Abstract
Integrin αEβ7 is expressed almost exclusively by mucosal T cells and mucosal dendritic antigen-presenting cells (APCs) and is thought to be induced locally by transforming growth factor-β (TGF-β). In mice, mRNA for the αE subunit was found to be abundant in mucosal T cells but absent from other tissues. Exposure of a T-cell line to TGF-β strongly up-regulated αE mRNA levels within 30 min, and nuclear run-on experiments established that regulation occurred at the level of transcription. The organization of the human αE gene and a very closely linked novel gene, ELG, was determined. The αE promoter was tested in T cells and fibroblasts and functioned equally well in both cell types and did not confer TGF-β responsiveness. Regions of the promoter providing enhancer activity and phorbol 12-myristate 13-acetate (PMA) responsiveness were identified by deletion studies. DNAse 1 hypersensitivity analysis of 36 kb of the αE gene revealed one hypersensitive site, found only in αE+ cells, located near the transcription start points. These results show that, unlike the situation with other integrins, lineage specificity and cytokine responsiveness of αE transcription are not conferred by the proximal promoter. Specificity may depend on distant control elements that have not yet been identified.
Introduction
The integrin αEβ7 (CD103) was discovered in the late 1980s,1,2 but many aspects of its function and regulation remain enigmatic.3 In healthy humans or mice, expression is restricted almost exclusively to T cells and dendritic antigen-presenting cells (APC) in the mucosal immune system, and the integrin is prominent on intraepithelial lymphocytes in the gut.4–6 In inflammatory diseases, αE expression can be detected elsewhere. For example, T cells in the rheumatoid joint express αE as do CD8 T cells infiltrating kidney tubular epithelium during acute rejection of renal allografts or associated with the ductal epithelium of salivary and lachrymal glands in Sjogren's syndrome.7–9 Expression of the integrin is induced by transforming growth factor-β (TGF-β)4,5 and this cytokine is prominent in all tissue microenvironments in which αE+ cells are found. The only known ligand for αEβ7 is E-cadherin,10,11 a homophilic cell adhesion molecule of the immunoglobulin superfamily that is expressed on the basolateral aspect of epithelial cells and in adherens junctions. Recent experiments with αE null/null mice suggest that the integrin plays a role in retention of T cells in or near the mucosal epithelia,12 supposedly by interacting with E-cadherin.
The restricted distribution of αEβ7 is attributable largely to expression of the αE subunit, because the beta chain, in association with α4, is widely distributed on T and B cells13,14 and forms the principal mucosal homing receptor. In previous reports, Northern blot analysis of a range of human tissues showed αΕ mRNA to be present in gut, lung, thymus and spleen15 and, for mouse tissues, in the thymus and gut.16 TGF-β is the only cytokine known to induce αE expression but it has not been clear in previous studies whether TGF-β directly influences transcription of the αE gene or modulates post-transcriptional events.
The organization of the αE gene in mice, downsteam from exon 2, has been described in connection with the preparation of a CD103 null/null mouse12 but the upstream region encompassing the promoter has not been investigated. There have been no previous studies carried out on the human αE gene or its promoter. Transcriptional regulation of most other integrin alpha subunit genes has been analysed using reporter constructs transiently transfected into appropriate cell lines. These experiments have generally been successful in revealing transcription control elements that determine cell specificity or responsiveness to cytokines or growth factors.17–20 The rationale for studying transcriptional control of αE embraces not only questions concerning its unique tissue specificity but also the possibility that cis-regulatory elements in the promoter or elsewhere in the gene could have practical utility in directing expression of transgenes specifically to mucosal T cells in mice. Such a genetic tool would be of considerable value in manipulating the mucosal immune system.
The present report addresses preliminary questions concerning the regulation of αE expression, specifically, whether αE is transcriptionally regulated by TGF-β and whether transcription control elements can be identified in the proximal promoter. We have determined the structure of the whole human αE gene, including the upstream promoter region, and in doing so have identified a novel gene, ELG, which is very closely linked to αE. Functional analysis of the human αE promoter, and experiments on induction of αE transcription by TGF-β, were conducted using mouse T-cell lines because suitable TGF-β-responsive human T-cell lines are not available. Similarly, the distribution of αE mRNA in vivo was investigated using mouse tissues.
Materials and methods
Cell lines and culture conditions
The mouse T-cell lymphoma, TK1, and the mouse T-cell hybridoma, MTC-1,4 were maintained in RPMI-1640 containing 10% fetal calf serum (FCS). When induction or maintenance of αEβ7 expression was required, the medium was supplemented with recombinant human (rh)TGF-β2 (5 ng/ml). In some experiments MTC-1 cells were treated with phorbol 12-myristate 13-acetate (PMA) (15 ng/ml) and ionomycin (400 ng/ml) 1 hr before transfection with promoter reporter constructs. The human T-cell lymphoma Molt16, which secretes TGF-β constitutively and expresses αEβ7, was cultured in Dulbecco's modified Eagle's minimal essential medium (DMEM) +10% FCS. Mouse L-cell fibroblasts and human MRC-5 fibroblasts were also cultured in this medium.
Northern blotting
Tissues and gut intraepithelial lymphocytes were prepared from 6-week-old BALB/c specific pathogen-free (SPF) mice. Total cellular RNA was extracted from these samples or from cultured cells by standard procedures involving homogenization in Trizol (Gibco, BRL, Life Technologies Ltd, Paisley, UK), treatment with choroform and precipitation with isopropanol. RNA was resolved on a 1% agarose formamide gel and transferred onto a nitrocellulose membrane. Blots were hybridized with denatured probe at 42° overnight in 5 × Denhardt's solution, 5 × sodium chloride/sodium citrate (SSC), 50% (w/v) formamide, 0·15% sodium dodecyl sulphate (SDS) and 100 µg/ml of salmon-sperm DNA.21 Membranes were washed twice at 50° in 1 × SSC + 0·1% SDS and autoradiographed. The following probes were generated by restriction digestion of plasmids or by the polymerase chain reaction (PCR): an 800-bp fragment from the 3′ end of mouse αE cDNA and a 250-bp fragment from the 5′ end of glyceraldehyde phosphate dehydrogenase (GAPDH) cDNA. Probes were labelled using the Prime-It II, Random Primer Labelling Kit (Stratagene Europe, Amsterdam, the Netherlands).
Nuclear run-on
TKI cells (5 × 107) were cultured overnight with or without TGF-β, washed in 10 mm Tris/HCl, pH 7·4, containing 3 mm CaCl2 and 2 mm MgCl2, and then lysed in the same buffer containing 1% Nonidet P-40 (NP-40). After homogenization with a Dounce homogeniser, nuclei were recovered by centrifugation (500 g at 4°) and resuspended in 200 µl of storage buffer [50 mm Tris/HCl (pH 8·3), 40% (w/v) glycerol, 5 mm MgCl2, 0·1 mm EDTA] and stored temporarily at −80°. To generate 32P-labelled unprocessed mRNA transcripts, nuclei were thawed and incubated for 30 min at 30° with 200 µl of reaction buffer: 10 mm Tris/HCl (pH 8·0), 5 mm MgCl2, 0·3 m KCl, 10 µl each of 100 mm ATP, 100 mm CTP and 100 mm GTP, and 10 µl of 10 mCi/ml [α-32P]UTP. Extraction, purification and hybridization of labelled RNA to cDNA adsorbed to nitrocellulose followed a standard protocol for nuclear run-on analysis.21 The membrane for hybridization was prepared as follows: maxipreps of pcDNA3 containing mouse αE cDNA were prepared using the Qiagen Maxiprep Kit. One-microgram samples in 200 µl of 6 × SSC were denatured at 100°, chilled on ice, diluted with 300 µl of 20 × SSC and then adsorbed to a nitrocellulose membrane using a dot-blot manifold. Control cDNA for β-actin was prepared similarly. The membrane was air-dried and the cDNA immobilized with a UV-Stratalinker.
Determination of the transcription start sites
The transcription start sites were identified by RNA Ligase-Mediated Rapid Amplification of cDNA Ends (RLM–RACE; Promega UK Ltd, Southampton, UK), according to the supplier's protocol. This technique depends on the selection of full-length and capped mRNA for analysis. Briefly, mRNA was prepared from Molt16 cells by adsorption to oligo (dT) cellulose (FastTrack 2·0; Invitrogen, Groningen, the Netherlands). It was then treated with calf intestinal phosphatase to dephosphorylate mRNA with incomplete, uncapped, 5′ ends, followed by treatment with Tobacco Acid Pyrophosphatatase (TAP) to remove the cap from full-length mRNA, leaving an intact 5′ phosphate. A synthetic RNA adaptor was then added using T4 RNA ligase. The product was reverse transcribed using random primers and the cDNA subjected to nested PCR using primers to the 5′ adaptor and outer and inner αE-specific reverse primers: outer, 5′-TGCATATCAAAACACCGTGGTGGC (323 bp downstream of the ATG initiation codon); inner, 5′-TGGGGACATGCTCTACAGGATGGC (260 bp from the initiation codon). In the αE gene, the primers were located in exon 4 (outer primer) and in the region spanning exons 3 and 4 (inner primer). The RACE products were cloned into pT-Adv (Clontech Labs, Basingstoke, Hampshire, UK) and sequenced. A negative control was performed in which treatment with TAP was omitted. In addition, conventional reverse transcription (RT)–PCR was performed on mRNA using a forward primer (5′-CCTCCTGGCTGAGGGGAAGCTG) encompassing a putative initiator sequence. This primer was used in combination with the inner reverse primer given above.
Promoter reporter assays
Genomic DNA was prepared from buffy coat human blood leucocytes using the Genomic G2 Kit (Helena Biosciences UK, Sunderland, UK). The region of the αE gene immediately adjacent to the translation start codon and extending to 4 kb upstream was prepared by PCR using Pfu polymerase. Deletion mutants were prepared from the full-length product by using a series of forward primers and a common reverse primer. Forward and reverse primers included a Kpn1 site and a HindIII site, respectively: −4 kb forward, 5′-TACGGTACCAACATGGTGAAACCCCGTCTCT-3′; −1909 bp forward, 5′-GGCGGTACCTGGGTTTGTGGGGAAAGCA-3′; −1510 bp forward, 5′-GGCGGTACCGAGGAGGAGGGTCACGAGC-3′; −1096 bp forward, 5′-GGCGGTACCAGACACTCATGTTAGAGAGC-3′; −516 bp forward, 5′-GGCGGTACCGGCTGGGGAGGCACTTCCTA-3′; −446 bp forward, 5′-GGCGGTACCTGGGCTGGAAGCTGGGCTGA-3′; −176 bp forward, 5′-GGCGGTACCTGTGTACGAGCCGGAGTGTC-3′ and reverse, 5′-GAGAAGCTTCCTTGCTGGAGCAGAGGCGG-3′.
PCR products were cloned into firefly luciferase reporter vectors pGL3-Basic and pGL3-Enhancer (Promega), according to standard procedures. The following control constructs were used: pGL3-Control containing the SV40 promoter/enhancer linked to firefly luciferase; pRL-CMV, a Renilla luciferase construct containing the cytomegalovirus (CMV) promoter used to normalize transfection efficiency; and (CAGA)12MLP-Luc containing the initiator sequence of an adenoviral minimal promoter plus a classical TGF-β response element linked to firefly luciferase (kindly supplied by Dr J. M. Gauthier, Glaxo Wellcome, 91951 Les Ulis Cedex, France).22 Promoter constructs (45 µg) were cotransfected with pRL-CMV (5 µg) into cell lines (4 × 106 cells) by electroporation (Gene Pulser; BioRad, Hemel Hempstead, Herts, UK). Twenty-four hours later, cell lysates were assayed sequentially for Firefly luciferase and Renilla luciferase using Promega reagents and a luminometer. Results are expressed as relative luciferase activity, representing the ratio Firefly: Renilla signals.
DNase hypersensitivity analysis
Buffers and procedures for preparation of nuclei and subsequent DNAse 1 digestion have been described in detail previously.21 Briefly, nuclei were prepared from 4 × 107 Molt16 T cells or MRC5 fibroblasts by lysis in 0·4% NP-40. Aliquots (200 µl) of nuclei suspension were treated with 0–20 U/ml of DNAse I (DNAse Grade 1; Boehringer Mannheim, Roche Diagnostics, Lewes, East Sussex, UK) for 10 min at 25° followed by addition of 200 µl of stop solution and overnight incubation at 50° in proteinase K (200 µg/ml). DNA was purified and incubated overnight with ClaI or with EspI plus MluI. Probes (459 bp to 744 bp) were generated by PCR and labelled using a Prime It Random Primer Labelling Kit (Stratagene). The αE gene was analysed in the region −6·1 kb upstream of the translation start codon to +8·7 kb downstream, encompassing exon 1 and the first half of intron 1. The segments of DNA and the primers used to generate probes specific for the 5′ or 3′ ends of the fragments are defined as follows, with numbering in accordance with the EMBL database entry AC002993: 37235–45145 (5′ upstream and exon 1), forward primer 5′-GTCGGTGAGGAAAGTGAGAGC-3′, reverse primer 5′-CAGAGGTGATGGTGACAATTTGAG-3′; 45046–52118 (5′ part of intron 1), forward primer 5′-CCATTACCCCCAAACAAGAACG-3′, reverse primer 5′-TCTTCACATCTCTGCCTCGGCCT-3′; Electrophoresis and hybridization were carried out as described above for Northern blotting.
Results
Tissue distribution of αE mRNA
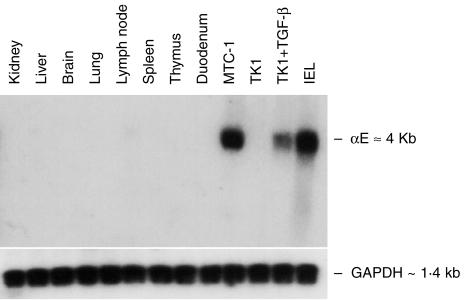
Northern blot analysis was conducted on a range of BALB/c mouse tissues (Fig. 1). The αE+ mouse T-cell hybridoma, MTC-1 (cultured in the presence of TGF-β), was included as a positive control, and the mouse T-cell lymphoma, TK1, was tested before and after overnight treatment with TGF-β. Intraepithelial lymphocytes (IEL) prepared from small intestine gave a strong αE signal at ≈4 kb, as did MTC-1 cells and TK1 cells after treatment with TGF-β. All tissue samples, including duodenum, were negative. Failure to detect αE transcripts in duodenum was a result of the low number of IELs present in the gut of 3-month-old barrier-reared mice.
Figure 1.
Northern blotting for αE mRNA in a range of mouse tissues. RNA extracted from BALB/c mouse tissues, from small intestinal intraepithelial lymphocytes (IEL) and from the T-cell lines MTC-1 and TK1, was tested by Northern blotting for αE mRNA. Blotting for glyceraldehyde phosphate dehydrogenase (GAPDH) verified equal loading.
TGF-β induces αE transcription
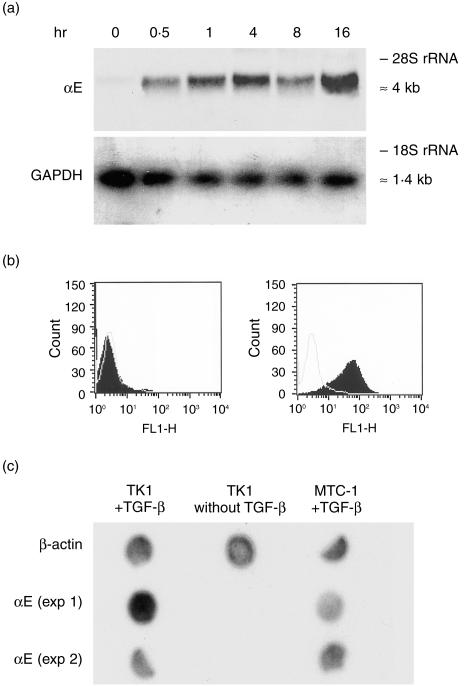
Mouse TK1 T lymphoma cells were cultured for 16 hr with TGF-β, and RNA was sampled at specific time intervals within this period. Northern blotting showed that a strong αE mRNA signal was detected after 30 min and was maximal at 16 hr (Fig. 2a). Cell-surface expression of αEβ7 at the beginning and end of this incubation period is shown (Fig. 2b). Nuclear run-on experiments were performed to determine whether regulation by TGF-β occurred at the level of transcription. Newly synthesized, unprocessed, nuclear RNA isolated from TK1 cells, before and after stimulation with TGF-β, was tested for nascent αE transcripts. Figure 2(c) shows that such transcripts were present after, but not before, exposure of the cells to TGF-β, clearly demonstrating regulation of αE transcription. Nuclear RNA from αE+ MTC-1 cells cultured continuously in the presence of TGF-β provided a positive control, and detection of β-actin mRNA in all samples validated the technique.
Figure 2.
Stimulation of transcription and expression of αE by transforming growth factor-β (TGF-β). (a) TK1 cells were treated for 16 hr with TGF-β and RNA was extracted at specific time-points and tested for αE transcripts. Probing for glyceraldehyde phosphate dehydrogenase (GAPDH) verified equal loading. (b) Cell-surface expression of αEβ7 by TK1 cells before (left panel) or after (right panel) treatment for 16 hr with TGF-β; black profiles show staining with monoclonal antibody (mAb) M290 (specific for αE), unfilled profiles show the negative control with isotyped-matched irrelevant mAb. (c) Nuclear run-on analysis to test for newly synthesized, nascent, αE transcripts in TK1 cells before and after treatment with TGF-β. 32P-labelled nuclear RNA was extracted from TK1 cells before or after overnight stimulation with TGF-β and from αE+ MTC-1 cells cultured in the presence of TGF-β. The RNA was tested for hybridization to αE cDNA. Detection of nascent mRNA for β-actin in all samples validated the technique.
Organization of the αE gene
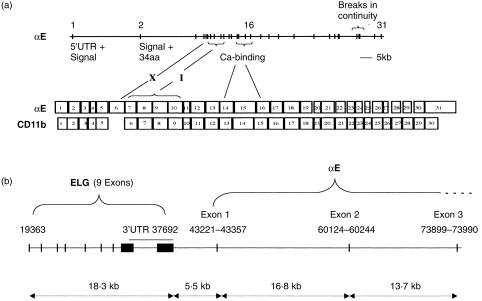
The human αE gene was identified in the EMBL database entry AC002993, which consists of a series of contiguous lengths of DNA from chromosome 17 with no previously identified affiliation to known genes. The entry was detected using BLAST searches with the published human αE cDNA sequence.15 The structure of the αE gene was deduced from the unordered contigs using LocalFASTA and reiterative searching; intron/exon boundaries were confirmed by known properties of splice junctions. Alpha E contains 31 exons spanning at least 85 kb (Fig. 3a). There were three gaps in continuity of sequence in the database entry. A gap in intron 1 was analysed by PCR and found to be 10 bp. Two gaps at the 3′ end have not yet been analysed. The alpha E protein contains a unique small acidic region, the X-domain, in which there is a post-translational cleavage site that has no clear functional role.23 It is encoded by exon 6. Immediately downstream, the I-domain is encoded by four exons, 7–10. In keeping with general principles governing the insertion of ‘functional modules’ into genes, both these regions have phase 1 intron/exon boundaries.24 Figure 3a shows an alignment of the αE gene with functionally homologous exons in CD11b, the most closely related integrin (15). Intron/exon boundaries were compared and shown to be closely similar, showing displacement of six amino acids or fewer. The seven repeats of the β propeller of αE are encoded by exons 2–17, the Ca2+-binding regions being in three consecutive exons, 14–16.
Figure 3.
The αE gene. (a) Organization of intron/exon boundaries and alignment of exons with those of the CD11b gene. Exons coding for the X-domain, the I-domain and the calcium-binding region are identified. (b) The proximal region of the human αE gene and the closely linked novel gene ELG. Numbers refer to nucleotide positions in the EMBL database entry accession no.: AC002993. Exons are marked as vertical bars or blocks.
We have identified a new gene closely linked to αE (Fig. 3b). The genomic sequence 5·5 kb upstream of αE matched four clusters of overlapping expressed sequence tags. The only possible open reading frame was identified and the amino acid sequence of a novel protein was deduced (acc. no.: AJ277841). The new gene has been named ELG. It contains nine exons and has an open reading frame of 1023 bp and a 3′ untranslanted region (UTR) of 3 kb. mRNA transcripts of ELG have been detected in a wide range of cell lines. Using multiple sequence alignment analysis, ELG was shown to have some characteristics in common with nucleosome assembly proteins, including possession of a nuclear localization signal and a highly acidic region. Taken together, these characteristics suggest that the new protein may be a nuclear chaperone.25 Because ELG is very closely linked to αE, and that we have analysed most of the 5·5 kb intervening region for promoter activity, the transcriptional regulation of this new gene was examined in parallel with αE. ELG was shown to be regulated independently of αE (data not included).
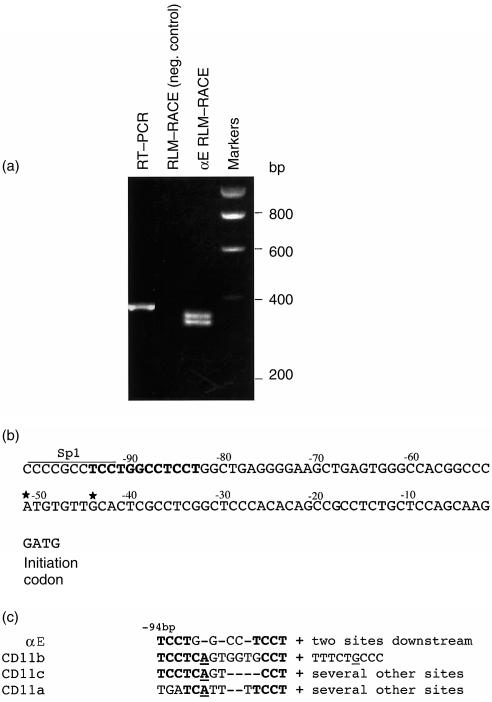
In keeping with most other integrin α-chain genes, the αE gene does not contain a TATA box. Analysis of full-length, capped, αE mRNA by 5′ RLM–RACE, identified two transcription start sites at 51 bp and 44 bp, respectively, upstream of the initial methionine codon (Fig. 4a, 4b). A region ≈40 bp further upstream contains a consensus initiator motif within close proximity of an SP1 site (Fig. 4c). It is probable that transcription starts here also, but we were not able to detect an RLM–RACE product of this length. Nevertheless, a longer mRNA encompassing the putative initiator was detected by conventional RT–PCR (Fig. 4a). RLM–RACE may have failed to reveal this longer species because it is present in low abundance or, alternatively, technical reasons may have prevented ligation of this mRNA to the RLM–RACE adaptor. It is notable that the sequence immediately downstream of the putative initiator was unusually GC-rich and may have displayed secondary structure, which would militate against its detection by RLM–RACE.
Figure 4.
Determination of αE transcription start sites. (a) RLM–RACE was performed on mRNA prepared from Molt16 cells. The track next to the markers shows two closely spaced products, of ≈330 bp in size, which define two transcription start points (note that the 5′ adaptor adds 38 bp to the 5′ end of the mRNA). The track on the left shows a longer product of 360 bp, which was generated by conventional RT–PCR using a forward primer spanning a consensus transcription-initiator sequence located ≈45 bp upstream of the other start sites. This product reflects the existence of a longer, probably minor, species of mRNA and an additional start point; it could not be detected by RML-RACE. (b) The αE gene immediately 5′ of the ATG methionine codon. A consensus initiator sequence is identified in bold type, closely adjacent to an Sp1 site. The two positively identified transcription start sites are identified with stars. (c) Comparison of initiation sequences among related integrin α-chain genes. Underlined bases indicate the start points.
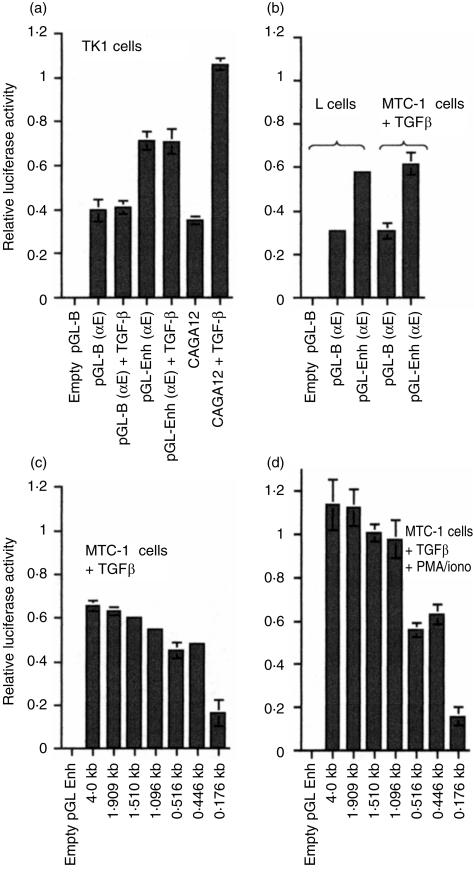
Activity of the αE promoter
Luciferase reporter constructs were prepared containing 1·9 kb of the αE promoter extending 5′ upstream from the initial methionine codon in exon 1 towards the ELG gene (see Fig. 3b). Two reporter vectors were used: pGL-B (a basic version) and pGL-Enh (which contains the SV40 enhancer incorporated downstream of luceriferase). Promoter constructs were tested in TK1 cells (a T-cell line) in the presence or absence of TGF-β (Fig. 5a). A construct containing a synthetic TGF-β response element (CAGA)12, linked to an adenoviral minimal promoter, was included as a positive control. Reporter constructs were also tested in mouse fibroblast L-cells or in the T-cell hybridoma MTC-1 (Fig. 5b). The promoter showed neither TGF-β responsiveness nor T-cell specificity, functioning equally well in T cells or fibroblasts regardless of the presence of TGF-β. In contrast, the (CAGA)12 construct responded strongly to TGF-β. The presence of the SV40 enhancer in the reporter vector doubled the activity of the αE promoter. To gain information about the strength of the αE promoter relative to that of a known strong viral promoter, we tested the SV40 early promoter in MTC-1 cells in parallel with αΕ. The viral promoter gave a relative luciferase value of 4·0 ± 0·3, ≈10-fold higher than the basic αE construct. The αE promoter construct was extended to 4 kb upstream of the open reading frame and a series of deletion mutants was prepared and tested in MTC-1 cells, which were cultured in the presence of TGF-β and were strongly αEβ7+ (Fig. 5c). The experiments were also performed in MTC-1 cells that had been treated with PMA and ionomycin immediately before transfection (Fig. 5d). The region −176 bp to −446 bp displayed enhancer activity, and the region −516 bp to −1096 bp was required for responsiveness to PMA/ionomycin. Inspection of the nucleotide sequence in these two regions showed potentially significant transcription-factor binding motifs, including GATA-1 sites in the putative enhancer region and a series of AP1 sites in the PMA-responsive region, but these have not yet been analysed for functional activity. Motifs for transcription factors of particular significance to T cells,26 e.g. GATA3, TCF1, LEF1 and Sox4, were not present in the promoter. The 4-kb promoter construct was tested also in fibroblasts (L-cells) and gave values similar to those obtained with the 1·9-kb promoter, as shown in Fig. 5(b).
Figure 5.
Alpha E promoter analysis. (a) The 1·9-kb αE promoter was incorporated into a basic luciferase reporter construct (pGL-B) or into a similar vector containing the SV40 enhancer (pGL-Enh) and transiently transfected into TK1 cells, which were cultured for 24 hr in the presence or absence of TGF-β. A positive control containing a TGF-β response element (CAGA)12 was tested in parallel. (b) Similar constructs were tested in L-cells in the absence of TGF-β or in MTC-1 cells in the presence of TGF-β. (c) deletions prepared from a 4-kb αE promoter construct incorporated into the pGL-Enh vector were tested in MTC-1 cells cultured in the presence of TGF-β. (d) The deletion series was tested in MTC-1 cells that had been stimulated, in addition, with phorbol 12-myristate 13-acetate (PMA) and ionomycin.
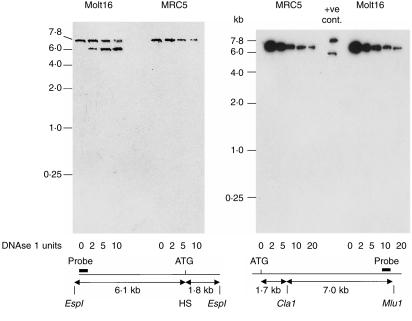
DNase 1 hypersensitivity
Because response elements conferring TGF-β responsiveness or T-cell specificity were not detected in promoter constructs, we sought evidence of more distant control elements by testing for hypersensitivity to DNAse 1. Human Molt16 cells, which were actively transcribing αE, were compared with human MRC5 fibroblasts, which cannot transcribe αE. Figure 6 shows results from the 5′ upstream region (left panel) and the first half of intron 1 (right panel). A hypersensitive region is clearly identified, ≈1·9kb 5′ of the EspI restriction site. This corresponds to a position near the ATG translation start codon and probably reflects the position of the transcription start sites. Analyses of the remainder of intron 1, as well as exon 2 and intron 2, gave negative results closely similar to those in the right panel of Fig. 6, and are not shown. Therefore, no additional DNAse 1 hypersensitive sites were detected over the entire 36·5 kb analysed. Detection of the hypersensitivity site near the first methionine codon was used as a positive control in all experiments. It is striking in Fig. 6 that the susceptibility of the αE gene to degradation by increasing concentrations of DNase was identical in Molt16 and MRC5 cells. This suggests that the chromatin organization in this region was similar in both cell types.
Figure 6.
DNAse 1 hypersensitivity analysis of the proximal region of the human αE gene. DNA from Molt16 cells that were transcribing αE was compared with DNA from MRC5 fibroblasts. The left panel shows the region flanking the initial methionine codon and the right panel extends the analysis through half of intron 1. HS indicates the position of a hypersensitive site. +ve cont., positive control.
Discussion
Our objectives were to affirm that transcription of the αE integrin gene is normally restricted to mucosal T cells and to gain insight into the regulation of transcription. In BALB/c mice, αE mRNA was detected only in IELs. The data extend and largely agree with previous results in human and mouse for the tissue distribution of αE mRNA,15,16 taking into account differences in the sensitivity of the detection systems used. The distribution of αE mRNA therefore reflects the tissue distribution of the αEβ7 heterodimer.1,2 Results from nuclear run-on and Northern blotting show unequivocally that TGF-β acts within 30 min to induce αE transcription. Most other TGF-β-responsive genes are also regulated at the level of transcription. Nevertheless, in a small number of studies, TGF-β has been shown to elevate steady-state levels of mRNA by increasing message stability.27–29 The mechanism depends on recognition of nucleotide motifs in the 3′-UTR of the mRNA by cytoplasmic transacting factors. Recognition sequences are usually repeated or palindromic, and are U-rich. There are no such motifs in the 3′-UTR of αE mRNA and no conserved motifs common to the human and mouse 3′-UTR. Because we could not detect nascent or mature αE mRNA in unstimulated TK1 cells, it was considered unlikely that regulation of message stability was a significant factor in the TGF-β responsiveness we observed.
The human promoter constructs were tested in mouse T-cell lines. This was necessary because we have not yet identified a convenient human T-cell line that synthesizes αE in response to stimulation by an exogenous source of TGF-β. It should be emphasized that the tissue distribution of αE in vivo, and its induction in peripheral T cells in vitro by TGF-β, is essentially identical in human and mouse. Our αE promoter constructs showed neither T-cell specificity nor TGF-β responsiveness in transient transfection assays. Furthermore, TK1 cells that had been stably transfected with the 1·9-kb promoter linked to GFP expressed the reporter in a TGF-β-independent manner (data not shown). It is very unlikely that the use of mouse cell lines, as opposed to human cells, had any bearing on our findings. An extensive body of literature validates the use of heterologous mouse/human systems for promoter studies, and human integrin promoters are known to confer tissue-specific expression in transgenic mice.30–32 The absence of cell lineage specificity and TGF-β responsiveness in our experiments was unexpected in view of results with other integrin subunit promoters, many of which have revealed cell lineage-specific regulatory elements in transient transfection assays. For example, a 1-kb construct of the human α2 promoter conferred both cell type and differentiation-dependent expression during megakaryocytic differentiation.20 Studies on the α6 integrin, which is expressed mainly in epithelial and endothelial cells, showed that the promoter was functional in keratinocytes and breast carcinoma cells, but not in a leukaemia cell line, again demonstrating lineage specificity.19 Similarly, the proximal promoter for the CD11c integrin subunit was functional only in LFA-1+ cells.18
DNAse 1 hypersensitivity has been used extensively to identify control elements, particularly enhancers and locus control regions, within transcriptionally active DNA.33 In the present study we compared the αE gene in Molt16 cells that were actively transcribing αE mRNA with that in MRC5 fibroblasts which cannot transcribe αE. Surprisingly, only one hypersensitive site was detected in Molt16 cells, in the proximal part of the promoter. Apart from this site, the αE gene in Molt16 and MRC5 showed equal susceptibility to DNAse 1 digestion, suggesting that the chromatin may have been in an open configuration in both cell types and that transcriptional regulation of αE may not involve chromatin remodelling. DNAse 1 sensitivity of αE and the closely linked gene ELG has not yet been examined in non-dividing cells and it is possible that the open configuration is related to cell division or activation.
We have been unable to reveal a TGF-β response element in the promoter region of the αE gene or a DNAse 1 hypersensitive site that might contain one. Because our promoter constructs were active in all circumstances, we suggest that TGF-β responsiveness may be conferred by relief from transcriptional repression caused by a control element some distance away from the region analysed. De-repression by TGF-β receptor signalling has been reported in other circumstances.34,35 Testing larger constructs in transgenic mice will be required to reveal such an element and to explain the bias of αE transcription towards CD8+ cells.36 The stimulatory effect of PMA/ionomycin on our reporter constructs, and the presence of several AP1 sites in the responsive region of DNA, is consistent with the observation that T-cell activation is important in initiating αE transcription.36,37
The very rapid increase of αE mRNA in TK1 cells following exposure to TGF-β suggests that transcription is a direct and immediate consequence of TGF-β signalling. Recently, the principal components of the TGF-β signalling pathway have been identified.38–40 Serine phosphorylation of Smad 2 or Smad 3 by the TGF-β receptor is a key event in the initial stages of the pathway. We recently confirmed that induction of αE expression in TK1 cells is dependent on serine/threonine phosphorylation, but not tyrosine phosphorylation, and showed also that Smad 3 is not required for expression of αE in vivo (P. J. Kilshaw, unpublished). The significance of Smad 2 in this context awaits investigation. The possibility that stimuli other than TGF-β could induce αE synthesis cannot be entirely ruled out but, apart from one report suggesting that ligation of β1 integrins induces αE expression,41 there is no evidence for other mechanisms.
It is probable that transcriptional regulation of the αE gene will prove to be considerably more complex than that of other integrin genes. Nevertheless, our observation that the distribution of αE mRNA is exquisitely tissue specific and mainly, if not solely, regulated at the transcriptional level, offers promise that the αE gene could be used as a vehicle for targeting expression of transgenes to the mucosal immune system by homologous recombination.
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council, UK. We thank Dr P. Evans for his help in determining the transcription start sites.
References
- 1.Cerf-Bensussan N, Jarry A, Brousse N, Lisowska-Grospierre B, Guy-Grand D, Griscelli C. A monoclonal antibody (HML-1) defining a novel membrane molecule present on human intestinal lymphocytes. Eur J Immunol. 1987;17:1279–85. doi: 10.1002/eji.1830170910. [DOI] [PubMed] [Google Scholar]
- 2.Kilshaw PJ, Baker KC. A unique surface antigen on intraepithelial lymphocytes in the mouse. Immunol Lett. 1988;18:149–54. doi: 10.1016/0165-2478(88)90056-9. [DOI] [PubMed] [Google Scholar]
- 3.Kilshaw PJ. Alpha E beta 7. J Clin Pathol Mol Pathol. 1999;52:203–7. doi: 10.1136/mp.52.4.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kilshaw PJ, Murant SJ. A new surface antigen on intraepithelial lymphocytes in the intestine. Eur J Immunol. 1990;20:2201–7. doi: 10.1002/eji.1830201008. [DOI] [PubMed] [Google Scholar]
- 5.Parker CM, Cepek K, Russell GJ, Shaw SK, Posnett DN, Schwarting R, Brenner MB. A family of beta 7 integrins on human mucosal lymphocytes. Proc Natl Acad Sci USA. 1992;89:1924–8. doi: 10.1073/pnas.89.5.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kilshaw PJ. Expression of the mucosal T cell integrin αM290β7 by a major subpopulation of dendritic cells in mice. Eur J Immunol. 1993;23:3365–8. doi: 10.1002/eji.1830231246. [DOI] [PubMed] [Google Scholar]
- 7.Baumgart M, Witt C, Huge W, Muller B. Increase in the expression of alpha E beta 7, characteristic of intestinal intraepithelial lymphocytes, on cells in the lung epithelium of patients with interstitial lung diseases and in synovial fluid of patients with rheumatic diseases. Immunobiology. 1996;196:415–24. doi: 10.1016/s0171-2985(96)80063-5. [DOI] [PubMed] [Google Scholar]
- 8.Robertson H, Wong WK, Burt AD, Kirby JA. Renal allograft rejection: does CD103 hold the clue to tissue-specific cytotoxicity? Immunology. 1999;98(Suppl. 1):125. [Google Scholar]
- 9.Fujihara T, Fujita H, Tsubota K, Saito K, Tsuzaka K, Abe T, Takeuchi T. Preferential localisation of CD8+ alpha E beta 7+ T cells around acinar epithelial cells with apoptosis in patients with Sjogren's syndrome. J Immunol. 1999;163:2226–35. [PubMed] [Google Scholar]
- 10.Cepek KL, Shaw SK, Parker CM, Russell GJ, Morrow JS, Rimm DL, Brenner MB. Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and alpha E beta 7 integrin. Nature. 1994;372:190–3. doi: 10.1038/372190a0. [DOI] [PubMed] [Google Scholar]
- 11.Karecla PI, Bowden SJ, Green SJ, Kilshaw PJ. Recognition of E-cadherin on epithelial cells by the mucosal T cell integrin alpha E beta 7. Eur J Immunol. 1995;25:852–6. doi: 10.1002/eji.1830250333. [DOI] [PubMed] [Google Scholar]
- 12.Schon MP, Arya A, Murphy EA, et al. Mucosal T lymphocyte numbers are selectively reduced in integrin αE (CD103)-deficient mice. J Immunol. 1999;162:6641–9. [PubMed] [Google Scholar]
- 13.Kilshaw PJ, Murant SJ. Expression and regulation of β7 (βp) integrins on mouse lymphocytes: relevance to the mucosal immune system. Eur J Immunol. 1991;21:2591–7. doi: 10.1002/eji.1830211041. [DOI] [PubMed] [Google Scholar]
- 14.Andrew DP, Rott LS, Kilshaw PJ, Butcher EC. Distribution of α4β7 and αEβ7 on thymocytes, intestinal epithelial lymphocytes and peripheral lymphocytes. Eur J Immunol. 1996;26:897–05. doi: 10.1002/eji.1830260427. [DOI] [PubMed] [Google Scholar]
- 15.Shaw SK, Cepek KL, Murphy EA, Russell GJ, Brenner MB, Parker CM. Molecular cloning of the human mucosal lymphocyte integrin αE subunit. Unusual structure and restricted RNA distribution. J Biol Chem. 1994;269:6016–25. [PubMed] [Google Scholar]
- 16.Smith TJ, Ducharme LA, Shaw SK, Parker CM, Brenner MB, Kilshaw PJ, Weis JH. Murine M290 integrin expression modulated by mast cell activation. Immunity. 1994;1:393–03. doi: 10.1016/1074-7613(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 17.Nueda A, Lopez-Cabrera M, Vara A, Corbi AL. Characterization of the CD11α (αL, LFA-1α) integrin gene promoter. J Biol Chem. 1993;268:19305–11. [PubMed] [Google Scholar]
- 18.Corbi AL, Lopez-Rodriguez C. CD11c integrin gene promoter activity during myeloid differentiation. Leuk Lymphoma. 1997;25:415–25. doi: 10.3109/10428199709039028. [DOI] [PubMed] [Google Scholar]
- 19.Lin C-S, Chen Y, Huynh T, Kramer R. Identification of the human α6 integrin gene promoter. DNA Cell Biol. 1997;16:929–37. doi: 10.1089/dna.1997.16.929. [DOI] [PubMed] [Google Scholar]
- 20.Zutter MM, Santoro SA, Painter AS, Tsung YL, Gafford A. The human α2 integrin gene promoter. Identification of positive and negative regulatory elements important for cell-type and developmentally restricted gene expression. J Biol Chem. 1994;269:463–9. [PubMed] [Google Scholar]
- 21.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. USA: John Wiley and Sons Inc.; 1998. [Google Scholar]
- 22.Dennler S, Itoh S, Vivien D, ten Dijke P, Huet S, Gauthier J-M. Direct binding of Smad3 and Smad4 to critical TGFβ-inducible elements in the promoter of human plasminogen activator inhibitor-type 1 gene. EMBO J. 1998;17:3091–00. doi: 10.1093/emboj/17.11.3091. 10.1093/emboj/17.11.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higgins JM, Cernadas M, Tan K, Irie A, Wang J, Takada Y, Brenner MB. The role of alpha and beta chains in ligand recognition by β7 integrins. J Biol Chem. 2000;275:25652–64. doi: 10.1074/jbc.M001228200. [DOI] [PubMed] [Google Scholar]
- 24.Tuckwell D. Evolution of von Willibrand factor A (VWA) domains. Biochem Soc Trans. 1999;27:835–40. doi: 10.1042/bst0270835. [DOI] [PubMed] [Google Scholar]
- 25.Philpott A, Krude T, Laskey RA. Nuclear chaperones. Semin Cell Dev Biol. 2000;11:7–14. doi: 10.1006/scdb.1999.0346. [DOI] [PubMed] [Google Scholar]
- 26.Kuo CT, Leiden JM. Transcriptional regulation of T lymphocyte development and function. Annu Rev Immunol. 1999;17:149–87. doi: 10.1146/annurev.immunol.17.1.149. [DOI] [PubMed] [Google Scholar]
- 27.Amara FM, Entwistle J, Kuschak TI, Turley EA, Wright JA. Transforming growth factor beta-1 stimulates multiple protein interactions at a unique cis-element in the 3′-untranslated region of the hyaluronan receptor RHAMM mRNA. J Biol Chem. 1996;271:15279–84. doi: 10.1074/jbc.271.25.15279. [DOI] [PubMed] [Google Scholar]
- 28.Amara FM, Junaid A, Clough RR, Liang B. TGF-beta (1) regulation of alzheimer amyloid precursor protein mRNA expression in a normal human astrocyte line: mRNA stabilization. Brain Res Mol Brain Res. 1999;71:42–9. doi: 10.1016/s0169-328x(99)00158-8. 10.1016/s0169-328x(99)00158-8. [DOI] [PubMed] [Google Scholar]
- 29.Werkmeister JR, Blomme EA, Weckmann MT, et al. Effect of transforming growth factor on parathyroid hormone-related protein secretion and mRNA expression by normal human keratinocytes in vitro. Endocrine. 1998;8:291–9. doi: 10.1385/ENDO:8:3:291. [DOI] [PubMed] [Google Scholar]
- 30.Hirsch E, Balzac F, Pastore C, Tarone G, Silengo L, Altruda F. The beta 1 integrin distal promoter is developentally regulated in transgenic mice. Cell Adhes Commun. 1993;1:203–12. doi: 10.3109/15419069309097254. [DOI] [PubMed] [Google Scholar]
- 31.Ritchie KA, Aprikian A, Gollahon KA, Hickstein DD. The human leukocyte integrin CD11a promoter directs expression in leukocytes of transgenic mice. Blood. 1995;86:147–55. [PubMed] [Google Scholar]
- 32.Kwiatkowski BA, Embree LJ, Ritchie KA, Hickstein DD. Human leukocyte integrin CD18 promoter directs low levels of expression of a mutated human CD4 reporter gene in leukocytes of transgenic mice. Biochem Biophys Res Commun. 1996;222:601–6. doi: 10.1006/bbrc.1996.0790. 10.1006/bbrc.1996.0790. [DOI] [PubMed] [Google Scholar]
- 33.Krebs JE, Peterson CL. Understanding ‘active’ chromatin: an historical perspective of chromatin remodeling. Crit Rev Eukaryot Gene Expr. 2000;10:1–12. [PubMed] [Google Scholar]
- 34.Shi X, Yang X, Chen D, Chang Z, Cao X. Smad1 interacts with homeobox DNA-binding proteins in bone morphogenic protein signaling. J Biol Chem. 1999;274:13711–7. doi: 10.1074/jbc.274.19.13711. [DOI] [PubMed] [Google Scholar]
- 35.Verschueren K, Remacle JE, Collart C, et al. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5′-CACCT sequences in candidate target genes. J Biol Chem. 1999;274:20489–98. doi: 10.1074/jbc.274.29.20489. [DOI] [PubMed] [Google Scholar]
- 36.Brew R, West DC, Burthem J, Christmas SE. Expression of the human mucosal lymphocyte antigen, HML-1, by T cells activated with mitogen or specific antigen in vitro. Scand J Immunol. 1995;41:553–62. doi: 10.1111/j.1365-3083.1995.tb03607.x. [DOI] [PubMed] [Google Scholar]
- 37.Schieferdecker HL, Ullrich R, Weiss-Breckwoldt AN, Schwarting R, Stein H, Riecken EO, Zeitz M. The HML-1 antigen of intestinal lymphocytes is an activation antigen. J Immunol. 1990;144:2541–9. [PubMed] [Google Scholar]
- 38.Miyazono K, tenDijke P, Heldin CH. TGFβ signalling by Smad proteins. Adv Immunol. 2000;75:115–57. doi: 10.1016/s0065-2776(00)75003-6. [DOI] [PubMed] [Google Scholar]
- 39.Massague J, Wotton D. Transcriptional control by the TGFβ/Smad signalling system. EMBO J. 2000;19:1745–54. doi: 10.1093/emboj/19.8.1745. 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Attisano L, Wrana JL. Smads as transcriptional co-modulators. Curr Opin Cell Biol. 2000;12:235–43. doi: 10.1016/s0955-0674(99)00081-2. 10.1016/s0955-0674(99)00081-2. [DOI] [PubMed] [Google Scholar]
- 41.Rih S, Walker C, Virchow JC, Boer C, Kroegel C, Giri SN, Braun RK. Differential expression of alphaEβ7 integrin on bronchioalveolar lavage T lymphocyte subsets: regulation by α4β1 integrin cross linking and TGFβ. Am J Respir Cell Mol Biol. 1996;15:600–10. doi: 10.1165/ajrcmb.15.5.8918367. [DOI] [PubMed] [Google Scholar]