Abstract
Mutations in genes coding for connexin26 (Cx26) and/or Cx30 are linked to approximately half of all cases of human autosomal nonsyndromic prelingual deafness. Cx26 and Cx30 are the two major Cx isoforms found in the cochlea, and they coassemble to form hybrid (heteromeric and heterotypic) gap junctions (GJs). This molecular arrangement implies that homomeric GJs would remain in the cochlea if one of the coassembly partners were mutated resulting in null expression. We generated mice in which extra copies of the Cx26 gene were transgenically expressed from a modified bacterial artificial chromosome in a Cx30−/− background. In the absence of the Cx30 gene, Cx26 expressed from extra alleles completely restored hearing sensitivity and prevented hair cell death in deaf Cx30−/− mice. The results indicated that hybrid GJs consisting of Cx26 and Cx30 were not essential for normal hearing in mice and suggested that up-regulation of Cx26 or slowing down its protein degradation might be a therapeutic strategy to prevent and treat deafness caused by Cx30 mutations.
Keywords: gap junction, hearing rescue, hereditary deafness
Connexins (Cxs) are a family of membrane proteins constituting gap junctions (GJs), which facilitate intercellular communication. The importance of Cxs in hearing has been revealed by genetic studies showing that mutations in Cx genes are among the most common forms of human genetic defects, resulting in hearing impairments in millions of patients with either autosomal dominant or recessive deafness (1–3). In many ethnic populations, mutations in Cx26 (1, 4), Cx30 (5, 6), and other Cx genes (7, 8) have been linked to approximately half of inherited prelingual nonsyndromic deafness cases. The most commonly found mutations are deletions of either Cx26 (e.g., 35delG or 235delC; refs. 9 and 10) or Cx30 (e.g., GJB6-D13S1830; ref. 6) that effectively eliminate gene expression. Animal models of conditional knockout of the Cx26 gene (11) and targeted deletion of Cx30-coding DNA (12) are available, both of which result in deafness in homozygous mice. Because GJs in the cochlea are coassembled from Cx26 and Cx30 (13–15), we suspected that deletion of one major cochlear Cx gene may not eliminate GJ intercellular coupling in the cochlea. Consistent with this speculation, immunocytochemical results revealed that the cellular expression pattern of the remaining Cx in the cochleae of Cx26 and Cx30 mutant mice is unaltered compared with WT animals (11, 12). Apparently, homomeric GJs remain in the cochlea of these Cx mutant mice. These data raise the question why gene expression from a single Cx isoform is insufficient for normal hearing.
Heteromeric assembly of Cxs is required for normal functions in some tissues (e.g., the lens in the eye; see ref. 16). These data obtained from a nonauditory system suggest that inappropriate biophysical properties of the homomeric GJ channels in the cochlea may underlie deafness caused by Cx mutations. It is also possible that the two major cochlear Cxs are coregulated at either the transcriptional and/or the translational level, such that the absence of one Cx gene significantly affects the expression of the other. Another simple explanation would be haploinsufficiency caused by gene deletion of one of the two coassembly partners. If an insufficient number of functional GJ channels causes deafness, we hypothesized that deafness might be corrected by increasing the expression of the remaining coassembly partner. Therefore, we investigated whether the hearing of deaf Cx30−/− mice could be restored by genetically overexpressing the Cx26 gene.
Results
Generation of Transgenic Mice in Which Cx26 Was Overexpressed in a Cx30−/− Background.
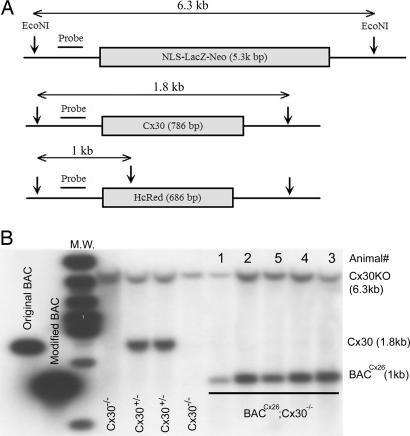
The overexpression of Cx26 in Cx30−/− mice was accomplished by using a genetic approach in two steps. First, we generated transgenic mice (BACCx26 mice), in which the Cx26 gene was overexpressed from the original locus in a modified BAC in the WT background. We produced BAC1 for overexpression of Cx26 and BAC2 as a control [supporting information (SI) Fig. 6A]. The successful modification of BACs in each step of homologous recombination (17) was confirmed by Southern blot hybridization (examples are shown in SI Fig. 6). Injections of BAC1 and BAC2 produced transgenic BACCx26 and BACNeoR-HcRed mice, respectively. Results obtained from three founder mice in each group are presented. Hearing sensitivities of both BACCx26 and BACNeo-HcRed mice in a WT background were normal, as determined by measuring auditory brainstem responses (ABRs; data not shown). In the second step, two generations of cross-breeding between BACCx26 and Cx30−/− mice yielded mice that carried extra alleles of Cx26 but without the Cx30 gene (BACCx26;Cx30−/− mice). These mice were born at the expected Mendelian frequency, and offspring was PCR-genotyped for further analyses (SI Fig. 7). Southern blotting was used for both genotyping and estimating the copy number of the BAC in the BACCx26;Cx30−/− mouse genome. Using a probe near the coding sequences (Fig. 1A), BACCx26;Cx30−/− mice displayed two bands at 6.3 (LacZ) and 1 kb (BAC1), respectively. The WT Cx30 band (1.8 kb; Fig. 1 B), which was seen in Cx30+/− mice, was absent in both Cx30−/− and BACCx26;Cx30−/− mice. Hybridization densities of the BAC1 transgene (1-kb band) were similar to the two chromosomal copies of Cx30 (LacZ knockin of the 6.3-kb band) in BACCx26;Cx30−/− mice (Fig. 1B). The ratio of hybridization density of the two bands was 1.08 ± 0.15 (n = 10), suggesting that two extra copies of the Cx26 gene were incorporated in the genome of these mice.
Fig. 1.
Structures of Cx30 loci in WT, Cx30−/−, and BACCx26;Cx30−/− mice. (A) Illustration of probe location and expected size of the DNA fragments treated by EcoNI for Cx30−/− (Top), WT (Middle), and BACCx26 (Bottom) mice. (B) Results of Southern blot hybridizations obtained with original and modified BAC, as well as with WT, Cx30+/−, Cx30−/−, and BACCx26;Cx30−/− mice.
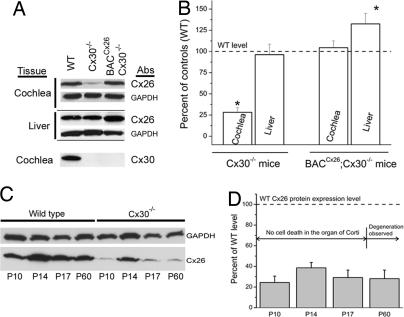
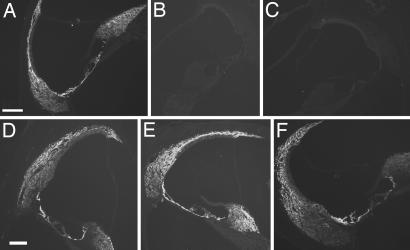
Consistent with the result of Southern blot hybridization indicating that the Cx30 gene was absent in the BACCx26;Cx30−/− mice (the 1.8-kb band in Fig. 1 B), Western blot analyses confirmed the absence of Cx30 protein in the cochlea of BACCx26;Cx30−/− mice (Fig. 2A). In addition, immunolabeling of cochlear sections obtained from BACCx26;Cx30−/− mice failed to detect immunopositive signals for Cx30 in the cochlea (Fig. 3A–C). The cellular expression pattern of Cx26 in the cochlea, however, was unaltered in WT, Cx30−/−, and BACCx26;Cx30−/− mice (Fig. 3 D–F). These results further confirmed the absence of Cx30 expression and supported the presence of homomeric Cx26 GJs in the cochlea of BACCx26;Cx30−/− mice. Unaltered cellular patterns of Cx26 immunolabeling in the cochlea of BACCx26;Cx30−/− mice (Fig. 3F) also suggested there was no ectopic Cx26 expression in these transgenic mice.
Fig. 2.
Restoration of Cx26 protein levels in the cochlea by transgenic expressions of BACCx26 in Cx30−/− mice. (A) Western blots showing Cx26 protein expression levels in the cochlea and liver. Results obtained with GADPH are shown as controls for the amount of protein loading in each lane. (B) Quantification of the relative Cx26 protein levels normalized to the WT level in the cochlea and liver in Cx30−/− and BACCx26;Cx30−/− mice. (C) Cx26 protein levels in the cochlea of WT and Cx30−/− mice at different postnatal days. (D) The Cx26 protein expression levels in Cx30−/− mice at different developmental stages were quantified by normalizing to their corresponding WT level.
Fig. 3.
Comparisons of immunolabeling patterns of Cx30 (A–C) and Cx26 (D–F) in the cochlea of WT (A and D), Cx30−/− (B and E), and BACCx26;Cx30−/− (C and F) mice. These are not confocal images. The intensities of immunolabeling in different images are not quantitatively comparable because they were obtained from different cochlear samples, and images were acquired with different exposure times. (Scale bar, ≈200 μm.)
Characterization of Cx26 Gene Expression Showed That Cx26 Protein Translation Was Restored to the WT Level in BACCx26;Cx30−/− Mice.
We compared changes in gene transcriptions in the cochlea resulting from Cx30 gene deletion (n = 9) by using both microarray and real-time PCR approaches. The ratio of hybridization density in the arrays for Cx26 was 0.82 ± 0.19 when WT and Cx30−/− mice were compared (P > 0.05), suggesting that the level of Cx26 mRNA in the cochlea of Cx30−/− mice was not significantly changed by Cx30 gene deletion. Comparing Cx26 transcription levels in BACCx26;Cx30−/−, and Cx30−/− mice gave a ratio of 1.52 ± 0.21 (P < 0.05), which was consistent with up-regulation of the Cx26 transcription in the cochlea as a result of transgenic BAC1 expression of Cx26 in BACCx26;Cx30−/− mice. In addition, microarray data demonstrated there were no significant changes in the transcription of other candidate Cxs in the cochlea, including Cx43, Cx45, Cx31, and Cx32 (data not shown). Microarray assay also identified significant changes in the expressions of apoptosis-related genes and specific markers for supporting cells. However, a full characterization of the microarray data is beyond the scope of this paper. Changes in Cx26 transcription level were further measured by real-time PCR amplifications. Normalized to the level of Cx30−/− mice, Cx26 mRNA in WT mice was expressed at 1.1 ± 0.2 (n = 6) and 1.02 ± 0.1 (n = 6) in the cochlea and liver, respectively. In BACCx26;Cx30−/− mice that harbored extra copies of the Cx26 gene, the Cx26 transcription level was expressed at 1.7 ± 0.4 (n = 6), 2.04 ± 0.2 (n = 6) of that of the Cx30−/− mice, respectively (SI Fig. 8A). These data confirmed results obtained by microarray analyses.
We next compared changes of the Cx26 protein level in WT, Cx30−/−, and BACCx26;Cx30−/− mice. It is known that the total amount of stable Cx protein may be affected by heteromeric protein–protein interaction (18). To avoid the potential complication caused by interactions between Cx26 and Cx30, we used Western blot results of Cx26 obtained from liver homogenates as controls because Cx26 coassemble with Cx32 in the liver, and the Cx32 gene is intact in both Cx30−/− and BACCx26;Cx30−/− mice. As expected, the Cx26 protein level in the liver of Cx30−/− mice was statistically the same as in WT (96.2 ± 12.4% compared with WT, n = 5; Fig. 2 A and B). In contrast, the Cx26 protein level was significantly higher in the liver of BACCx26;Cx30−/− mice (132.5 ± 12.5% compared with WT, n = 5; Fig. 2 A and B), suggesting that the Cx26 gene was indeed overexpressed in these genetically modified mice. In the cochlea, the Cx26 protein level in Cx30−/− mice was significantly reduced to 28.2 ± 5.4% of the WT level (n = 5, P < 0.05). The reduction in the Cx26 protein level in the cochlea of Cx30−/−, Cx30+/−, and Cx30+/+ mice appeared to depend on the copy number of Cx30 as well (SI Fig. 8B). In BACCx26;Cx30−/− mice, Cx26 protein expressions in the cochlea were restored to the WT level (104.4 ± 8.1% of the WT level, n = 5; see also Fig. 2).
Deletion of the Cx30 gene in mice causes degeneration of hair cells, apparently by an apoptotic mechanism (12). To differentiate whether reduced Cx26 protein expression was due to cell deaths in the organ of Corti or decreases in the stable cellular protein translation of Cx26, we compared the Cx26 protein levels in the cochlea at a series of developmental stages (Fig. 2 C and D), which span a period before and after the onset of Cx30 deletion-induced cell death in the organ of Corti (12). The reduction in the Cx26 protein level was observed at all stages we measured (postnatal days 10–60), suggesting the observed reduction in Cx26 translation was not due to selective cell death in the organ of Corti.
Restoration of Cx26 Protein Level in the Cochlea Prevented Hair-Cell Death in the Organ of Corti and Completely Rescued the Hearing of Cx30−/− Mice.
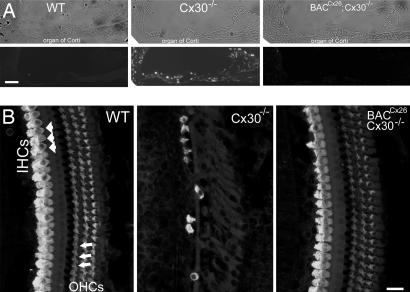
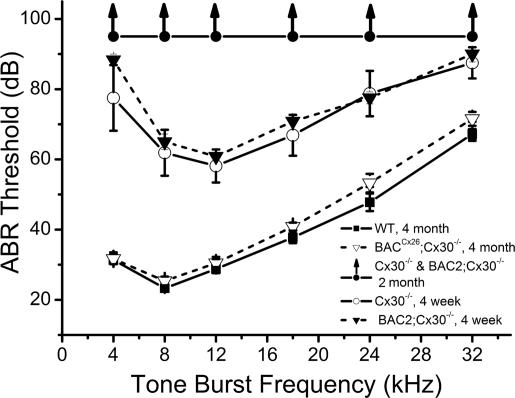
Cx30−/− mice show severe hearing loss at the onset of hearing and are completely deaf 2 months after they are born (12). Examinations of the cochlear morphology of adult mice revealed that apoptosis, as well as degeneration of the organ of Corti, in BACCx26;Cx30−/− mice was completely prevented (Fig. 4). In addition, measurements of both ABR (n = 10 for each group) and endolymphatic potentials (EPs; n = 5 in each group) showed a dramatic rescue of hearing sensitivity and large positive EP in BACCx26;Cx30−/− mice. The hearing thresholds of Cx30−/− and BACNeoR−HcRed;Cx30−/− mice measured at 3–4 weeks both showed significant hearing losses across a frequency range of 4–32 kHz, and these mice were deaf 2 months after birth (Fig. 5). Their hearing thresholds were all >90 dB, the loudest sound we could produce in experiments (curve with upward arrows in Fig. 5). In contrast, hearing thresholds were indistinguishable between BACCx26;Cx30−/− and WT mice measured at 4 weeks (data not shown) and 4 months after birth (Fig. 5, P > 0.05 for all frequencies). Examples of raw ABR waveforms are given in SI Fig. 9A. Average EPs (±standard error) measured in WT, Cx30−/−, and BACCx26;Cx30−/− were 93.6 ± 7.8 (n = 5), 8.6 ± 2.9 (n = 5), and 91.4 ± 5.6 (n = 5), respectively. Examples of EP recording are given in SI Fig. 9B. The recovery of EPs in BACCx26;Cx30−/− mice further supported that hearing functions were normal in the rescued mice. The failure of BAC2 to rescue the hearing of Cx30−/− mice indicated that the restoration of normal cochlear histology and hearing sensitivities in BACCx26;Cx30−/− mice was due specifically to the overexpression of Cx26 from its original locus in the BAC1.
Fig. 4.
Prevention of apoptosis and hair-cell degeneration by transgenic expression of BACCx26 in Cx30−/− mice. Apoptosis TUNEL staining pattern and morphology of the organ of Corti are given for WT (Left), Cx30−/− (Center), and BACCx26;Cx30−/− (Right) mice. (A) (Upper) Cochlear sections imaged with conventional phase-contrast optics. (Lower) TUNEL staining of corresponding cochlear sections comparing apoptosis in the cochlea of WT, Cx30−/−, and BACCx26;Cx30−/− mice. (Scale bar, ≈200 μm.) (B) Confocal views of whole-mount cochleae obtained from WT, Cx30−/−, and BACCx26;Cx30−/− mice (2 months old). The whole-mount cochlear samples were immunolabeled with a hair-cell-specific antibody against myosin VI. Examples of labeled inner and outer hair cells are indicated by arrowheads and arrows, respectively (Left). Outer hair cells were completely missing in the 2-month-old Cx30−/− mice, and inner hair cells in these mice were degenerated (Center). The cochlear morphology of BACCx26;Cx30−/− mice was indistinguishable from WT mice (comparing Left and Right). (Scale bar, ≈60 μm.)
Fig. 5.
Restoration of hearing sensitivities in Cx30−/− mice by transgenic expression of BACCx26. Hearing thresholds of WT, Cx30−/−, BACCx26;Cx30−/−, and BACNeoR−HcRed;Cx30−/− mice were measured by ABR tests across a frequency range of 4–32 kHz. Legends indicate the grouping and age of mice. Upward arrows indicate that the hearing threshold is >90 dB, the loudest sound level that could be generated by the sound transducer used in our experiments.
Discussion
Cx30 is required for normal hearing because homozygous human Cx30 deletion mutation results in deafness (6), and Cx30−/− homozygous mice are also deaf (12). However, it is not clear at all why Cx30 is required for hearing. There are at least two contrasting scenarios: (i) Cx30 is required for the assembly of hybrid GJ channels. In the cochlea of WT mice, current data support that GJs are coassembled from Cx26 and Cx30 (13–15). It is speculated that hybrid GJs are required for normal hearing based on the result obtained from nonauditory systems (16). (ii) Cx30 is required for producing sufficient quantities of GJs in the cochlea. In this scenario, the hybrid molecular configuration of GJs is not required. Comprehensive ABR and EP measurements (Fig. 5 and SI Fig. 9B), together with the data showing a total recovery of cochlear morphology in BACCx26;Cx30−/− mice (Fig. 4), demonstrated clearly that both hearing sensitivities and cochlear morphology are rescued in BACCx26;Cx30−/− mice. It is established that Cx26 and Cx30 are colocalized in the supporting cells and cochlear fibrocytes in the lateral wall and spiral limbus. No other Cxs are expressed in regions where Cx26 and Cx30 are coexpressed (13–15). In the absence of the Cx30 gene, Cx26 is the only Cx expressed in the cochlear regions normally expressing both Cx26 and Cx30. Therefore, our results demonstrated that hybrid GJs consisting of Cx26 and Cx30 were not essential for normal cochlear functions. Homomeric cochlear GJs consisting of Cx26 expressed at the appropriate level was sufficient for normal hearing in mice.
Our observation that the Cx26 mRNA level in the cochlea was similar in WT and Cx30−/− mice did not support that Cx26 and Cx30 were coregulated at the transcriptional level. In the cochlea of BACCx26;Cx30−/− mice, for which hearing sensitivity was totally restored, Cx26 protein expression was not significantly higher compared with WT mice, although it was overexpressed in the liver (Fig. 2 A and B). Because the Cx26 mRNA level was not significantly affected in Cx30−/− mice, the reduced Cx26 protein in the cochlea was presumably caused by accelerated degradation of less stable homomeric Cx26 GJs (note that both Cx30−/− and WT mice have two Cx26 alleles). These data suggested posttranscriptional interactions of the two Cxs may have a stabilization effect on functional cochlear GJs. The results also indicated that the cellular degeneration and deafness observed in Cx30−/− mice (12) were caused by a ≈3-fold reduction in Cx26 protein level (Fig. 2 A and B) when Cx30 is not expressed. Reduced availability of GJs may hinder the proposed K+ recycling (19) or intercellular diffusion of other molecules important for cellular activities and survival, ultimately resulting in the death of supporting and hair cells. Our results also imply that a 3-fold increase in Cx26 expression over the level expressed in patients with Cx30 deletion mutations could be therapeutic. It is also possible that smaller increases could still reduce disease severity and progression. Our data showing that BACCx26 mice had normal hearing also suggested that altering ratio of gene copies between Cx26 and Cx30 did not critically affect normal cochlear functions. Because the level of protein is not controllable by gene therapy-initiated exogenous expressions based on current technologies, these are encouraging data for further pursuing therapeutic designs using the gene therapy approach.
Cx26 deafness cases are linked to homozygous Cx26 mutations (3) and double-heterozygous Cx26 and Cx30 mutations (20), as well as homozygous Cx30 deletion mutations (20). Because mice heterozygous for Cx26 or Cx30 mutations show normal hearing, it is possible that double-heterozygous Cx mutated patients would benefit from up-regulation of either Cx26 or Cx30, or both. Up-regulating the expression of Cx26 in patients with homozygous Cx30 mutations but an intact Cx26 gene (20) may be a new therapeutic strategy worth further exploration, although investigations are still needed to find out whether increasing the Cx30 protein level in Cx26−/− background could restore hearing. In summary, our results open routes for investigating mechanism-based therapeutic strategies for treating patients suffering from Cx mutation-linked deafness. The results suggest that either elevating the expression or slowing down the protein degradation of the healthy copy of cochlear Cx in patients may be therapeutically beneficial.
Materials and Methods
BAC Modifications and Their Confirmations by PCR and Southern Blotting.
We obtained a BAC clone (RP23–332J19.F) from The Institute of Genomic Research (Rockville, MD) that contains both Cx26 and Cx30 genes approximately in the center of the BAC (SI Fig. 6A). The total length of mouse genomic DNA contained in this BAC is 205 kb, with 108.5 kb upstream and 76.5 kb downstream of the Cx26 and Cx30 genes, respectively. We first modified this BAC to obtain BAC1 by replacing the Cx30 gene with the HcRed sequence. The correct modification of BAC1 was confirmed by PCR and Southern blot hybridization (SI Fig. 6B). To check for the specific effect of overexpressing Cx26, BAC1 was further modified to obtain BAC2 by replacing the Cx26 coding sequence with the Neomycin resistance gene and its promoter (NeoR). The modification of the BAC2 was also confirmed by PCR and Southern blot hybridization (SI Fig. 6E Left). Germ-line transmissions of BACs were confirmed by PCR genotyping of mice (SI Fig. 6E Right). Primer sequences used for PCR genotyping on tail DNAs, probe synthesis for Southern blots, and approximate locations of primer pairs are given in SI Fig. 6.
Generation of BACCx26;Cx30−/− and BACNeoR−HcRed Mice and Mouse Genotyping by PCR and Southern Blotting.
The experimental protocol for animal use was approved by the Institutional Animal Care and Use Committee of Emory University. Linearized BAC DNA at the PI-SceI site (≈3 ng/μl) was sent to the Emory University School of Medicine mouse transgenic and gene targeting core facility for injections into fertilized mouse oocytes (strain FVB) to obtain BAC transgenic mice by the standard method (18). The pups born from BAC injections were screened for the presence of targeted modification by PCR from genomic DNA extracted from mouse tail tips using the Puregene mouse tail kit (Gentra, Minneapolis, MN). The mouse genotypes were further confirmed by Southern blot hybridization, with a specific probe to DNA fragments digested with EcoNI (SI Fig. 6D). After one more generation of breeding to confirm germ-line transmission, the BACCx26 and BACNeoR-HcRed mice were crossed with Cx30−/− mice (the same strain as those used in ref. 12) to create BACCx26;Cx30−/− and BACNeoR-HcRed;Cx30−/− mice, respectively.
Western Blots Analyzing Protein Expression of Cxs.
Details of experimental procedures were given in a previous publication (21). Briefly, total proteins were extracted by using RIPA lysis buffer, following the manufacturer's instructions (Upstate Biotechnology Cell Signaling Systems, Lake Placid, NY). Protein concentrations were measured by using a bicinchoninic acid protein assay kit (Pierce, Rockford, IL). Proteins were separated by electrophoresis on a 12% SDS polyacrylamide gel. After transferring to nitrocellulose membrane, Cxs were detected by Western blotting by using polyclonal antibodies against Cx26 (0.5 μg/ml) and/or Cx30 (1 μg/ml; Zymed, South San Francisco, CA). The same antibodies as those used in our previous publication (21) were used. The antibodies to Cx30 (catalog no. 33-2500, Zymed) and Cx26 (catalog no. 71-0500, Zymed) have lot numbers of 10966936 and 50393497, respectively. An equal amount of protein (5 μg) was loaded in each lane. The amount of loading was further checked by Western blotting of a housekeeping protein (GAPDH, dilution factor 1:2,000; Chemicon International, Temecula, CA). Protein bands on the blots were visualized by enhanced chemiluminescence (SuperSignal; Pierce) exposed to x-ray films (Hyper Film; Amersham Biosciences, Piscataway, NJ).
Microarray and Real-Time PCR Analyses of Gene Transcriptions in WT, Cx30−/−, and BACCx26;Cx30−/− Mice.
Total RNA was isolated from cochleae of WT, Cx30−/−, and BACCx26;Cx30−/− mice by using the PicoPure RNA Isolation Kit (Arcturus Bioscience, Mountain View, CA). The integrity and concentration of total RNA were assessed by using an Agilent Bioanalyzer 2100 with RNA 6000 Naco LabChips (Agilent, Palo Alto, CA). RNA (0.2 μg) was used to synthesize cDNA, and biotinylated antisense cRNA was generated by in vitro transcription in the presence of Biotin-11-UTP (PerkinElmer, Boston, MA). cRNA (10 μg) was hybridized to CodeLink Mouse Genome Bioarray (CodeLink Systems; GE Healthcare, Piscataway, NJ). The microarray slides were scanned by using a GenePix 4000B scanner (Molecular Devices, Palo Alto, CA). The signal intensities of spots were extracted from the scanned images by using CodeLink Expression Analysis Software (version 4.1; GE Healthcare) and imported into GeneSpring 7.3 (Silicon Genetics, Redwood City, CA) for further data analyses. Details of real-time PCR amplifications were given in our previous publication (22). The starting amount of mRNA in samples was quantified relatively by setting the threshold cycle (Ct) at a linear increasing phase of PCR amplifications. The relative change in transcription level of the gene for PCR product 1 (Q1) vs. product 2 (Q2) was calculated by the equation: Q1/Q2 = 2Ct2-Ct1. Differences in the mRNA level were examined by Student's t test, and the significance level was set at P < 0.005.
Immunolabeling of Cochlear Samples Obtained from WT, Cx30−/−, and BACCx26;Cx30−/− Mice.
Details of immunolabeling protocol were given in our previous publication (21). Briefly, decalcified cochlear sections along the modiolar axis and whole-mount cochlear samples were labeled with antibodies against Cx26 (1:200), Cx30 (1:100; both purchased from Zymed Laboratories, South San Francisco, CA), or Myosin VI (1:200; Proteus Biosciences, Ramona, CA). After washing three times with PBS, the samples were labeled with appropriate secondary antibodies conjugated to either Cy2 or Cy3 (1:500 dilution; Jackson Immunoresearch, West Grove, PA). Labeled cochlear samples were mounted in an antifade medium (Molecular Probes, Eugene, OR) and examined with either a conventional (Zeiss Axiovert 135; Carl Zeiss USA, Shrewsbury, PA) optical or confocal microscope (Zeiss LSM). Negative controls were processed similarly with primary antibodies substituted by equal volume of PBS.
Apoptotic cells in the cochlea were detected by using an in situ cell death detection kit (Roche, Oberkochen, Germany). Apoptotic cleavage of genomic DNA was identified by TUNEL, following the manufacturer's instructions. Positive controls for apoptosis were obtained by treating cochlear sections with DNase I.
ABR and EP Measurements for Objectively Determining Hearing Sensitivities of Mice.
Details of the ABR testing protocol are given in our previous publication (23). Briefly, mice were anesthetized with ketamine (80 mg/kg) and xylazine (10 mg/kg). Tone bursts of various frequencies ranging from 4 to 32 kHz (10 ms in duration and a rise–fall time of 0.5 ms) were generated by a Tucker–Davis system II hardware and software (Tucker–Davis Technologies, Alachua, FL). The ABR threshold was measured visually based on the appearance of wave II in a series of repeatable ABR responses obtained at various sound intensities (SI Fig. 9A). The genotype of mice was kept confidential to the ABR tester. Data acquisition and signal averaging were accomplished by Tucker–Davis system II hardware and software. Standard procedures for recording EP (11, 12) were used. Glass electrodes with a resistance of ≈10 MΩ were mounted on a manual micromanipulator. The electrodes were back-filled with 150 mM K+ solution. The electrical potential was zeroed when electrodes were touching the outside of the lateral wall of the cochlea before they were advanced to penetrate into the endolymphatic space. The direct current potential in the middle turn of the endolymphatic space was amplified with a high-resistance (1012 MΩ) amplifier (DAM70; World Precision Instrument, Sarasota, FL) and recorded by using pClamp7 software (SI Fig. 9B).
Supplementary Material
Acknowledgments
We thank Criss Hartzell for contributing critical comments for the manuscript. This study was supported by the National Institute on Deafness and Other Communicative Disorders (Grants R01-DC04709 and RO1-DC006483, to X.L.) and the Woodruff Foundation (to X.L.). Work at the University of Bonn was supported by grants from the German Research Association and the Fritz Thyssen Foundation (to K.W.).
Abbreviations
- Cx
connexin
- GJ
gap junction
- ABR
auditory brainstem response
- EP
endolymphatic potential
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0606855104/DC1.
References
- 1.Kelsell DP, Dunlop J, Stevens HP, Lench NJ, Liang JN, Parry G, Mueller RF, Leigh IM. Nature. 1997;387:80–83. doi: 10.1038/387080a0. [DOI] [PubMed] [Google Scholar]
- 2.Estivill X, Fortina P, Surrey S, Rabionet R, Melchionda S, D'Agruma L, Mansfield E, Rappaport E, Govea N, Mila M, et al. Lancet. 1998;351:394–398. doi: 10.1016/S0140-6736(97)11124-2. [DOI] [PubMed] [Google Scholar]
- 3.Chang EH, Van Camp G, Smith RJ. Ear Hear. 2003;24:314–323. doi: 10.1097/01.AUD.0000079801.55588.13. [DOI] [PubMed] [Google Scholar]
- 4.Guilford P, Ben Arab S, Blanchard S, Levilliers J, Weissenbach J, Belkahia A, Petit C. Nat Genet. 1994;6:24–28. doi: 10.1038/ng0194-24. [DOI] [PubMed] [Google Scholar]
- 5.Grifa A, Wagner CA, D'Ambrosio L, Melchionda S, Bernardi F, Lopez-Bigas N, Rabionet R, Arbones M, Monica MD, Estivill X, et al. Nat Genet. 1999;23:16–18. doi: 10.1038/12612. [DOI] [PubMed] [Google Scholar]
- 6.del Castillo I, Villamar M, Moreno-Pelayo MA, del Castillo FJ, Alvarez A, Telleria D, Menendez I, Moreno F. N Engl J Med. 2002;346:243–249. doi: 10.1056/NEJMoa012052. [DOI] [PubMed] [Google Scholar]
- 7.Xia JH, Liu CY, Tang BS, Pan Q, Huang L, Dai HP, Zhang BR, Xie W, Hu DX, Zheng D, et al. Nat Genet. 1998;20:370–373. doi: 10.1038/3845. [DOI] [PubMed] [Google Scholar]
- 8.Liu XZ, Xia XJ, Xu LR, Pandya A, Liang CY, Blanton SH, Brown SD, Steel KP, Nance WE. Hum Mol Genet. 2000;9:63–67. doi: 10.1093/hmg/9.1.63. [DOI] [PubMed] [Google Scholar]
- 9.Liu XZ, Xia XJ, Ke XM, Ouyang XM, Du LL, Liu YH, Angeli S, Telischi FF, Nance WE, Balkany T, Xu LR. Hum Genet. 2002;111:394–397. doi: 10.1007/s00439-002-0811-6. [DOI] [PubMed] [Google Scholar]
- 10.Denoyelle F, Weil D, Maw MA, Wilcox SA, Lench NJ, Allen-Powell DR, Osborn AH, Dahl HH, Middleton A, Houseman MJ, et al. Hum Mol Genet. 1997;6:2173–2177. doi: 10.1093/hmg/6.12.2173. [DOI] [PubMed] [Google Scholar]
- 11.Cohen-Salmon M, Ott T, Michel V, Hardelin JP, Perfettini I, Eybalin M, Wu T, Marcus DC, Wangemann P, Willecke K, Petit C. Curr Biol. 2002;12:1106–1111. doi: 10.1016/s0960-9822(02)00904-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Teubner B, Michel V, Pesch J, Lautermann J, Cohen-Salmon M, Sohl G, Jahnke K, Winterhager E, Herberhold C, Hardelin JP, et al. Hum Mol Genet. 2003;12:13–21. doi: 10.1093/hmg/ddg001. [DOI] [PubMed] [Google Scholar]
- 13.Lautermann J, ten Cate WJ, Altenhoff P, Grummer R, Traub O, Frank H, Jahnke K, Winterhager E. Cell Tissue Res. 1998;294:415–420. doi: 10.1007/s004410051192. [DOI] [PubMed] [Google Scholar]
- 14.Ahmad S, Chen S, Sun J, Lin X. Biochem Biophys Res Commun. 2003;307:362–368. doi: 10.1016/s0006-291x(03)01166-5. [DOI] [PubMed] [Google Scholar]
- 15.Forge A, Becker D, Casalotti S, Edwards J, Marziano N, Nevill G. J Comp Neurol. 2003;467:207–231. doi: 10.1002/cne.10916. [DOI] [PubMed] [Google Scholar]
- 16.White TW. Science. 2002;295:319–320. doi: 10.1126/science.1067582. [DOI] [PubMed] [Google Scholar]
- 17.Yang XW, Model P, Heintz N. Nat Biotechnol. 1997;15:859–865. doi: 10.1038/nbt0997-859. [DOI] [PubMed] [Google Scholar]
- 18.Nelles E, Butzler C, Jung D, Temme A, Gabriel HD, Dahl U, Traub O, Stumpel F, Jungermann K, Zielasek J, et al. Proc Natl Acad Sci USA. 1996;93:9565–9570. doi: 10.1073/pnas.93.18.9565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kikuchi T, Kimura RS, Paul DL, Takasaka T, Adams JC. Brain Res Brain Res Rev. 2000;32:163–166. doi: 10.1016/s0165-0173(99)00076-4. [DOI] [PubMed] [Google Scholar]
- 20.Del Castillo I, Moreno-Pelayo MA, Del Castillo FJ, Brownstein Z, Marlin S, Adina Q, Cockburn DJ, Pandya A, Siemering KR, Chamberlin GP, et al. Am J Hum Genet. 2003;73:1452–1458. doi: 10.1086/380205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun J, Ahmad S, Chen S, Tang W, Zhang Y, Chen P, Lin X. Am J Physiol. 2005;288:C613–C623. doi: 10.1152/ajpcell.00341.2004. [DOI] [PubMed] [Google Scholar]
- 22.Huang D, Chen P, Chen S, Nagura M, Lim DJ, Lin X. Hearing Res. 2002;165:85–95. doi: 10.1016/s0378-5955(02)00288-5. [DOI] [PubMed] [Google Scholar]
- 23.Peng BG, Ahmad S, Chen S, Chen P, Price MP, Lin X. J Neurosci. 2004;24:10167–10175. doi: 10.1523/JNEUROSCI.3196-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.