Abstract
Different transcription factors have been shown to control the transition of naive T cells into T helper 1 (Th1)/Th2 subsets. The T-cell-specific transcription factor GATA-3 is known to be selectively expressed in murine developing Th2 cells and to exert a positive action on Th2-specific cytokine production. Investigating GATA-3 gene regulation in human T cells we have found that naive T cells highly express GATA-3, and during early T2 or T1 polarization, respectively, they either maintain or quickly down-regulate expression. In developing T2 cells, as well as in committed Th2 cell lines and clones, we found a positive correlation among GATA-3, interleukin (IL)-5 and IL-4 gene expression kinetics, supporting the positive action of GATA-3 on Th2-specific cytokine production. A possible relationship between GATA-3 gene expression and the down-regulation of the IL-12 receptor (β2-chain; IL-12Rβ2) gene was evident only in the early phases of T2 polarization (within 24 hr), and not demonstrated at later times. During T-cell commitment the presence of IL-4 in the culture was essential to maintain or enhance GATA-3 transcription, while IL-12 was not necessary for full repression of GATA-3. Finally, we showed selective GATA-3 up-regulation in human Th2 cell lines and clones and the maintainance of a low basal level of GATA-3 expression in Th1 cells upon activation.
Introduction
As they develop, T cells can become ‘polarized’ and restricted to producing either T1 or T2 patterns of cytokines.1 T-cell subsets were first described for helper T cells, which bear the cell-surface marker CD4·2,3 Two mutually exclusive patterns of cytokine gene expression were observed:3–5 T helper 1 (Th1) cells secrete interleukin (IL)-2, interferon-γ (IFN-γ) and tumour necrosis factor-β (TNF-β), which promote cellular immune responses against intracellular pathogens and viruses and are clinically associated with inflammation and autoimmune diseases; Th2 cells produce IL-4, IL-5, IL-6, IL10 and IL-13, which promote humoral immunity and are characteristic of allergic responses and asthma.6–9 Subsequently, functionally polarized responses were also shown in CD8 cytotoxic T cells (Tc1/Tc2).10,11 In general, activated CD8+ T cells exhibit a Tc1 cytokine profile but they can express a Tc2 profile in some pathological conditions.12
Both Th1 and Th2 cells derive from a common naive precursor cell whose differentiation pathway is determined by a number of factors, including cytokines, dose and form of antigens, antigen-presenting cells, costimulators and the genetic background of the responding host.6,9,13 Indeed, the most effective inducer of differentiation is the cytokine environment present during priming of the precursor cells: IL-12, produced by activated macrophages and dendritic cells14–16 and IL-4, whose initial source is elusive, play a dominant role in driving the development of Th1 and Th2 cells, respectively.17–19 Both cytokines promote the growth–differentiation of their subset and inhibit the growth–differentiation of the opposing subset; when both cytokines are present in the same culture the IL-4 effect is predominant such that Th2 cells develop in the presence of IL-12·18,20
Numerous studies have focused on identifying of specific transcription factors that control the transition of naive T cells to Th1–Th2 subsets.21,22 GATA-3, a zinc finger protein that belongs to the GATA family of transcription factors23 was shown to be selectively expressed in murine Th2, but not Th1 cells.24,25 GATA-3 was initially cloned as a T cell-specific transcription factor that can bind to the T-cell receptor (TCR) α and δ genes.26,27 Subsequently, it was found to be critical in regulating the expression of several T-cell-specific genes in addition to TCR and Th2 cytokine genes.28–30 Furthermore, GATA-3 was found to be essential for normal embryonic development as well as for the generation of the T-cell lineage.31,32 While a GATA-3-positive action on Th2-specific cytokine gene expression has been demonstrated,24,25,33,34 less is known about how GATA-3 expression is regulated. It was shown that IL-4 can induce early expression of GATA-3 in a Stat-6-dependent manner and that IL-12 can inhibit GATA-3 expression in a Stat-4-dependent manner.35 Subsequently, it was found that GATA-3 can exert a Stat-6-independent autoactivation creating a feedback pathway stabilizing Th2 commitment.36
Up to date, little is known about GATA-3 gene expression and regulation in human peripheral T-cell subsets. The only indirect evidence that GATA-3 is expressed by human Th2 cells derives from studies of human asthma, where Th2 cells are known to play a critical role.37–39 Human GATA-3 mRNA was shown to be significantly increased in asthmatic patients40 as well as in patients with allergic rhinitis,41 and so poses this transcription factor as a potential therapeutic target for the treatment of asthma and allergy.41–43 For the first time this study analyses the expression kinetics of GATA-3 during early human T-cell differentiation towards the T1 or T2 pathways as well as in committed Th cells, and its correlation with cytokine–receptor gene expression.
Materials and methods
Media and reagents
Medium RPMI-1640 supplemented with 2 mm l-glutamine, 1% non-essential amino acids, 1% pyruvate, 50 µg/ml kanamycin (Gibco BRL, Gaithersburg, MD), 5 × 10−5 2-mercaptoehtanol (Sigma Chemical Co, St. Louis, MO), and 5% human serum (EuroClone Ltd, Wetherby, West York, UK) was used throughout. Human recombinant IL-2 and IL-4 were produced in our laboratory by polymerase chain reaction (PCR) cloning and expression in the myeloma expression system.44
Cells, T-cell lines and clones
Blood samples were obtained from healthy volunteers and peripheral blood mononuclear cells (PBMC) were separated by Ficoll-HyPaque (Amersham Pharmacia-Biotech AB, Uppsala, Sweden) density gradient centrifugation. Cord blood T lymphocytes were sorted using anti-CD3 conjugated magnetic microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) following the manufacturer's instructions. The kinetics analysis was performed on CD3+ cells stimulated with phytohaemagglutinin (PHA; 1 µg/ml; Gibco BRL) and 20 U/ml rIL-2 in complete RPMI-1640 supplemented with 5% human serum (EuroClone) under T1- ((2 ng/ml human rIL12; R&D Systems, Inc., Minneapolis, MN) plus 200 ng/ml neutralizing monoclonal antibody (mAb) to human IL-4 (PharMingen, San Diego, CA)), or T2-polarizing conditions (200 U/ml human rIL-4 plus 2·5 µg/ml neutralizing mAb to human IL-12 (R&D Systems)) or in the absence of exogenous cytokines (PHA-line). After 1 day of polarization aliquots of the cells were washed, plated in opposing culture conditions and harvested after a further 24 and 48 hr.
Cell lines were generated from sorted CD4+ naive T cells, by stimulating the cells with PHA (1 µg/ml; Gibco), irradiated allogeneic PBMC (3000 rad from a 137Cs source) and rIL2 (40 U/ml) under Th1- or Th2-polarizing conditions. The cultures were weekly restimulated in the same polarizing conditions and analysed after a further 10 days. T-cell blasts were cloned by limiting dilution and maintained by periodic restimulation with PHA, irradiated allogeneic PBMC and rIL-2 as previously described.45
In some experiments the cells were activated for 4 hr with 10−7 m phorbol 12-myristate 13-acetate (PMA) plus 1 µg/ml ionomycin (Sigma; PI-activation), in others they were activated with anti-CD3 mAb (TR66; immunoglobulin G1, IgG1)-coated plates (5 µg/ml) and harvested at the indicated time points.
Cytokine detection at the single cell level
T cells were stimulated with 10−7 m PMA plus 1 µg/ml ionomycin (Sigma) for 4 hr. Brefeldin A (10 µg/ml, Sigma) was added during the last 2 hr. Cells were fixed with 2% paraformaldehyde, permeabilized with phosphate-buffered saline containing fetal calf serum (FCS 1%) and saponin (0·5%) and stained with fluoroscein isothiocyanate (FITC)-labelled anti-IFN-γ (IgG1), PE-labelled anti-IL-4 (IgG2b) (Becton Dickinson, Mountain View, CA), FITC-labelled anti-IL-5 (IgG2a), and PE-labelled anti-IL-10 (IgG1; BioSource International, Camarillo, CA)
Fluorescence-activated cell sorting (FACS) analysis
Indirect double-staining was performed using anti-human-CD3 (OKT3, IgG2a), anti-human-CD4 (6D10, IgG1), anti-CD45RA (IgG1; Southern Biotechnology Associates, Birmingham, AL) and anti-CD45RO (IgG2a; Southern Biotechnology Associates) mAbs. Secondary antibodies were PE-labelled goat anti-mouse IgG2a and FITC-labelled goat anti-mouse IgG1 (Southern Biotechnology Associates). The stained cells were analysed by flow-cytometry on a FACScalibur (Becton Dickinson) with the CELLQUEST software.
RNA extraction, reverse transcription (RT)–PCR and oligotyping
Total RNA was extracted from T cells using TRIZOL (Gibco) following the manufacturer's instructions. First-strand cDNA was synthesized using oligo d(T) and Moloney murine leukaemia virus (MMLV)–RT (Promega Corp., Madison, WI) in 100 µl final volume. Serial dilutions of template cDNAs were subjected to low cycle-PCR using β-actin-specific primers β-actin 5′, ACACTGTGCCCATCTACGAGGGG; β-actin 3′, ATGATGGAGTTGAAGGTAGTTTCGTGGAT. Amount of normalized templates were used in subsequent gene-specific PCR reactions. PCR cycles were always kept in the linear portion of the amplification curve. Specific primers were as follows: GATA-3 forward, TGTCTGCAGCCAGGAGAGC; GATA-3 reverse, ATGCATCAAACAACTGTGGCCA; IFN-γ forward, TGTTACTGCCAGGACCCAT; IFN-γ reverse, GCGTTGGACATTCAAGTCAG; IL-12Rβ2 forward, AACATCACAGGACACACCTCCT; IL-12Rβ2 reverse, CCTTGCAGACAAAATTCCCTCTC; IL-4 forward, ACAAGTGCGATATCACCTTAC; IL-4 reverse, CAACGTACTCTGGTTGGCT; IL-5 forward, GTGAAAGAGACCTTGGCACTG; IL-5 reverse, GGCAAAGTGTCAGTATGCCTG.
GATA-3 PCR products were alkali-blotted onto Hybond-N+ membrane (Amersham, Arlington Heights, IL). Filters were prehybridized in BLOTTO solution (6× sodium saline citrate (SSC), 1% milk, 5 mm ethylenediamine tetra-acetic acid (EDTA), 0·1% sodium dodecyl sulphate (SDS)) at 42° for 3 hr and hybridized overnight at 42° with the 32P-labelled forward oligonucleotide.
Results
Kinetics of human GATA-3 and cytokine–receptor gene expression during early T1 and T2 differentiation
We purified human neonatal T lymphocytes by sorting cord blood cells with anti-CD3 mAb-conjugated magnetic beads; the cells obtained were >99% CD3+ and they showed a naive phenotype (>97% CD45RA+, not shown). GATA-3 and cytokine–receptor gene expression kinetics were analysed by semiquantitative RT–PCR in these naive T cells stimulated under T1 and T2 polarizing conditions (IL-12 and anti-IL-4 mAb for T1; IL-4 and anti-IL-12 mAb for T2) (Fig. 1). To allow comparison between mRNAs transcribed at different time points, serial dilutions of template cDNAs were subjected to low-cycle PCR using β-actin-specific primers. This pilot experiment allowed us to normalize the templates to be used in subsequent gene-specific PCR reactions (not shown). PCR cycles were always kept in the linear portion of the amplification curve.
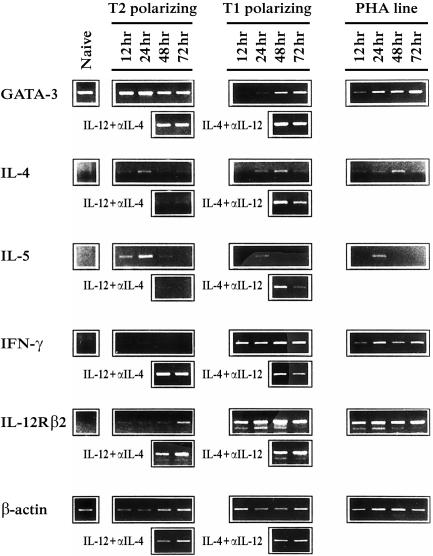
Figure 1.
Kinetics of gene expression during early human T1/T2 commitment. Cord blood derived, human naive T cells were cultured in T2 and T1 polarizing conditions (IL-4 and anti-IL-12 mAb for T2 cells; IL-12 and anti-IL-4 mAb for T1 cells), and GATA-3, IL-4, IL-5, IFN-γ and IL-12Rβ2 gene expression was analysed by RT–PCR at different time points (12, 24, 48, 72 hr). After 24 hr of polarization, part of the polarizing cells were washed, plated in the opposite culture conditions (IL-12 and anti-IL-4 mAb for T2 cells; IL-4 and anti-IL-12 mAb for T1 cells), and analysed after further 24 and 48 hr. As control, we plated naive T cells in the absence of exogenous cytokines (PHA-line). The different templates were normalized through β-actin RT–PCR experiments using serially diluted cDNAs. In the lower part of the figure the β-actin RT–PCR (30 cycles) with the normalized templates is shown.
Naive T cells transcribed GATA-3 efficiently and this high expression level was maintained during T2 polarizing conditions with a slight increase at 24 hr. During T1 commitment, a drastic GATA-3 down-regulation was observed as early as after 12 hr of polarization. At 24 hr GATA-3 mRNA remained very low and returned to basal levels at 48 hr. As control, we cultured sorted naive T cells in the absence of exogenous cytokines (PHA-line). GATA-3 gene transcription in these cells appeared to be similar to the T1 line, with a rapid decrease at 12 hr and a trend to restore basal GATA-3 transcription levels at the following time points.
After 24 hr of polarization, part of the cells were washed and plated in opposing stimulation conditions (IL-12 and anti-IL-4 mAb for T2 cells; IL4 and anti-IL-12 mAb for T1 cells). GATA-3 mRNA expression was then analysed after a further 24 and 48 hr of culturing. Inversion of the polarizing conditions caused a strong GATA-3 transcription in the ‘T1-reverted’ cells, comparable to that found in T2 cells; in the ‘T2-reverted’ cells GATA-3 transcription was not lost.
We next analysed the expression kinetics of T2-(IL-4 and IL-5) and T1-(IFN-γ and IL-12Rβ2) specific cytokine–receptor genes. Transcription of these four genes was undetectable in sorted naive T cells. In T2-polarized cells, the kinetics of IL-4 and IL-5 mRNA expression correlated with that of GATA-3, peaking at 24 hr. IFN-γ gene expression could not be observed, while an IL-12Rβ2-specific signal was evidenced at 72 hr. Inversion of polarizing conditions at 24 hr of T2 commitment induced a clear increase of IL-12Rβ2 and IFN-γ gene expression. T1 polarizing cells showed strong IL-12Rβ2 and IFN-γ gene expression already after 12 hr of culture and transcription of both genes was maintained at every time point, peaking at 48 hr. T1 ‘reverted’ cells showed slightly diminished IFN-γ and IL-12Rβ2 transcription levels and they expressed both IL-4 and IL-5 mRNAs. Surprisingly, the IL-4 expression level was higher compared to that in T2-polarized cells.
Basal expression of human GATA-3 in resting, committed Th cell lines and its differential regulation in activated cells
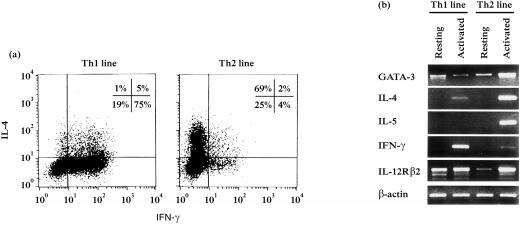
To examine transcription of GATA-3 gene in human committed Th cells, we generated Th1 and Th2 cell lines from CD4+ naive cells by three-round stimulation under polarizing conditions. Purity of the cell lines was assessed by analysing IL-4 and IFN-γ production at the single cell level by intracellular staining (Fig. 2a). Resting (at least 20 days after the last stimulation) and PI-activated cells were harvested and transcription of the five specific genes tested by RT–PCR (Fig. 2b).
Figure 2.
Expression of GATA-3 gene in human committed Th1 and Th2 cell lines. Human CD4+ naive T cells were polarized by consecutive cycles of stimulation in the presence of IL-12 and anti-IL-4 mAb (Th1) or IL-4 and anti-IL-12 mAb (Th2). The committed Th lines were analysed after at least three cycles of stimulation. (a) IFN-γ and IL-4 production was measured at the single cell level by intracellular staining. (b) Resting (at least 20 days after final stimulation) and PI-activated cells were harvested and GATA-3 and cytokine–receptor gene transcription was tested by RT–PCR. β-actin PCR (30 cycles) is shown as a control of the relative amount of cDNA loaded in the different lanes.
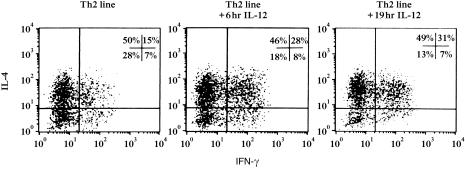
Both Th1 and Th2 resting cell lines showed a basal level of GATA-3 gene expression. When we activated the lines, Th2 cells up-regulated GATA-3 expression as well as the specific cytokine genes IL-4 and IL-5, while Th1 cells maintained the same basal levels of GATA-3 expression and up-regulated IFN-γ mRNA. IL-12Rβ2 gene was highly transcribed by both resting and activated Th1 cells, but unexpectedly it was also expressed by Th2 cells following activation. To test whether the IL-12Rβ2 mRNA could correspond to functional IL-12R expressed on the surface of Th2 cells, IL-12 responsiveness was evaluated by culturing Th2 cells in the presence of 25 ng/ml IL-12 for 6 and 19 hr. IL-12-induced IFN-γ production was assessed by intracellular staining (Fig. 3). Following IL-12 treatment, we found a two-fold increase in the percentage of cells coproducing IFN-γ and IL-4 (from 15% before IL-12 treatment to 31% after IL-12 addition), suggesting the presence of an IL-12-responsive population within the line. Interestingly, we noticed that the percentages of both IL-4 producing (Th2-polarized cells, 50%) and IFN-γ producing (contaminating cells, 7%) cells inside the line did not change following IL-12 treatment. The increase of IFN-γ and IL-4 coproducing cells, instead, corresponded to a decrease of the IFN-γ–/IL-4– cell population.
Figure 3.
Analysis of IL-12-responsiveness by an activated Th2 line. Polarized Th2 cells were plated in the presence of 25 ng/ml IL-12 in medium without IL-2, for 6 and 19 hr. During the last 4 hr of incubation, the cells were PI-stimulated and cytokine production (IL-4 and IFN-γ) was tested by intracellular staining in both untreated and IL-12 treated cells.
Preferential expression and selective GATA-3 up-regulation in human Th2 clones
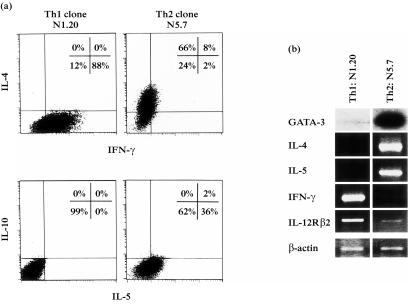
Th1 and Th2 clones were derived by limiting dilution from human CD4+ Th1/Th2 polarized cell lines. Seven days after stimulation with IL-2 and irradiated feeder cells, cytokine production (IL-4, IFN-γ, IL-10, IL-5) was checked by intracellular staining while the GATA-3 and cytokine–receptor gene expression were examined by RT–PCR in parallel. To better display the differential GATA-3 expression, few PCR cycles were done and the products blotted and hybridized with a GATA-3-specific, internal oligonucleotide (PCR-oligotyping).
The two representative clones shown in Fig. 4 presented a highly specific cytokine production at the single cell level (Fig. 4a) and the molecular analysis (Fig. 4b) evidenced a strong GATA-3 transcription in the Th2 clone (N5.7) which corresponded to high IL-4 and IL-5 mRNA levels. On the contrary, very low GATA-3 expression was detectable in the Th1 clone (N1.20). Th1 cells highly transcribed IFN-γ and IL-12Rβ2 genes, while IL-4 and IL-5 gene expression was not evident.
Figure 4.
Preferential GATA-3 expression in human Th2 clones. Human CD4+ clones were derived by limiting dilution from Th1 and Th2 polarized cell lines. (a) Cytokine production (IL-4, IFN-γ, IL-10, IL-5) was measured by intracellular staining and (b) GATA-3 and cytokine–receptor gene expression were examined by RT–PCR. PCR-cycles of GATA-3 gene were kept very low and the products were blotted and hybridized with a GATA-3-specific, internal oligonucleotide (PCR-oligotyping) to better evidence the preferential expression. β-actin PCR (20 cycles) is also shown.
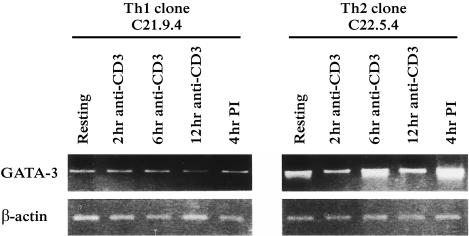
Next, we analysed the kinetics of GATA-3 gene expression following anti-CD3 activation (Fig. 5). The weak basal GATA-3 transcription level found in the resting Th1 clone (C21.9.4) was maintained during the whole time-course; the resting Th2 cells (C22.5.4) presented a higher GATA-3 gene expression and its transcription increased following activation with a peak at 6 hr, which corresponded to the IL-4 and IL-5 kinetics (not shown). The strong GATA-3 up-regulation in the Th2 clone and the maintenance of basal levels in the Th1 clone were further confirmed following 4 hr of PI stimulation.
Figure 5.
Selective up-regulation of GATA-3 gene in human Th2 clones after TCR activation. One human Th1 (C21.9.4) and one Th2 (C22.5.4) clones were activated either with anti-CD3 mAb (5 µg/ml) coated on plate or with PI. To analyse the GATA-3 expression kinetics, the cells were harvested at different time points (2, 6, 12 hr) after anti-CD3 activation. GATA-3 and cytokine–receptor gene expression were examined by RT–PCR. β-actin PCR (30 cycles) is shown as control.
Discussion
In this study we analyse the expression of GATA-3, a multifunctional transcription factor essential for T-cell development and Th differentiation, in human naive T cells, during early T1/T2 commitment and in Th polarized cells. We show high amounts of GATA-3 mRNA in naive T cells and maintenance of this high transcription level in T2 polarizing cells, with a peak of transcription at 24 hr. On the contrary, during T1 commitment we found a drastic and rapid GATA-3 down-regulation and a trend to restore the basal transcription level at 48 hr of polarization. Our data are in accordance with the murine model, where GATA-3 gene was strongly up-regulated in Th2 conditions with a peak of expression at 48 hr, while it decreased during the Th1 commitment.24,25,35 However, low levels of GATA-3 gene expression were described in murine naive T cells, at variance with the high transcription level we found in human cord blood cells. In long-term polarized Th cell lines and clones, a basal expression of GATA-3 gene was always detected in resting cells. GATA-3 transcription appeared to be differentially regulated only after activation: while Th2 cells strongly up-regulated GATA-3 transcription, Th1 cells maintained basal expression.
We found a peak of IL-4 and IL-5 mRNAs by 24 hr following polarization, concomitantly with the highest expression of GATA-3. Similarly, GATA-3 correlated with IL-4 and IL-5 gene up-regulation in activated Th2 cell lines and clones. Thus, the known positive action of GATA-3 gene on Th2-specific cytokine production is reflected by their expression kinetics. While a direct interaction of GATA-3 with the IL-5 promoter was demonstrated,24,46 a similar direct role for GATA-3 in IL-4 promoter activation was not evident.34 However, a GATA-3-dependent enhancer activity has been found in several regions surrounding the IL-4 gene.33 Because in murine T cells, the Th1–Th2 commitment appears to occur between 24 and 48 hr after the initial T-cell activation,47 we decided to revert the T1 and T2 phenotype after 24 hr of polarizing conditions. Interestingly, T1 cells were still able to up-regulate GATA-3 upon addition of IL-4 in the culture and this expression corresponded to high IL-4 and IL-5 transcription. These data provide further evidence for the positive correlation between GATA-3 and Th2-specific cytokine gene expression. Recently, Lee et al.48 showed that committed Th1 cells expressing ectopic GATA-3 could change their phenotype by inducing Th2 cytokine expression and chromatin remodelling.
The IL-12Rβ2 chain is lost in Th2 cells but maintained in Th1 cells.49,50 GATA-3 was proposed as an inhibitory factor of IL-12Rβ2 expression and therefore as a possible blocking agent of the IL-12-induced responses.35 If an inhibitory influence of GATA-3 on IL-12Rβ2 transcription exists, from our data it appeared to be effective only before 24 hr of polarization. In early human T2 commitment (12 hr), strong expression of GATA-3 corresponded to a complete block of IL-12Rβ2 transcription while after 24 hr of polarization, the presence of GATA-3 transcription did not inhibit IL-12Rβ2 gene expression upon IL-12 addition (T2 ‘reverted’ cells). Moreover, after 24 hr of polarization the induced GATA-3 expression in T1 ‘reverted’ cells did not cause loss of IL-12Rβ2 transcription. These data suggest that the major role of GATA-3 at the initial phases of T-cell commitment might be to influence the polarization by inhibiting IL-12 responsiveness, while in later steps, may be mainly to enhance Th2 cytokine production. This could explain why the strong GATA-3 transcription we observed at 48 hr in T1 ‘reverted’ cells correlated with a very high IL-4 expression, which exceeded that found in T2 cells, but not with IL-12Rβ2 inhibition. We also observed transcription of IL-12Rβ2 in a long-term-polarized activated Th2 cell line and we demonstrated the presence of an IL-12 responsive population inside the line. It is possible that the IL-12-responsive population originated from a fraction of uncommitted cells. However, the alternative possibility that some IL-4-producing cells can up-regulate IL-12Rβ2 and modify their phenotype towards Th1 pathway cannot be ruled out.
In murine developing Th1 cells it was found that the ectopic expression of GATA-3 induces Th2-specific cytokines and suppresses IFN-γ production in part by down-regulating IL-12Rβ2·25,35,51 Although we observed a slightly diminished IFN-γ production in T1 ‘reverted’ cells, this could suggest a repressing activity of GATA-3 on IFN-γ promoter, we favour the possibility that the IFN-γ decrease was simply due to the absence of IL-12 in the culture medium. Accordingly, Ouyang et al.35 showed that GATA-3 repressed IFN-γ only when continuously expressed during initial naive T-cell differentiation but not when reintroduced into Th1 cells after IL-12-induced Th1 development. Altogether, these data indicate that GATA-3 behaves more likely as a regulator in early steps of Th commitment, rather then as a repressor of the IFN-γ promoter.
It is well known that IL-12 and IL-4 play a dominant role in driving the development of Th1 and Th2 cells, respectively,17–19 and that IL-4 can induce early expression of GATA-3 in a Stat6-dependent manner.35 Accordingly with the results obtained for murine T cells,25,35 here we show that during the early phases of polarization the presence of IL-4 in the culture was essential to maintain or enhance human GATA-3 transcription. First, we observed that human naive T cells cultured in the absence of exogenous cytokines (PHA-line) showed a rapid decrease of GATA-3 transcription and maintained low levels of GATA-3 gene expression at the following time points, analogously to that observed in T1 cells. Second, the finding that T1 cells after 24 hr of polarization were still able to up-regulate GATA-3 upon addition of IL-4 in the culture strongly supports the importance of IL-4 on GATA-3 transcription at early stages of T-cell commitment. In long-term-polarized Th2 cells, we observed an IL-4-independent GATA-3 up-regulation, and this might indicate that GATA-3 could have an active role in the maintainance of the Th2 phenotype that was demonstrated to become increasingly stable over time.50,52 A positive GATA-3 autoactivation was observed in Stat6-deficient T cells, suggesting a role for GATA-3 in maintaining cell fate commitment.36
An important and still debated point is the influence of IL-12 on GATA-3 transcription. Requirement of IL-12 for full repression of GATA-3 was reported in developing naive T cells.35 On the contrary, our data indicated that IL-12 was not necessary to down-regulate GATA-3 whose expression naturally decreased in the absence of exogenous cytokines in culture (PHA-line). During T1 polarization, the presence of IL-12 had the only effect of delaying the restoration of the basal GATA-3 expression level (at 48 hr instead of 24 hr) when compared to the PHA-line. Moreover, we observed that the addition of IL-12 and anti-IL-4 after 24 hr of T2 polarization could not block GATA-3 transcription. In agreement with this view, a recent work of Nishikomori et al.53 showed that in IL-12Rβ2 transgenic Th2 cells, the addition of IL-12 did not shut down the expression of GATA-3 mRNA previously induced by IL-4.
The demonstration of a selective GATA-3 expression along the human Th2 pathway and of its strong up-regulation in Th2-polarized cells following activation could contribute to clarify the origin of GATA-3 overexpression in asthma.40,41 This is most probably due to locally activated Th2 cells, as also supported by the observation that the number of local T lymphocytes did not differ between normal and asthmatic airways.40,54
Acknowledgments
We thank Dr M. De Marchi for careful critical reading and Dr F. Novelli for helpful discussion.
Abbreviations
- IL-12Rβ2
IL-12 receptor (β2-chain)
- mAb
monoclonal antibody
- PBMC
peripheral blood mononuclear cells
- PI
phorbol 12-myristate 13-acetate and ionomycin
References
- 1.Swain SL. T-cell subsets. Who does the polarizing? Curr Biol. 1995;5:849. doi: 10.1016/s0960-9822(95)00170-9. [DOI] [PubMed] [Google Scholar]
- 2.Mosmann TR, Cherwinski H, Bond MW, Giedlin MA, Coffman RL. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J Immunol. 1986;136:2348. [PubMed] [Google Scholar]
- 3.Mosmann TR, Coffman RL. TH1 and TH2 cells: different patterns of lymphokine secretion lead to different functional properties. Annu Rev Immunol. 1989;7:145. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 4.Paul WE, Seder RA. Lymphocyte responses and cytokines. Cell. 1994;76:241. doi: 10.1016/0092-8674(94)90332-8. [DOI] [PubMed] [Google Scholar]
- 5.Murphy KM. T lymphocyte responses and cytokines. Current Opin Immunol. 1999;10:226. [Google Scholar]
- 6.Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 7.Seder RA, Paul WE. Acquisition of lymphokine-producing phenotype by CD4+ T cells. Annu Rev Immunol. 1994;12:635. doi: 10.1146/annurev.iy.12.040194.003223. [DOI] [PubMed] [Google Scholar]
- 8.Romagnani S. The Th1/Th2 paradigm. Immunol Today. 1997;18:263. doi: 10.1016/s0167-5699(97)80019-9. 10.1016/s0167-5699(97)01070-0. [DOI] [PubMed] [Google Scholar]
- 9.O'Garra A. Cytokines induce the development of functionally heterogeneous T helper cell subsets. Immunity. 1998;8:275. doi: 10.1016/s1074-7613(00)80533-6. [DOI] [PubMed] [Google Scholar]
- 10.Croft M, Carter L, Swain SL, Dutton RW. Generation of polarized antigen-specific CD8 effector populations: reciprocal action of interleukin (IL)-4 and IL-12 in promoting type 2 versus type 1 cytokine profiles. J Exp Med. 1994;180:1715. doi: 10.1084/jem.180.5.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erard F, Wild MT, Garcia-Sanz JA, Le Gros G. Switch of CD8 T cells to noncytolytic CD8– CD4– cells that make TH2 cytokines and help B cells. Science. 1993;260:1802. doi: 10.1126/science.8511588. [DOI] [PubMed] [Google Scholar]
- 12.Romagnani S. Th1 and Th2 in human diseases. Clin Immunol Immunopathol. 1996;80:225. doi: 10.1006/clin.1996.0118. 10.1006/clin.1996.0118. [DOI] [PubMed] [Google Scholar]
- 13.Constant SL, Bottomly K. Induction of Th1 and Th2, CD4+ T cell responses: the alternative approaches. Annu Rev Immunol. 1997;15:297. doi: 10.1146/annurev.immunol.15.1.297. [DOI] [PubMed] [Google Scholar]
- 14.MacAtonia SE, Hsieh CS, Murphy KM, O'Garra A. Dendritic cells and macrophages are required for Th1 development of CD4+ T cells from alpha beta TCR transgenic mice: IL-12 substitution for macrophages to stimulate IFN-gamma production is IFN-gamma-dependent. Int Immunol. 1993;5:1119. doi: 10.1093/intimm/5.9.1119. [DOI] [PubMed] [Google Scholar]
- 15.MacAtonia SE, Hosken NA, Litton M, et al. Dendritic cells produce IL-12 and direct the development of Th1 cells from naive CD4+ T cells. J Immunol. 1995;154:5071. [PubMed] [Google Scholar]
- 16.de Saint-Vis B, Fugier-Vivier I, Massacrier C, et al. The cytokine profile expressed by human dendritic cells is dependent on cell subtype and mode of activation. J Immunol. 1998;160:1666. [PubMed] [Google Scholar]
- 17.Swain SL, Weinberg AD, English M, Huston G. IL-4 directs the development of Th2-like helper effectors. J Immunol. 1990;145:3796. [PubMed] [Google Scholar]
- 18.Hsieh CS, MacAtonia SE, Tripp CS, Wolf SF, O'Garra A, Murphy KM. Development of TH1, CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science. 1993;260:547. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- 19.Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. The IL-4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol. 1999;17:701. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 20.Seder RA, Gazzinelli R, Sher A, Paul WE. Interleukin 12 acts directly on CD4+ T cells to enhance priming for interferon gamma production and diminishes interleukin 4 inhibition of such priming. Proc Natl Acad Sci USA. 1993;90:10188. doi: 10.1073/pnas.90.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rincon M, Flavell RA. T-cell subsets: transcriptional control in the Th1/Th2 decision. Curr Biol. 1997;7:R729. doi: 10.1016/s0960-9822(06)00368-x. [DOI] [PubMed] [Google Scholar]
- 22.Kuo CT, Leiden JM. Transcriptional regulation of T lymphocyte development and function. Annu Rev Immunol. 1999;17:149. doi: 10.1146/annurev.immunol.17.1.149. [DOI] [PubMed] [Google Scholar]
- 23.Weiss MJ, Orkin SH. GATA transcription factors: key regulators of hematopoiesis. Exp Hematol. 1995;23:99. [PubMed] [Google Scholar]
- 24.Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J Biol Chem. 1997;272:21597. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- 25.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 26.Ho IC, Vorhees P, Marin N, Oakley BK, Tsai SF, Orkin SH, Leiden JM. Human GATA-3: a lineage-restricted transcription factor that regulates the expression of the T cell receptor alpha gene. EMBO J. 1991;10:1187. doi: 10.1002/j.1460-2075.1991.tb08059.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ko LJ, Yamamoto M, Leonard MW, George KM, Ting P, Engel JD. Murine and human T-lymphocyte GATA-3 factors mediate transcription through a cis-regulatory element within the human T-cell receptor delta gene enhancer. Mol Cell Biol. 1991;11:2778. doi: 10.1128/mcb.11.5.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landry DB, Engel JD, Sen R. Functional GATA-3 binding sites within murine CD8 alpha upstream regulatory sequences. J Exp Med. 1993;178:941. doi: 10.1084/jem.178.3.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leiden JM. Transcriptional regulation of T cell receptor genes. Annu Rev Immunol. 1993;11:539. doi: 10.1146/annurev.iy.11.040193.002543. [DOI] [PubMed] [Google Scholar]
- 30.Merika M, Orkin SH. DNA-binding specificity of GATA family transcription factors. Mol Cell Biol. 1993;13:3999. doi: 10.1128/mcb.13.7.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pandolfi PP, Roth ME, Karis A, Leonard MW, Dzierzak E, Grosveld FG, Engel JD, Lindenbaum MH. Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat Genet. 1995;11:40. doi: 10.1038/ng0995-40. [DOI] [PubMed] [Google Scholar]
- 32.Ting CN, Olson MC, Barton KP, Leiden JM. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature. 1996;384:474. doi: 10.1038/384474a0. [DOI] [PubMed] [Google Scholar]
- 33.Ranganath S, Ouyang W, Bhattacharya D, Sha WC, Grupe A, Peltz G, Murphy KM. GATA-3-dependent enhancer activity in IL-4 gene regulation. J Immunol. 1998;161:3822. [PubMed] [Google Scholar]
- 34.Zhang DH, Yang L, Ray A. Differential responsiveness of the IL-5 and IL-4 genes to transcription factor GATA-3. J Immunol. 1998;161:3817. [PubMed] [Google Scholar]
- 35.Ouyang W, Ranganath SH, Weindel K, Bhattacharya D, Murphy TL, Sha WC, Murphy KM. Inhibition of Th1 development mediated by GATA-3 through an IL-4-independent mechanism. Immunity. 1998;9:745. doi: 10.1016/s1074-7613(00)80671-8. [DOI] [PubMed] [Google Scholar]
- 36.Ouyang W, Lohning M, Gao Z, Assenmacher M, Ranganath S, Radbruch A, Murphy KM. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity. 2000;12:27. doi: 10.1016/s1074-7613(00)80156-9. [DOI] [PubMed] [Google Scholar]
- 37.Robinson DS, Hamid Q, Ying S, et al. Predominant TH2-like bronchoalveolar T-lymphocyte population in atopic asthma. N Engl J Med. 1992;326:298. doi: 10.1056/NEJM199201303260504. [DOI] [PubMed] [Google Scholar]
- 38.Maggi E, Romagnani S. Role of T cells and T-cell-derived cytokines in the pathogenesis of allergic diseases. Ann N Y Acad Sci. 1994;725:2. doi: 10.1111/j.1749-6632.1994.tb39784.x. [DOI] [PubMed] [Google Scholar]
- 39.Umetsu DT, Dekruyff RH. Th1 and Th2, CD4+ cells in the pathogenesis of allergic diseases. Proc Soc Exp Biol Med. 1997;215:11. doi: 10.3181/00379727-215-44109. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura Y, Ghaffar O, Olivenstein R, Taha RA, Soussi-Gounni A, Zhang DH, Ray A, Hamid Q. Gene expression of the GATA-3 transcription factor is increased in atopic asthma. J Allergy Clin Immunol. 1999;103:215. doi: 10.1016/s0091-6749(99)70493-8. [DOI] [PubMed] [Google Scholar]
- 41.Nakamura Y, Christodoulopoulos P, Cameron L, et al. Upregulation of the transcription factor GATA-3 in upper airway mucosa after in vivo and in vitro allergen challenge. J Allergy Clin Immunol. 2000;105:1146. doi: 10.1067/mai.2000.107045. 10.1067/mai.2000.107045. [DOI] [PubMed] [Google Scholar]
- 42.Zhang DH, Yang L, Cohn L, Parkyn L, Homer R, Ray P, Ray A. Inhibition of allergic inflammation in a murine model of asthma by expression of a dominant-negative mutant of GATA-3. Immunity. 1999;11:473. doi: 10.1016/s1074-7613(00)80122-3. [DOI] [PubMed] [Google Scholar]
- 43.Ray A, Cohn L. Th2 cells and GATA-3 in asthma: new insights into the regulation of airway inflammation. J Clin Invest. 1999;104:985. doi: 10.1172/JCI8204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Traunecker A, Olivieri F, Karjalainen K. Myeloma based expression system for production of large mammalian proteins. Trends Biotechnol. 1991;9:109. doi: 10.1016/0167-7799(91)90038-j. [DOI] [PubMed] [Google Scholar]
- 45.Lanzavecchia A. Antigen-specific interaction between T and B cells. Nature. 1985;314:537. doi: 10.1038/314537a0. [DOI] [PubMed] [Google Scholar]
- 46.Lee HJ, O'Garra A, Arai K, Arai N. Characterization of cis-regulatory elements and nuclear factors conferring Th2-specific expression of the IL-5 gene: a role for a GATA-binding protein. J Immunol. 1998;160:2343. [PubMed] [Google Scholar]
- 47.Nakamura T, Lee RK, Nam SY, Podack ER, Bottomly K, Flavell RA. Roles of IL-4 and IFN-gamma in stabilizing the T helper cell type 1 and 2 phenotype. J Immunol. 1997;158:2648. [PubMed] [Google Scholar]
- 48.Lee HJ, Takemoto N, Kurata H, Kamogawa Y, Miyatake S, O'Garra A, Arai N. GATA-3 induces T helper cell type 2 (Th2) cytokine expression and chromatin remodeling in committed Th1 cells. J Exp Med. 2000;192:105. doi: 10.1084/jem.192.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rogge L, Barberis-Maino L, Biffi M, Passini N, Presky DH, Gubler U, Sinigaglia F. Selective expression of an interleukin-12 receptor component by human T helper 1 cells. J Exp Med. 1997;185:825. doi: 10.1084/jem.185.5.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szabo SJ, Jacobson NG, Dighe AS, Gubler U, Murphy KM. Developmental commitment to the Th2 lineage by extinction of IL-12 signaling. Immunity. 1995;2:665. doi: 10.1016/1074-7613(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 51.Ferber IA, Lee HJ, Zonin F, Heath V, Mui A, Arai N, O'Garra A. GATA-3 significantly downregulates IFN-gamma production from developing Th1 cells in addition to inducing IL-4 and IL-5 levels. Clin Immunol. 1999;91:134. doi: 10.1006/clim.1999.4718. 10.1006/clim.1999.4718. [DOI] [PubMed] [Google Scholar]
- 52.Murphy E, Shibuya K, Hosken N, Openshaw P, Maino V, Davis K, Murphy K, O'Garra A. Reversibility of T helper 1 and 2 populations is lost after long-term stimulation. J Exp Med. 1996;183:901. doi: 10.1084/jem.183.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishikomori R, Ehrhardt RO, Strober W. T helper type 2 cell differentiation occurs in the presence of interleukin 12 receptor beta2 chain expression and signaling. J Exp Med. 2000;191:847. doi: 10.1084/jem.191.5.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ying S, Durham SR, Corrigan CJ, Hamid Q, Kay AB. Phenotype of cells expressing mRNA for Th2-type (interleukin-4 and interleukin-5) and Th1-type (interleukin-2 and IFNγ) cytokines in bronchoalveolar lavage and bronchial biopsies from atopic asthmatic and normal control subjects. Am J Respir Cell Mol Biol. 1995;12:477. doi: 10.1165/ajrcmb.12.5.7742012. [DOI] [PubMed] [Google Scholar]