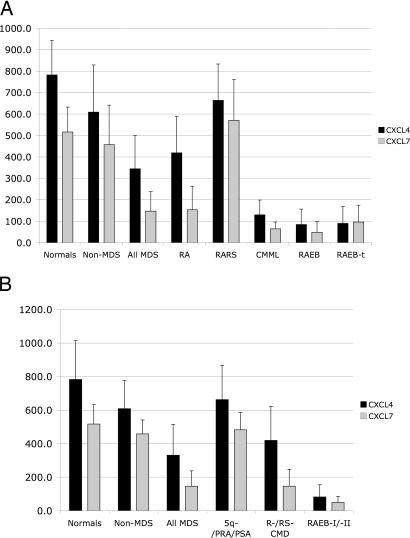
Given the heterogeneity of MDS, we performed subtype analyses and observed that CXCL4 and CXCL7 levels vary significantly among different MDS types. Specifically, early disease forms according to French–American–British (FAB) classification (RA and RA with ring sideroblasts), as well as World Health Organization (WHO) classification (5q-, PSA, and PRA) show levels as high as healthy donors or of non-MDS cytopenia patients (Fig. 5). In contrast, advanced disease forms according to FAB- [RA with excess of blasts (RAEB) and RAEB in transformation], as well as WHO classification (RAEB I and II), show median CXCL4 serum levels between 7.0- and 7.5-fold, respectively, lower than in non-MDS cytopenia patients (Mann–Whitney test, P < 0.001; Fig. 5A). Similarly, for CXCL7, advanced forms of MDS according to FAB and WHO classification have median serum levels between 6.3- and 9.3-fold, respectively, lower than in non-MDS cytopenia patients (Mann–Whitney test, P < 0.001; Fig. 5B).
Fig. 5.
Serum levels as shown by antibodies against CXCL4 and CXCL7 in normal healthy donors, patients with non-MDS cytopenia, and patients with different MDS types according to FAB (A) and WHO classification (B). For both proteins, differences between normal healthy donors and patients with non-MDS cytopenia are statistically not significant. However, advanced MDS patients according to FAB (RAEB, and RAEB in transformation), and WHO (RAEB I and II) show median CXCL4 serum levels that are 7.0-fold (Mann–Whitney test, P < 0.001) and 7.5-fold (Mann–Whitney test, P < 0.001) lower than in patients with non-MDS cytopenia, respectively, and for CXCL7 median serum levels that are 6.3-fold (Mann–Whitney test, P < 0.001) and 9.3-fold (Mann–Whitney test, P < 0.001) lower than in patients with non-MDS cytopenia, respectively.
Blast cells in the BM and peripheral blood, elevated serum LDH levels, and chromosomal aberrations are known markers of advanced disease. We investigated whether decreased CXCL4 and -7 levels were correlated with these known markers. CXCL4 levels were significantly correlated with serum LDH (Pearson correlation = −0.501, P = 0.0001), but logistic regression analysis revealed that CXCL4 is associated with advanced MDS independently of serum LDH levels. In contrast, CXCL7 levels were significantly correlated with blast cell counts and serum LDH levels but were not an independent predictor of disease stage.
Discussion
In this study, we generated serum protein mass spectra from 218 individuals and discovered a proteome profile strongly associated with MDS. Our profile distinguished MDS from non-MDS cytopenia patients independent of potential confounding by Hb, peripheral blood blast counts, age, and gender bias due to serum sampling by different hospitals or in vivo and in vitro hemolysis. Several reports have determined that class prediction results obtained from genomics or proteomics studies should be corroborated in independent sample sets (20–23). Therefore, we tested our predictive profile in two separate independent validation sets run 5 months apart, rarely done before in proteomic or genomic studies (24–26). Our profile showed high accuracy in the learning and both validation sample sets (80.5 ± 1.5%).
Many patients with autoimmune disorders are treated with cytotoxic drugs such as azathioprine or methotrexate and are therefore cytopenic. When cytopenia deteriorates, those patients are often suspected of developing MDS due to an increased risk (27, 28). Consequently, these patients may undergo several BM biopsies. Our profile displayed consistently high sensitivity, ranging between 80.8% and 91.6%; thus, serum proteomics profiling may serve as a noninvasive aid to reduce the number of BM biopsies in these patients by supporting the decision of whether or when to perform a BM examination.
The limitations of current MDS diagnostic approaches have been outlined elsewhere (29). In our study, we ensured the correctness of the diagnosis of MDS by subjecting all BM samples to a central morphology review and by ascertaining a followup of at least 2 years, thus confirming the validity of the morphological diagnosis. Interestingly, the only exception was one patient who was “erroneously” predicted by our profile as non-MDS, and whose clinical diagnosis of MDS had later been reversed by clinicians without any previous knowledge of our study results. Albeit anecdotal, this observation exemplifies how proteomic approaches may one day complement standard tools in the management of this disease.
We identified two differential markers, CXCL4 and CXCL7. These CXC chemokines, originating from the α-granules of platelets, showed decreased serum levels in MDS patients corroborated with immunoassays, indicating potential pathophysiologic implications. Upon platelet activation and release of CXCL7 into serum, CXCL7 is processed to neutrophil-activating peptide 2, a potent activator of neutrophil granulocytes (30). Similarly, CXCL4 promotes degranulation of neutrophil granulocytes (31). In MDS, neutrophils have lower microbicidal activity, rendering patients, even with normal neutrophil count, more prone to infection (32, 33). Conceivably, this immune deficiency is partly related to lower serum levels of CXCL4 and CXCL7 in patients with MDS.
One prominent feature in the majority of MDS patients is BM hypercellularity. A decrease in CXCL4 serum levels in MDS patients may contribute to this hypercellularity, because CXCL4 inhibits hematopoiesis by promoting adhesion and cell cycle inhibition of hematopoietic stem and progenitor cells (34). CXCL4 also supports survival of hematopoietic cells (35), and CXCL7 has been implicated in megakaryocytopoiesis (36). Further research is warranted to explore whether reduced levels of CXCL4 and CXCL7 represent a new therapeutic target in MDS.
CXCL4 is already expressed in maturating megakaryocytes (37). Disturbed maturation of megakaryocytes and α-granule defects in platelets are frequent features of MDS (38–40) and might be reflected by either low CXCL4 expression or a release deficiency in MDS platelets. Our Western blot analyses of platelet lysates showed CXCL4 protein expression in platelet lysates from 9 of 10 non-MDS cytopenia patients but a lack in 7 of 10 MDS platelet lysates. Our observation, therefore, represents a molecular correlate of structural platelet defects and suggests that additional platelet-derived markers may be revealed in the serum of MDS patients.
Given the notable relationship between CXCL4 and CXCL7, we scrutinized whether they may represent a specific feature inherent to a subtype of MDS. In fact, we found that advanced forms of MDS, independent of classification (RAEB and RAEB in transformation, according to FAB or RAEB-I and -II according to WHO), show lowest serum levels of CXCL4 and CXCL7 (Fig. 5). In conclusion, we propose CXCL4 and CXCL7 as markers addressing the paucity of molecular markers in advanced MDS. Our results warrant further studies on the utility of proteomic profiling for biomarker discovery and diagnostic evaluation of hematological malignancies that can ultimately be translated into refined patient management.
Materials and Methods
Additional details are provided in SI Text, SI Tables 4 and 5, and SI Spectral Data.
Patients.
We analyzed serum samples from 250 individuals from three different blood collection sets. The first set was from 137 individuals (74 MDS, 39 non-MDS cytopenia, and 24 healthy), a second prospectively collected set was from 81 patients (48 MDS and 33 non-MDS cytopenia), and a third prospectively collected set was from 32 patients (16 MDS and 16 non-MDS cytopenia). All samples were collected at the Beth Israel Deaconess Medical Center; the University Hospital of Düsseldorf, Düsseldorf, Germany; and St. Johannes Hospital, Duisburg, Germany. The study was approved by the Institutional Review Boards of the participating sites, and study subjects gave written informed consent.
All MDS patients were diagnosed by BM examination and classified according to FAB and WHO classification (41, 42) (SI Table 4). All BM smears underwent central morphology review (by U.G.). The diagnoses of non-MDS cytopenia patients were made according to standard criteria, e.g., American College for Rheumatology criteria to classify systemic lupus erythematosus (43) (SI Table 4). Fifty-four percent of all non-MDS cytopenia patients also had a BM examination to exclude the diagnosis of MDS.
Serum Preparation and Chromatographic Fractionation.
Samples were collected and processed, following the same protocol (18) at all participating centers. Samples were aliquoted into 96-well microtiter plates by using a robotic liquid-handling system (Biomek FX; Beckman Coulter, Fullerton, CA), thus ensuring that samples were subjected to only one freeze–thaw cycle. Subsequently, serum samples were fractionated by anion-exchange chromatography, as published (24, 44). In brief, 20 μl of serum was mixed with 30 μl of denaturating 9 M urea/2% CHAPS/50 mM Tris·HCl, pH 9.0, for 20 min at 4°C. The serum-buffer mixture was transferred to a filter-bottom 96-well microplate prefilled with Q Ceramic Hyper D F sorbent beads (Biosepra, Marlborough, MA), rehydrated with 50 mM Tris·HCl, pH 9.0, and equilibrated with 1 M urea/0.2% CHAPS/50 mM Tris·HCl, pH 9.0. After incubation for 30 min at 4°C, the flowthrough and a subsequent wash with 100 μl of 0.1% octyl-β-glucoside (OGP)/50 mM Tris·HCl, pH 9.0, for 10 min at room temperature were collected into a microtiter plate designated “pH 9.” The filtration plate was incubated with 2 × 100 μl of the following buffers to yield the following fractions: 0.1% OGP/50 mM Hepes, pH 7.0 (“pH 7”); 0.1% OGP/100 mM Na acetate, pH 5.0 (“pH 5.0”); 0.1% OGP/100 mM Na acetate, pH 4.0 (“pH 4”); 0.1% OGP/50 mM Na citrate, pH 3.0 (“pH 3”); and 33.3% isopropanol/16.7% acetonitrile/0.1% trifluoroacetic acid (“organic fraction”).
Proteomic Analyses and Quality Control.
In contrast to previous work (17), three rigorous steps preceded each proteomic experiment in our study to optimize reproducibility and reliability: (i) frequent assessment and adjustment of MS detector voltage to standardize sensitivity across experiments, (ii) frequent determination of MS resolution, and (iii) application of a highly standardized procedure to dry protein arrays before MS (refs. 18 and 19; available in SI Text).
Reversed-phase arrays (H50; Ciphergen, Fremont, CA) and weak cationic exchange protein arrays (CM10; Ciphergen) were preprocessed as published (14, 18, 44). Subsequently, we spotted the pH 9 and pH 5 fractions to cationic exchange arrays and the organic fraction and unfractionated serum to reversed-phase arrays. The matrix molecule sinapinic acid (SPA) was prepared and applied as published (refs. 14, 18, and 44; see also SI Text), again by using the Biomek FX. The arrays were air-dried again and immediately analyzed.
In each experiment, six randomly selected samples among 24 serum samples from healthy donors as reference serum controls were included to determine assay reproducibility. Equal proportions of samples from healthy individuals and patients with MDS and non-MDS cytopenia were processed in each experiment and analyzed in duplicate. The entire array processing procedure was performed by using the Biomek FX. Protein arrays were analyzed by SELDI-TOF-MS (PBS II; Ciphergen). Resulting mass spectra were processed and protein peaks detected as published (15, 16, 18, 45), by using a signal-to-noise ratio of 3.
Enrichment and identification of proteins of interest were performed by using anion-exchange and size-exclusion chromatography followed by 1D polyacrylamide gel electrophoresis, tryptic band digestion, and quadrupole TOF MS with collision-induced dissociation (SI Fig. 7 and SI Text), as published (14).
Preparation of Platelets and Platelet-Free Plasma.
Platelets and platelet-free plasma preparation are described in SI Text.
Antibody-Based Assays.
For our immunoassays, rabbit anti-human CXCL4 antibody (AB1488; Chemicon, Temecula, CA) and rabbit anti-human neutrophil-activating peptide 2 antibody (AB1484P; Chemicon), which cross-reacts with CXCL7, were conjugated to protein arrays, as published (46), or were used for Western blot analysis of plasma and platelet protein extracts. Briefly, 100 μg of protein per sample was loaded on a 12% SDS polyacrylamide gel. Chemiluminescence was detected by using the Super Signal West Pico Kit (Pierce, Rockford, IL). Free-serum Hb was measured with the Human Hemoglobin ELISA Quantitation Kit (Bethyl Laboratories, Montgomery, TX).
Statistical Analyses.
A supervised pattern recognition algorithm (Genes@Work 2.0; IBM, Yorktown Heights, NY) was used to identify subsets of proteomic markers whose serum levels are tightly clustered within MDS or non-MDS cytopenia, respectively (47–50). Those differential markers were further evaluated by using signal-to-noise metric and permutation tests (51). Class predictions were performed by using the k-nn algorithm (50, 52).
To develop and test a predictive profile, we randomly split the first blood collection set into learning sets (MDS = 48, non-MDS cytopenia = 24) and first-validation sets (MDS = 26, non-MDS cytopenia = 15). Randomization was stratified for the MDS subtypes and for the different categories within the non-MDS cytopenia group. In the learning set, we built a predictive proteomic profile with pattern recognition and class prediction algorithms (described above). The predictive accuracy was calculated by leave-one-out cross-validation (51), and the statistical significance of the predictor was estimated with Fisher's test on the confusion matrix and a class permutation test (52). Then, the proteomic profile with optimal learning accuracy was applied on the first-validation set and subsequently on a second prospectively collected independent validation set. Confidence intervals for predictive accuracy were constructed by using the normal approximation to the binomial distribution for proportions. Differences in median expression levels among sample groups were assessed by the Mann–Whitney test. Multivariate analysis for confounding factors was performed with logistic regression.
Supplementary Material
Acknowledgments
We thank Susan Hankinson, Bärbel Junge, Sabine Haase, and Lena Kikuchi for helpful comments, serum preparation, and technical assistance; Carlo Aul for support; and Else-Marie Meyer for inspiration. This study was supported by Leukämie-Liga e.V., Düsseldorf, Germany, and by a Dr. Mildred Scheel Cancer Foundation scholarship (to M.A.), the Whitaker Foundation (G.A.), and National Institutes of Health Grants U24 DK 58739 and 1RO1 CA 85467 (to T.A.L.).
Abbreviations
- MDS
myelodysplastic syndrome
- CXCL
CXC chemokine ligand
- BM
bone marrow
- FAB
French–American–British
- SELDI-TOF-MS
surface-enhanced laser desorption/ionization TOF MS
- WHO
World Health Organization
- k-nn
k nearest neighbor
- RA
refractory anemia
- RAEB
RA with excess of blasts
- LDH
lactate dehydrogenase.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 1109.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610330104/DC1.
References
- 1.Aul C, Gattermann N, Schneider W. Br J Haematol. 1992;82:358–367. doi: 10.1111/j.1365-2141.1992.tb06430.x. [DOI] [PubMed] [Google Scholar]
- 2.Williamson PJ, Kruger AR, Reynolds PJ, Hamblin TJ, Oscier DG. Br J Haematol. 1994;87:743–745. doi: 10.1111/j.1365-2141.1994.tb06733.x. [DOI] [PubMed] [Google Scholar]
- 3.Germing U, Gattermann N, Strupp C, Aivado M, Aul C. Leukemia Res. 2000;24:983–992. doi: 10.1016/s0145-2126(00)00088-6. [DOI] [PubMed] [Google Scholar]
- 4.Ramos F, Fernandez-Ferrero S, Suarez D, Barbon M, Rodriguez JA, Gil S, Megido M, Ciudad J, Lopez N, del Canizo C, Orfao A. Leukemia Res. 1999;23:283–290. doi: 10.1016/s0145-2126(98)00166-0. [DOI] [PubMed] [Google Scholar]
- 5.Heaney ML, Golde DW. N Engl J Med. 1999;340:1649–1660. doi: 10.1056/NEJM199905273402107. [DOI] [PubMed] [Google Scholar]
- 6.Steensma DP, Tefferi A. Leukemia Res. 2003;27:95–120. doi: 10.1016/s0145-2126(02)00098-x. [DOI] [PubMed] [Google Scholar]
- 7.Greenberg P, Cox C, LeBeau MM, Fenaux P, Morel P, Sanz G, Sanz M, Vallespi T, Hamblin T, Oscier D, et al. Blood. 1997;89:2079–2088. [PubMed] [Google Scholar]
- 8.Lubbert M, Jonas D, Miller CW, Herrmann F, Mertelsmann R, McCormick F, Koeffler HP. Oncogene. 1990;5:583–587. [PubMed] [Google Scholar]
- 9.Ruter B, Wijermans PW, Lubbert M. Int J Hematol. 2004;80:128–135. doi: 10.1532/ijh97.04094. [DOI] [PubMed] [Google Scholar]
- 10.Westwood NB, Mufti GJ. Curr Hematol Rep. 2003;2:186–192. [PubMed] [Google Scholar]
- 11.Meyers CA, Albitar M, Estey E. Cancer. 2005;104:788–793. doi: 10.1002/cncr.21234. [DOI] [PubMed] [Google Scholar]
- 12.Hsu HC, Lee YM, Tsai WH, Jiang ML, Ho CH, Ho CK, Wang SY. Oncology. 2002;63:64–69. doi: 10.1159/000065722. [DOI] [PubMed] [Google Scholar]
- 13.Alexandrakis MG, Passam FH, Pappa CA, Damilakis J, Tsirakis G, Kandidaki E, Passam AM, Stathopoulos EN, Kyriakou DS. Int J Immunopathol Pharmacol. 2005;18:287–295. doi: 10.1177/039463200501800211. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, Yu W, He T, Yu J, Caffrey RE, Dalmasso EA, Fu S, Pham T, Mei J, Ho JJ, et al. Science. 2002;298:995–1000. doi: 10.1126/science.1076185. [DOI] [PubMed] [Google Scholar]
- 15.Adam BL, Qu Y, Davis JW, Ward MD, Clements MA, Cazares LH, Semmes OJ, Schellhammer PF, Yasui Y, Feng Z, Wright GL., Jr Cancer Res. 2002;62:3609–3614. [PubMed] [Google Scholar]
- 16.Kozak KR, Amneus MW, Pusey SM, Su F, Luong MN, Luong SA, Reddy ST, Farias-Eisner R. Proc Natl Acad Sci USA. 2003;100:12343–12348. doi: 10.1073/pnas.2033602100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Petricoin EF, Ardekani AM, Hitt BA, Levine PJ, Fusaro VA, Steinberg SM, Mills GB, Simone C, Fishman DA, Kohn EC, Liotta LA. Lancet. 2002;359:572–577. doi: 10.1016/S0140-6736(02)07746-2. [DOI] [PubMed] [Google Scholar]
- 18.Aivado M, Spentzos D, Alterovitz G, Otu HH, Grall F, Giagounidis AA, Wells M, Cho JY, Germing U, Czibere A, et al. Clin Chem Lab Med. 2005;43:133–140. doi: 10.1515/CCLM.2005.022. [DOI] [PubMed] [Google Scholar]
- 19.Semmes OJ, Feng Z, Adam BL, Banez LL, Bigbee WL, Campos D, Cazares LH, Chan DW, Grizzle WE, Izbicka E, et al. Clin Chem. 2005;51:102–112. doi: 10.1373/clinchem.2004.038950. [DOI] [PubMed] [Google Scholar]
- 20.Sullivan Pepe M, Etzioni R, Feng Z, Potter JD, Thompson ML, Thornquist M, Winget M, Yasui Y. J Natl Cancer Inst. 2001;93:1054–1061. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- 21.Zhu W, Wang X, Ma Y, Rao M, Glimm J, Kovach JS. Proc Natl Acad Sci USA. 2003;100:14666–14671. doi: 10.1073/pnas.2532248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simon R, Radmacher MD, Dobbin K, McShane LM. J Natl Cancer Inst. 2003;95:14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- 23.Ransohoff DF. Nat Rev Cancer. 2004;4:309–314. doi: 10.1038/nrc1322. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Z, Bast RC, Jr, Yu Y, Li J, Sokoll LJ, Rai AJ, Rosenzweig JM, Cameron B, Wang YY, Meng XY, et al. Cancer Res. 2004;64:5882–5890. doi: 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
- 25.van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 26.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. N Engl J Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 27.Rosenthal NS, Farhi DC. Am J Clin Pathol. 1996;106:676–679. doi: 10.1093/ajcp/106.5.676. [DOI] [PubMed] [Google Scholar]
- 28.Knipp S, Hildebrandt B, Richter J, Haas R, Germing U, Gattermann N. Haematologica. 2005;90:691–693. [PubMed] [Google Scholar]
- 28.List AF, Vardiman J, Issa JP, DeWitte TM. Hematology (Am Soc Hematol Educ Program. 2004:297–317. doi: 10.1182/asheducation-2004.1.297. [DOI] [PubMed] [Google Scholar]
- 30.Walz A, Baggiolini M. J Exp Med. 1990;171:449–454. doi: 10.1084/jem.171.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petersen F, Ludwig A, Flad HD, Brandt E. J Immunol. 1996;156:1954–1962. [PubMed] [Google Scholar]
- 32.Ruutu P, Ruutu T, Vuopie P, Kosunen TU, de la Chapelle A. Nature. 1977;265:146–147. doi: 10.1038/265146a0. [DOI] [PubMed] [Google Scholar]
- 33.Greenberg PL. Blood. 1983;61:1035–1044. [PubMed] [Google Scholar]
- 34.Dudek AZ, Nesmelova I, Mayo K, Verfaillie CM, Pitchford S, Slungaard A. Blood. 2003;101:4687–4694. doi: 10.1182/blood-2002-08-2363. [DOI] [PubMed] [Google Scholar]
- 35.Han ZC, Lu M, Li J, Defard M, Boval B, Schlegel N, Caen JP. Blood. 1997;89:2328–2335. [PubMed] [Google Scholar]
- 36.Gewirtz AM, Zhang J, Ratajczak J, Ratajczak M, Park KS, Li C, Yan Z, Poncz M. Blood. 1995;86:2559–2567. [PubMed] [Google Scholar]
- 37.Vinci G, Tabilio A, Deschamps JF, Van Haeke D, Henri A, Guichard J, Tetteroo P, Lansdorp PM, Hercend T, Vainchenker W, et al. Br J Haematol. 1984;56:589–605. doi: 10.1111/j.1365-2141.1984.tb02184.x. [DOI] [PubMed] [Google Scholar]
- 38.Pamphilon DH, Aparicio SR, Roberts BE, Menys VC, Tate G, Davies JA. Scand J Haematol. 1984;33:486–491. doi: 10.1111/j.1600-0609.1984.tb00730.x. [DOI] [PubMed] [Google Scholar]
- 39.Blockmans D, Heynen MJ, Verhoef GE, Vermylen J. Haematologia (Budapest) 1995;26:159–172. [PubMed] [Google Scholar]
- 40.Zeidman A, Sokolover N, Fradin Z, Cohen A, Redlich O, Mittelman M. Hematol J. 2004;5:234–238. doi: 10.1038/sj.thj.6200364. [DOI] [PubMed] [Google Scholar]
- 41.Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick HR, Sultan C. Br J Haematol. 1982;51:189–199. [PubMed] [Google Scholar]
- 42.Bennett JM. Int J Hematol. 2000;72:131–133. [PubMed] [Google Scholar]
- 43.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, Schaller JG, Talal N, Winchester RJ. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 44.Poon TC, Yip TT, Chan AT, Yip C, Yip V, Mok TS, Lee CC, Leung TW, Ho SK, Johnson PJ. Clin Chem. 2003;49:752–760. doi: 10.1373/49.5.752. [DOI] [PubMed] [Google Scholar]
- 45.Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Clin Chem. 2002;48:1296–1304. [PubMed] [Google Scholar]
- 46.Lichtenthaler SF, Beher D, Grimm HS, Wang R, Shearman MS, Masters CL, Beyreuther K. Proc Natl Acad Sci USA. 2002;99:1365–1370. doi: 10.1073/pnas.032395699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Califano A. Bioinformatics. 2000;16:341–357. doi: 10.1093/bioinformatics/16.4.341. [DOI] [PubMed] [Google Scholar]
- 48.Klein U, Tu Y, Stolovitzky GA, Mattioli M, Cattoretti G, Husson H, Freedman A, Inghirami G, Cro L, Baldini L, et al. J Exp Med. 2001;194:1625–1638. doi: 10.1084/jem.194.11.1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lepre J, Rice JJ, Tu Y, Stolovitzky G. Bioinformatics. 2004;20:1033–1044. doi: 10.1093/bioinformatics/bth035. [DOI] [PubMed] [Google Scholar]
- 50.Spentzos D, Levine DA, Ramoni MF, Joseph M, Gu X, Boyd J, Libermann TA, Cannistra SA. J Clin Oncol. 2004;22:4648–4658. doi: 10.1200/JCO.2004.04.070. [DOI] [PubMed] [Google Scholar]
- 51.Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 52.Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.