Abstract
Macrophages (Mφ) play a key role in innate and acquired immunity. The study of Mφ biology has been hampered by the absence of suitable gene regulatory sequences for the overexpression of heterologous genes in Mφ. The human CD68 gene encodes a glycoprotein that is expressed in monocytes and Mφ, and therefore represents an attractive candidate gene for the generation of a Mφ-specific gene-targeting vector. A transgene expression cassette that combines 2·9 kb of CD68 5′ flanking sequence with the 83-bp first intron (IVS-1) of the CD68 gene, directed high-level, long-lasting expression of class A human scavenger receptor (hSR-A) isoforms in the murine Mφ cell line, RAW-264. By using this CD68 expression cassette to generate Mφ cell lines that overexpress a soluble secreted form of the extracellular portion of type I human SR-A, we were able to purify significant quantities of this protein and show its ability to inhibit SR-A-mediated endocytosis. Analysis of two independent lines of transgenic mice that expressed type III human SR-A under the control of the CD68 gene sequences revealed transgene mRNA expression in elicited Mφ populations and in mouse tissues in a pattern that was consistent with Mφ-specific gene targeting. These data show that CD68 transcriptional regulatory sequences can be used to direct high-level transgene expression in Mφ in vitro and in vivo.
Introduction
Tissue macrophages (Mφs) play an important role in tissue homeostasis and are important in innate and acquired immune responses.1 Moreover, monocyte recruitment and Mφ differentiation are important in the pathogenesis of human diseases such as atherosclerosis.2 The class A scavenger receptors (SR-A) are trimeric integral membrane glycoproteins that have been implicated in various Mφ functions, including endocytosis,3,4 adhesion,5 phagocytosis6 and intracellular signalling.7 There are three forms of the receptor, which are derived by alternative splicing of a single gene.8–10 The three isoforms each contain six predicted structural domains and differ only at the C-terminus.11–13 Type I SR-A has the 110-amino acid scavenger receptor cysteine-rich domain (SRCR), type II SR-A has a short C-terminal domain and type III SR-A has a truncated form of the SRCR domain and has been shown to act as a dominant negative receptor.10 Both type I and type II SR-As bind a diverse array of macromolecules, including modified lipoproteins, bacterial surface lipids (endotoxin and lipoteichoic acid), proteins modified by advanced glycation and β-amyloid fibrils.3,14–18 A number of in vivo roles for SR-A have been proposed, based on the diverse binding properties and cellular functions of this Mφ scavenger receptor. These include lipid accumulation by Mφs in developing atherosclerotic lesions, clearance of apoptotic cells and host defence.4,6,19 The development of SR-A-deficient mice has allowed many questions regarding the role of SR-A to be addressed.6,18,20 However, numerous questions regarding the in vivo function of SR-A isoforms remain unanswered. The development of a system to overexpress SR-A isoforms in Mφs in vitro and in vivo would be of great benefit for addressing these unanswered questions.
Mφ cell lines repress the expression of genes under the control of the human cytomegalovirus (CMV) major immediate-early promoter, the most widely used promoter in mammalian expression vectors.21 To study SR-A function in vivo, through its overexpression in Mφs of transgenic mice, requires a gene-targeting vector that directs high-level SR-A expression in a Mφ-specific manner. Several cis-acting DNA elements have been tested for their ability to direct Mφ-specific gene expression (for a recent review see ref. 22). A number of transgenic mouse lines have been generated using promoter fragments from the human CD11b, c-fms, lysozyme and SR-A genes to drive expression of exogenous genes in Mφs.23–28 These promoters are not ideal for overexpression of SR-A in Mφs in vivo as they can either give rise to expression in non-Mφ cell types or only direct expression in a subset of Mφs, or have subsequently been shown to give inconsistent results in transgenic mice. Hence, other candidates for potential Mφ-specific promoters were considered.
Human CD68 and macrosialin, its murine homologue, are both heavily glycosylated type I transmembrane proteins that belong to the lysosomal/endosomal-associated membrane glycoprotein (LAMP) family.29–33 Both CD68 and macrosialin are expressed in the endosomal compartment of all cells of the mononuclear phagocyte lineage, including monocytes, Mφs, microglia, osteoclasts and, to a lesser extent, immature dendritic cells.34–38 CD68 expression has also been reported in other haematopoietic cell types, although this may merely reflect antibody recognition of shared, non-protein epitopes on other antigens.34,36,39,40 The human CD68 gene lies 667 bp downstream of the EIF4A1 gene, which encodes eukaryotic initiation factor 4A1 (eIF-4A1).41 A 666-bp fragment of the human CD68 promoter, corresponding to the eIF4A1/CD68 intergenic region, has been shown to direct CAT reporter gene expression in Mφ cell lines, at levels equal to or higher than the human CD11b and lysozyme promoters.42 The 83-bp first intron (IVS-1) of the human CD68 gene can act as a Mφ-specific enhancer when added to the 666-bp CD68 promoter fragment, and this combination generated higher levels of CAT enzyme activity than the SV40 promoter/enhancer sequences.42
We show that an expression cassette combining 2·9 kb of the CD68 5′ flanking sequence with the 83-bp first intron of the CD68 gene is able to give high-level, long-lasting expression of human SR-A (hSR-A) in the murine Mφ cell line, RAW-264. We have used this CD68 expression cassette to generate stable cell lines that secrete a soluble form of the extracellular portion of type I hSR-A (shSR-AI). The potential utility of CD68 gene sequences to direct Mφ-specific expression in vivo was demonstrated in two lines of transgenic mice that express type III hSR-A in elicited Mφ populations and in mouse tissues, in a pattern that is consistent with Mφ-specific targeting. These data show that CD68 gene regulatory elements offer a new tool for the study of Mφ gene function in vitro and in vivo.
Materials and methods
Cell culture and transfection
RAW-264 cells were maintained in RPMI-1640 supplemented with 50 IU/ml of penicillin G, 50 µg/ml of streptomycin, 2 mm glutamine (PSG) (all from Invitrogen Life Technologies, Paisley, UK) and 10% fetal calf serum (FCS) (Sigma-Aldrich, Poole, UK). Transfection of the RAW-264 cell line was achieved by electroporation, as described previously.42,43 Briefly, cells were harvested, washed twice in Opti-MEM (Invitrogen Life Technologies) and resuspended at 4 × 107 cells/ml in Opti-MEM. A 0·5-ml aliquot of cells was mixed with 50 µg of plasmid DNA, added to a 0·4-cm electrode gap electroporation cuvette (Bio-Rad, Hemel Hempstead, UK) and shocked in a BioRad GenePulser (300 V, 960 µFD) at room temperature. Immediately postshock, cells were resuspended in 1 ml of prewarmed culture medium prior to transfer to a 9-cm tissue culture plastic Petri dish containing 10 ml of growth medium. For the generation of stable cell lines, cells were grown in culture medium containing 0·5 mg/ml of G418.
Generation of constructs
A BstXI fragment corresponding to the 2940 bp 5′ of the ATG initiation codon of the CD68 gene was excised from cosmid cosCD68C1,43 rendered blunt ended and cloned into EcoRV-restricted pBluescript SK- (Stratagene, La Jolla, CA). The first intron of the CD68 gene was polymerase chain reaction (PCR) amplified using primers 5′-CCGGAATTCTGCTGGGGCTACTGGCAG-3′ and 5′-TGATCTAGAGTCCCCTGGGCTTTTGGCAG-3′, which resulted in the addition of EcoRI and XbaI sites (the underlined bases in the sequences). Following digestion with EcoRI and XbaI, the first intron fragment was cloned into a similarly digested pBluescript vector containing the BstXI CD68 promoter fragment to generate plasmid pBSCD68. Restriction fragments containing hSR-A cDNA and bovine growth hormone polyadenylation sequences (bGHpA) were excised from pcDNA3 (Invitrogen Life Technologies) plasmids using HindIII and AvrII and cloned into plasmid pBSCD68, which had been digested with NotI and blunt ended using the Klenow fragment of Escherichia coli DNA polymerase I (Fig. 1a). Three such constructs were created using cDNA fragments encoding full-length, FLAG-epitope tagged type I (pBSCD68hSR-AI) and type III (pBSCD68hSR-AIII) hSR-A isoforms, and a soluble secreted form of type I hSR-A (pBSCD68shSR-AI), using inserts excised from pcDNA3-based plasmids, as described previously.10,44 To create a plasmid with a selectable marker for the generation of stable cell lines expressing hSR-A under the control of the CD68 promoter, fragments containing the CD68 promoter, hSR-A cDNA and bGHpA were excised from pBluescript vectors by digestion with KpnI and BsaAI, blunt ended and cloned into a pcDNA3 expression vector in which the CMV promoter, multiple cloning site and bGHpA sequence had been removed by digestion with NruI and BbsI and blunt ended using Klenow fragment. These plasmids were designated pC3CD68hSR-AI, pC3CD68hSR-AIII and pC3CD68shSR-AI.
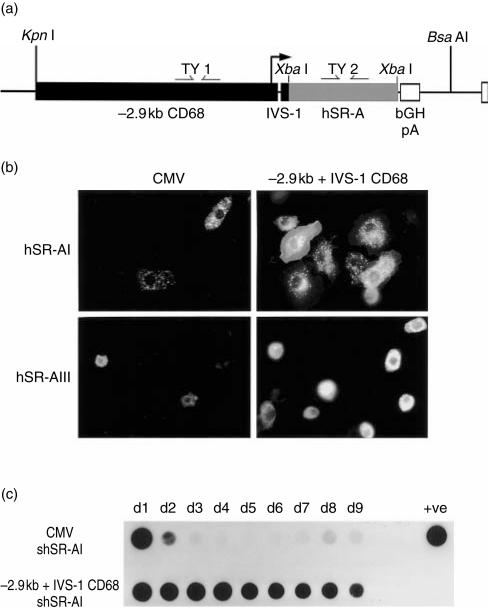
Figure 1.
Comparison of the levels and longevity of class A scavenger receptor (SR-A) expression provided by cytomegalovirus (CMV) and CD68 promoter-driven constructs. (a) The CD68 promoter constructs used in these studies. The 2940 bp of sequence 5′ to ATG and the 83 bp first intron (IVS-1) sequence were cloned upstream of cDNAs encoding FLAG epitope-tagged full-length type I or type III human (h)SR-A isoforms or a soluble secreted version of type I hSR-A,10,44 and a bovine growth hormone polyadenylation cassette (bGHpA). For the generation of transgenic mice and pcDNA3 backbone-based expression constructs, the CD68/SR-A cassette was excised using KpnI and BsaAI. The bent arrow represents the multiple transcription initiation sites of the CD68 promoter, and the straight arrows represent the polymerase chain reaction (PCR) primer pairs TY1 and TY2 used for screening of genomic DNA prepared from tail biopsies. The XbaI sites allow the cloning of other cDNAs for expression under the control of the CD68 promoter. (b) Comparison of CMV and −2·9 kb + IVS-1 CD68 promoters in RAW-264 cells by immunofluorescence microscopy. RAW-264 cells were transfected with expression constructs encoding FLAG epitope-tagged full-length type I or type III hSR-A protein under the control of either the CMV or −2·9 kb + IVS-1 CD68 promoter and plated into 10-cm plates containing 11-mm glass coverslips. Twenty-four hours post-transfection, cells were fixed in 4% paraformaldehyde, stained with the anti-FLAG antibody, M2, and viewed under fluorescence microscopy. Each photograph was taken using the same exposure time. Each panel contained approximately equal numbers of cells, as judged by phase-contrast microscopy. Original magnification × 150. (c) Comparison of CMV and CD68 promoter-driven type I soluble secreted (s)hSR-AI secretion in RAW-264 cells by dot-blotting. RAW-264 cells were transfected with the indicated expression constructs encoding FLAG epitope-tagged shSR-A under the control of either the CMV promoter or the −2·9kb + IVS-1 CD68 sequence. Supernatants were harvested and the media replaced every 24 hr for a total of 9 days. One-hundred microlitres of each supernatant was spotted onto a nitrocellulose membrane and probed with anti-FLAG M2 antibody. Positive control supernatant (+ ve) was produced by transfection of Chinese hamster ovary (CHO)-K1 cells and had previously been shown to contain shSR-AI protein by Western blotting. Data are from a single experiment and are representative of three similar experiments.
Immunofluorescence microscopy of transfected RAW-264 cells
RAW-264 cells were grown on 11-mm glass coverslips for 24 hr after electroporation prior to fixing in a 4% paraformaldehyde solution in phosphate-buffered saline (PBS). Visualization of hSR-A expression was achieved by staining of permeabilized cells with anti-FLAG antibody M2 and fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse immunoglobulin G (IgG) F(ab′)2, as previously described.10
Immunoblotting
Cell lysates were prepared by washing adherent monolayers three times in PBS prior to lysis on ice in lysis buffer (150 mm NaCl, 10 mm EDTA, 10 mm NaN3, 10 mm Tris, pH 8·0, 1 mm phenylmethylsulphonyl fluoride [PMSF], 5 mm iodoacetamide and 1% Nonidet P-40 [NP-40]) and centrifugation at 15 000 g for 10 min to remove debris. Lysates were boiled for 5 min in non-reducing Laemmli sample buffer and resolved by 6% sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE)45 with protein lysate from an equal number of cells loaded per lane. Separated proteins were transferred to nitrocellulose membranes (Hybond-C; Amersham Pharmacia Biotech, Little Chalfont, UK). For dot-blot analysis, 100 µl of tissue culture supernatant was spotted onto nitrocellulose membrane using a 96-well vacuum blot apparatus. Membranes were blocked for 1 hr at room temperature in PBS containing 3% (w/v) powdered milk and 0·1% Tween-20 prior to the addition of primary antibody, at the indicated dilution, in blocking buffer. Binding was detected by incubation with an appro priate peroxidase-conjugated anti-primary species of IgG (Sigma) diluted 1 : 1000 in blocking buffer and visualised by enhanced chemiluminescence (ECL; Amersham Pharmacia Biotech).
Generation, purification and characterization of soluble secreted type I SR-A
RAW-264 cells, stably expressing shSR-AI, were generated by transfection with plasmid pC3CD68shSR-AI and selection in medium containing G418, as described above. Single-cell clones were generated by plating cells at a density of one cell per three wells in 96-well plates in medium containing 1 mg/ml of G418, and were screened by a combination of dot-blot and fluorescence-activated cell sorter (FACS) analysis to isolate clone RAW-1F7, which secreted the highest levels of shSR-AI protein.
Purification of shSR-AI was performed using a modified version of the protocol used by Krieger and colleagues and is summarized in Fig. 2(a).46 RAW-1F7 cells were seeded into 1400-cm2 roller bottles in normal growth medium at a density of 5 × 107 cells/bottle. Cells were grown to confluence, washed once in PBS and re-fed with 250 ml of RPMI-1640 supplemented with PSG and allowed to secrete for 5 days, after which cells were similarly re-fed and allowed to secrete for a further 9 days. Conditioned media were pooled, clarified by centrifugation and sodium azide was added to a final concentration of 10 mm.
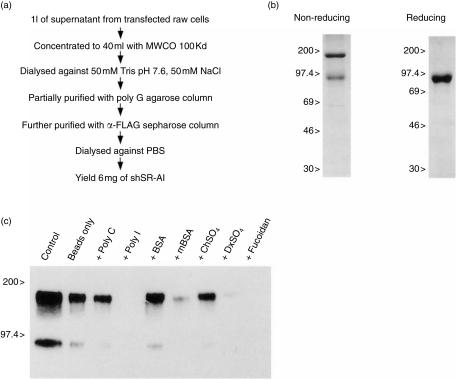
Figure 2.
Generation and characterization of the soluble secreted form of type I human class A scavenger receptor (shSR-AI). (a) Scheme for purification of the shSR-AI protein. The flow diagram shows the important steps involved in the purification of shSR-AI and highlights the 6-mg yield obtained per litre of conditioned tissue culture supernatant. (b) Analysis of purified shSR-AI protein by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) and Coomassie Brilliant Blue staining. Purified type I shSR-AI protein (2·5 µg) was separated by 10% SDS–PAGE under non-reducing and reducing conditions. Protein visualization was achieved by staining with Coomassie R-250. (c) Analysis of shSR-AI binding properties by poly G–agarose. The binding of purified type I shSR-AI protein was performed in the presence of the following inhibitors: polycytidylic acid (poly C), polyinosinic acid (poly I) (25 µg/ml), bovine serum albumin (BSA), maleylated BSA (mBSA) (100 µg/ml), chondroitin sulphate (ChSO4), dextran sulphate (DxSO4) and fucoidan (150 µg/ml). The control sample lane was loaded with 25 µl of unpurified conditioned medium.
All subsequent steps were carried out at 4°, or at room temperature using ice-cold reagents. Supernatants were concentrated using an Amicon stirred ultrafiltration cell, pressurized liquid reservoir and a 100-kDa MWCO cellulose-ester membrane (Millipore, Watford, UK). After concentration of 5 l of conditioned media to ≈ 75 ml, the supernatant was dialysed against 50 mm Tris (pH 7·6) and 50 mm NaCl (wash buffer) before passing through a 0·45-µm filter. The supernatant was then applied overnight to a 6 × 1·5-cm column containing 10 ml of poly G resin (Sigma) at a rate of 1 ml/min using a peristaltic pump and allowing the supernatant to recycle. After washing the column with 100 ml of wash buffer, bound proteins were eluted with 50 mm Tris (pH 7·6) and 1 m NaCl, and 1-ml fractions were collected. Fractions containing shSR-AI protein were determined by measurement of absorbance at 280 nm in a spectrophotometer and by SDS–PAGE with Coomassie Brilliant Blue staining, and were subsequently pooled and dialysed against PBS. Soluble hSR-A protein was further purified by twice passing the supernatant over a 5-ml anti-FLAG antibody M2 sepharose-affinity column (Sigma), washing with 50 ml PBS and eluting with 15 ml of 0·1 m glycine (pH 2·9), collecting 1-ml fractions. Fractions containing shSR-AI were determined as described above, prior to being pooled, dialysed against PBS and the final protein concentration measured using the BCA assay (Pierce & Warriner, Chester, UK). The yield of shSR-AI protein was typically 30 mg from 5 l of conditioned supernatant, and showed no contaminating protein bands when stained with Coomassie Brilliant Blue.
The binding properties of purified shSR-AI were analysed using a poly G bead binding assay, as described previously.47 Briefly, 0·1 µg of purified shSR-AI protein was added to 1 ml of 150 mm NaCl, 50 mm Tris (pH 7·6) buffer and incubated with poly G agarose beads for 2 hr at 4°, in the presence or absence of a panel of SR-A ligands and their cognate non-ligand controls. The beads were washed three times in 150 mm NaCl, 50 mm Tris (pH 7·6), and bound protein was eluted by boiling the beads in protein gel loading buffer for 5 min. The samples were separated by SDS–PAGE and the binding of shSR-AI was determined by Western blotting using anti-FLAG antibody M2.
Quantification of acetylated low-density lipoprotein uptake
To assay the uptake of acetylated low-density lipoprotein (AcLDL), cells in 24-well plates were washed twice in PBS and then preincubated with inhibitors at the indicated concentrations for 30 min prior to labelling for 90 min with 2 µg/ml of DiI (1,1′-dioctadecyl-1–3,3,3′,3′-tetramethylindocarbocyanine perchlorate)-labelled-AcLDL (DiI-AcLDL) (Biogenesis, Poole, UK) for 90 min at 37° in the presence of inhibitors. Cells were washed five times in PBS, resuspended in PBS containing 5 mm EDTA and 10 mg/ml Lidocaine-HCl (Sigma), fixed in a 4% solution of paraformaldehyde in PBS and analysed on a fluorescence-activated cell sorter (FACScan; BD Pharmingen, San Diego, CA) using the FL2 photomultiplier.
Generation of transgenic lines
A linear DNA fragment for the generation of transgenic mice, shown in Fig. 1(a), was generated by digesting plasmid pBSCD68hSR-AIII with KpnI, BsaAI and DrdI, and purifying the separated fragment from an 0·8% agarose gel using a QIAEX II gel extraction kit (Qiagen, Crawley, UK) and an Elutip-D column (Schleicher & Schuell, London, UK). Microinjection of CBA/B6 eggs with purified DNA and their subsequent transfer into pseudopregnant foster mothers was performed using standard techniques.48 Transgenic founders were identified by the PCR using genomic DNA extracted from 1-cm tail clips and two primer pair combinations, as shown in Fig. 1(a): TY1: 5′-TTCTCGGCTCTGTGAATGACA-3′ and 5′-CAGCCCTCTCTTGGAAAGGAGG-3′; TY2: 5′-AAGTGGGAAACGAAGAATTGC-3′ and 5′-CTCTTGTTGTTTGAAGGTATTCTC-3′. Founder mice positive for the transgene were mated with C57BL/6 mice, and all subsequent analyses were performed using the resulting F1 progeny.
Isolation of primary Mφ populations
Peritoneal cells were isolated by peritoneal lavage with PBS at the indicated time-points after intraperitoneal injection of 1 ml of 4% thioglycollate broth. Bone marrow-derived Mφs (BMDM) were generated by the culture of bone marrow cells in RPMI-1640 containing PSG, 10% FCS and 15% L-cell conditioned medium as a source of macrophage-colony stimulating factor (M-CSF).49 All animal procedures were carried out in accordance with the Animals (Scientific Procedures) Act 1986, and in accordance with Departmental Ethical Review.
Analysis of transgene RNA expression
Total RNA was isolated from Mφ populations and murine organs using RNAzol (Biogenesis), according to the manufacturer's recommended protocol, and a Polytron tissue homogenizer (Brinkmann Instruments, Westbury, NY) to ensure complete disruption of tissues. For Northern blot analyses, total RNA (5–10 µg) was fractionated on a 1% formaldehyde/agarose gel and transferred, by capillary blotting, to nylon membrane (Hybond N+ Amersham Pharmacia Biotech). Membranes were baked at 85° for 3 hr prior to prehybridization in ExpressHyb hybridization solution (Clontech, Palo Alto, CA) at 68° for 1 hr. Probes for β-actin (a 2-kb fragment of human β-actin; Clontech) and hSR-AIII (a 1·2-kb fragment generated by digestion of the expression plasmid, pcDNA3,10 with HindIII and XbaI) were labelled by random priming with [α-32P]dATP and then incubated with blots in ExpressHyb for 2 hr at 68°. Blots were washed according to the manufacturer's recommendations and then subjected to autoradiography at −70° with intensifying screens.
For reverse transcription–PCR (RT–PCR) analyses, total RNA was reverse transcribed using Moloney murine leukaemia virus reverse transcriptase and an Oligo-dT primer (Life Technologies), with reactions set up in the presence and absence of reverse transcriptase to control for genomic DNA contamination. The cDNA obtained served as a template for PCR using the following oligonucleotide pairs: HPRT: 5′-GCTACCTGCTGGATTACAT-3′ and 5′-CCAGTTTCACTAATGACACAA-3′; macrosialin: 5′-TCCTTCACGATGACACCTACAG-3′ and 5′-GGACCAGGCCAATGATGAGAG-3′; hSR-A III transgene: 5′-GGGTGAGGCGGTTCAGCC-3′ and 5′-TTTCCTCTTCGCTGTCATTTC-3′. The PCR products were separated by electrophoresis on a 1·2% agarose gel and visualized by ethidium bromide staining.
Metabolic labelling and pulse-chase analysis
Bone marrow cells were plated in six-well plates at a density of 5 × 105 cells/well and cultured for 6 days in medium containing M-CSF, as described above. Adherent cells were washed three times with PBS and pulse-labelled for 30 min with 0·5 ml of methionine- and cysteine-deficient RPMI-1640 supplemented with 5% dialysed FCS and 500 µCi/ml of 35S-Trans label (ICN Biomedicals, Aurora, OH). Cells were washed three times with PBS and incubated in ‘chase’ medium consisting of RPMI-1640 supplemented with 10% FCS, PSG, 1 mm unlabelled methionine and 4 mm unlabelled cysteine. After incubation for the indicated times, cells were washed three times in PBS and lysates were prepared as described above. After preclearing samples by incubation with protein G–sepharose (Amersham Pharmacia Biotech) at 4°, antigen was precipitated by overnight incubation with 10 µg/ml of the anti-FLAG antibody, M2, and protein G–sepharose. Samples were washed six times, as described in ref. 10, and immunoprecipitated protein was collected by boiling the beads in reducing Laemmli sample buffer prior to separation by 10% SDS–PAGE.45 Gels were fixed and impregnated with EN3HANCE (NEN Life Science Products, Zaventen, Belgium) before drying and exposure to film at −70°.
Results
CD68 gene sequences direct high-level, long-lasting expression of hSR-A in RAW-264 cells
To characterize the usefulness of CD68 transcriptional regulatory sequences as a tool to overexpress hSR-A in Mφs, we generated constructs in which 2·9 kb of 5′ sequence and the IVS-1 from the CD68 gene (− 2·9 kb + IVS-1 CD68) were placed upstream of an hSR-A cDNA and bGHpA (Fig. 1a). Three plasmids with a pBluescript backbone were generated, containing cDNAs for membrane-bound, FLAG-epitope tagged type I or type III hSR-A (see ref. 10 for details) and a soluble secreted type I hSR-A (shSR-AI) (see ref. 44 for details).
The ability of these constructs to direct expression of hSR-A was tested by transient transfection of the murine Mφ cell line, RAW-264. Immunofluorescence microscopy of cells 24-hr post-transfection showed high-level expression of both type I and type III hSR-A, as assessed by staining with the anti-FLAG antibody, M2 (Fig. 1b). The levels of expression of both hSR-A I and III, as judged by the brightness of cells, was much greater in cells transfected with −2·9kb + IVS-1 CD68 promoter-driven constructs than in CMV-driven controls. The high levels of hSR-A expression produced by the −2·9kb + IVS-1 CD68 promoter-driven plasmids allowed for differences in functions of the type I and type III hSR-A isoforms to be appreciated. Cells overexpressing type I hSR-A spread out more on glass coverslips, displaying a large veil of membrane containing type I hSR-A protein at the point of adherence to the glass surface, while type III hSR-A showed a predominately perinuclear staining pattern in transiently transfected RAW cells. These data are consistent with a role for type I SR-A in Mφ adhesion to glass surfaces. In previous experiments we have shown that type III SR-A becomes trapped in the endoplasmic reticulum where it acts as a dominant negative regulator of SR-A function.10 The data of Fig. 1 highlight the usefulness of CD68 promoter-mediated overexpression for studying SR-A biology in transfected Mφ cell lines.
To test whether CD68 gene sequences could give sustained expression of hSR-A, we examined the secretion of shSR-AI (over a 9-day period) in RAW-264 cells transiently transfected with either CMV- or −2·9kb + IVS-1 CD68-driven expression constructs. Cell supernatants were harvested and replaced with fresh medium every 24 hr, and the presence of the shSR-AI protein in the supernatant was detected by dot-blot analysis (Fig. 1c). In the supernatant obtained 24 hr post-transfection, cells transfected with CD68 and CMV expression vectors produced approximately equal levels of shSR-AI. Secretion of shSR-AI by cells transfected with constructs containing the CMV promoter declined rapidly, while the CD68-based plasmid directed high levels of expression for the duration of the experiment. Hence, the −2·9kb + IVS-1 CD68 promoter fragment is able to generate high-level, long-lasting gene expression in transiently transfected RAW-264 cells.
Purification and characterization of soluble human SR-AI protein
In previous experiments we had been unable to produce milligram quantities of shSR-AI, using a variety of different eukaryotic expression systems. The ability of the −2·9kb + IVS-1 CD68 sequences to direct high-level secretion of shSR-AI in RAW-264 cells offered a new approach for the generation of large quantities of shSR-AI protein. We generated stable shSR-AI-expressing RAW-264 polyclonal populations using selection in medium containing 0·5 mg/ml of G418. Single-cell clones were generated in medium containing 1 mg/ml of G418 and were screened for high-level expression of shSR-AI through a combination of dot-blot and FACS analysis (data not shown). Clone RAW-1F7 was selected for further use and secreted shSR-AI at ≈ 5 µg/ml (data not shown). Purification of shSR-AI protein was performed using supernatants from bulk cultures of RAW-1F7 as the starting material, as outlined in Fig. 2(a) and described in detail in the Materials and methods. The purity of shSR-AI preparations was assessed by SDS–PAGE and Coomassie Brilliant Blue staining (Fig. 2b). Only the predicted bands corresponding to dimeric and monomeric shSR-AI protein (Mr 160 and 85 kDa, respectively) were detected under non-reducing conditions, and only a single band of 85 kDa, representing monomeric shSR-AI, was detected under reducing conditions. In two independent preparations of shSR-AI, yields of purified protein averaged 6 mg/l of conditioned culture supernatant.
To ensure that the shSR-AI protein produced by the RAW cells had binding properties that were similar to the cellular form of the receptor, the binding of shSR-AI to poly G–agarose was examined. This approach has been used previously to show that a radiolabelled, secreted version of the bovine receptor had the requisite binding properties.47 Binding of purified shSR-AI to poly G–agarose was performed in the presence or absence of a number of SR-A ligands and their cognate non-ligands, and was assessed by SDS–PAGE and Western blotting using the anti-FLAG antibody (Fig. 2c). Under non-reducing conditions, the shSR-AI protein migrated (as predicted) with bands corresponding to dimeric and monomeric shSR-AI (160 and 85 kDa, respectively), detected when binding occured in the absence of inhibitors. The binding of shSR-AI to the poly G beads was specifically inhibited by the SR-A ligands poly I, maleylated bovine serum albumin (BSA), dextran sulphate and fucoidan, but not by the appropriate non-ligands, indicating that the binding properties of shSR-AI closely resemble those of its cellular counterpart.
Soluble human SR-I protein inhibits SR-A-mediated endocytosis
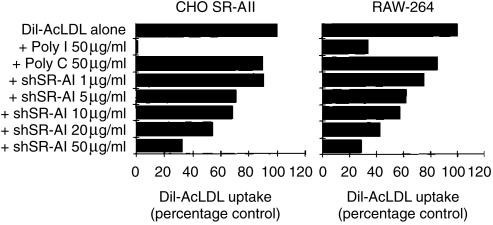
We examined the effect of purified shSR-AI protein on SR-A-mediated endocytosis. Chinese hamster ovary (CHO) -K1 cells expressing high levels of type II hSR-A10 and RAW-264 cells were labelled with the fluorescently labelled scavenger receptor ligand, DiI-AcLDL, in the presence or absence of poly I, poly C and shSR-AI (Fig. 3). The uptake of DiI-AcLDL was inhibited by poly I in both cell types, although this was incomplete in RAW-264 cells owing to the expression of a non-poly I inhibitable scavenger receptor. Increasing concentrations of shSR-AI protein produced a dose-dependent inhibition of DiI-AcLDL uptake in both cell types, with a 70% reduction at the maximal concentration of shSR-AI used in this experiment. This inhibition was specific for SR-A-mediated endocytosis as the uptake of fluorescently labelled LDL, a non-SR-A ligand, was unaffected and therefore shSR-AI protein did not induce a general suppression of endocytic activity in these cells (data not shown). Hence, purified shSR-AI protein can be used to inhibit SR-A-mediated endocytosis.
Figure 3.
Analysis of the ability of the soluble secreted form of type I human class A scavenger receptor (shSR-AI) to inhibit SR-A-mediated endocytosis. Comparison of the effects of shSR-AI on the uptake of 1,1′-dioctadecyl-1–3,3,3′,3′-tetramethylindocarbocyanine perchlorate-labelled acetylated low-density lipoprotein (DiI-AcLDL) by Chinese hamster ovary (CHO)-K1 cells expressing high levels of type II hSR-A and by RAW-264 cells. Cells were labelled with DiI-AcLDL (2·5 µg/ml) in the presence of inhibitors at the indicated concentrations for 90 min. Specific fluorescence intensity was calculated by subtracting autofluorescence from the fluorescence intensity of DiI-AcLDL-labelled cells using means derived by data analysis with CellQuest software (Becton-Dickinson). Data are expressed as the percentage of uptake by control cells labelled in the absence of inhibitors in the same experiment. Results are from a single experiment and are representative of three independent experiments. Poly C, polycytidylic acid; poly I, polyinosinic acid.
CD68 gene sequences direct expression of type III hSR-A mRNA in Mφ populations of transgenic mice
To test the ability of the −2·9 kb + IVS-1 CD68 gene sequences to direct Mφ-specific transgene expression in vivo, fragments containing the CD68 promoter, IVS-1 and hSR-A cDNA, were used to generate transgenic mice by pronuclear injection. Linear DNA fragments containing the −2·9 kb CD68 promoter + IVS-1, the coding sequence for type III membrane bound hSR-A and bovine growth hormone poly A+ sequences (Fig. 1a) were microinjected into fertilized mouse eggs. Potential transgenic founders were screened by using a combination of two PCR primer pairs, TY1 and TY2 (Fig. 1a), and confirmed by Southern blot analysis of restriction enzyme-digested genomic DNA (data not shown). Two founder mice were obtained and bred with C57BL/6 mice to establish the F1 animals used for all subsequent analyses. Both lines of mice generated using the CD68 type III hSR-A fragment, lines III.4 and III.11, showed germline transgene transmission to F1 offspring in an approximately Mendelian pattern (data not shown).
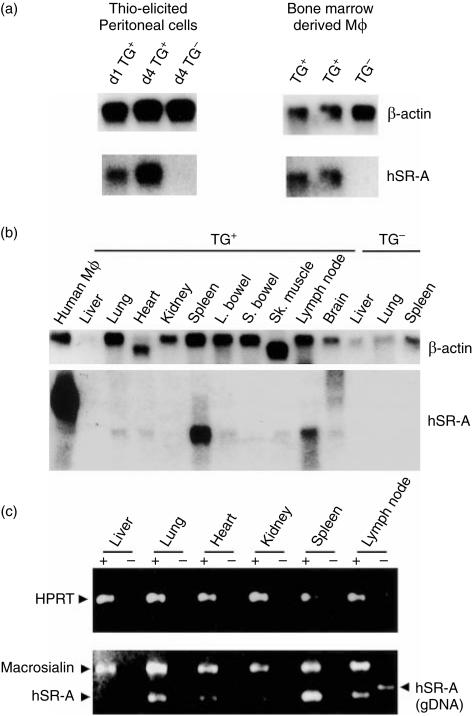
Expression of transgene mRNA in isolated Mφ populations from both transgenic lines was assessed by Northern blotting. Peritoneal cells elicited either 1 or 4 days after injection of III.11 mice with thioglycollate broth expressed type III hSR-A mRNA, while no signal was detectable from transgene negative (Tg–) littermates, despite the presence of equal amounts of RNA, as judged by probing with β-actin (Fig. 4a). The intensity of the hSR-A specific band was greater in the RNA sample from the cells elicited 4 days after injection with thioglycollate broth. The peritoneal cell population obtained 1 day after injection of thioglycollate broth was a mixture of newly recruited and resident peritoneal Mφs and recruited neutrophils. In contrast, by 4 days after thioglycollate injection, all the recruited neutrophils had died and were replaced with recruited monocytes and Mφs. Hence, the relative levels of type III hSR-A mRNA 1 or 4 days after thioglycollate injection was consistent with Mφ-restricted transgene expression. Expression of type III hSR-A mRNA in Mφs from III.11 transgenic mice was further tested by examining expression in BMDMs. Two independent BMDM preparations from transgenic (Tg+) mice showed type III hSR-A transgene mRNA expression, with no detectable expression from Tg– littermates. Similar results were obtained in an analysis of transgene mRNA expression by Mφs isolated from Tg+ III.4 mice (data not shown). These data show that the CD68 sequences can drive transgene expression in Mφ populations of transgenic mice.
Figure 4.
Analysis of human class A scavenger receptor (hSR-A) III transgene RNA expression. (a) Northern blot of isolated macrophage (Mφ) populations. Total RNA was prepared from peritoneal cells isolated from transgenic line III.11 (Tg+) mice and non-transgenic (Tg–) littermates, either 1 or 4 days after injection of 1 ml of thioglycollate broth. Bone marrow-derived Mφs (BMDMs) were generated by the culture of bone marrow precursors in medium containing macrophage colony-stimulating factor (M-CSF) and total RNA isolated from adherent cells cultured for 6 days. Total RNA (5 µg) was denatured and separated on a 1% formaldehyde–agarose gel prior to capillary transfer to a nylon membrane. Blots were hybridized with [α-32P]dATP-labelled DNA probes for β-actin (a 2-kb fragment of human β-actin; Clontech) or hSR-A III transgene (a 1·2-kb fragment of type III hSR-A coding sequence generated by digestion of expression plasmid pcDNA 3 with HindIII and XbaI) and exposed to film. (b) Northern blot of adult tissues. Total tissue RNA (10 µg) prepared using organs collected from transgenic line III.4 (Tg+) mice and non-transgenic (Tg–) littermates, and from human monocyte-derived Mφ total RNA (10 µg), was denatured and separated on a 1% formaldehyde–agarose gel prior to transfer and hybridization as described in (a). The different sizes of SR-A hybridizing bands in the human and transgenic RNA samples are caused by the absence of 3′ untranslated sequences in the CD68 SR-A type III transgene. (c) Reverse transcription–polymerase chain reaction (RT–PCR) of adult tissues. Total tissue RNA from organs collected from transgenic line III.4 (Tg+) mice was reverse transcribed in the presence (+) or absence (–) of Moloney murine leukemia virus reverse transcriptase. cDNA synthesized from 100 ng of total RNA served as a template for subsequent PCR using primers specific for hypoxanthine phosphoribosyl transferase (HPRT), macrosialin or the hSR-A III transgene. To enable a comparison of the relative levels of RNA for macrosialin and the hSR-A III transgene, PCR was performed with both oligonucleotide pairs in the same reaction tube. Primers for HPRT and macrosialin do not generate a detectable reaction product from contaminating genomic DNA (gDNA) under the conditions used, while the product corresponding to the amplification of the hSR-A III transgene genomic DNA is 83 bp larger owing to the presence of the first intron (IVS-1) sequence, which is spliced out during the processing of the primary RNA transcript. Reaction products were separated on a 1·2% agarose gel and visualized by ethidium bromide staining.
The pattern of type III hSR-A mRNA expression was examined in a variety of murine tissues from Tg+ mice. Northern blot analysis showed that transgene mRNA can be detected in all of the organ samples from Tg+ III.4 mice (except for liver where the β-actin probed control showed low levels of intact mRNA), with lung, spleen and lymph nodes showing the highest levels of expression (Fig. 4b). No bands of the appropriate size could be detected in RNA samples from a Tg– mouse. This pattern of transgene expression is consistent with Mφ-restricted targeting as the spleen and lymph nodes are organs that contain the largest proportion of Mφs as a percentage of the total cell population. Comparison of the intensity of SR-A-specific bands between RNA samples from human monocyte-derived Mφs, which express high levels of SR-A mRNA,50 and transgenic spleen, where Mφs constitute ≈ 10–20% of the total cell number, shows that CD68 sequences direct significant levels of transgene RNA expression.
To further examine the specificity of expression generated by CD68 gene sequences, the expression of transgene RNA was compared to that of macrosialin, the murine homologue of human CD68, using RT–PCR. Each of the samples generated from organs from III.4 Tg+ mice had approximately equal amounts of cDNA, as judged by the intensity of the bands produced using an oligonucleotide pair specific for the housekeeping gene, hypoxanthine phosphoribosyl transferase (HPRT) (Fig. 4c). The highest levels of macrosialin expression were seen in the lung, spleen and lymph nodes, although all tissues expressed some mRNA. The pattern of mRNA expression seen with hSR-A transgene specific primers was largely similar to that for macrosialin, although there were variations in the relative intensities of the bands for the two mRNA species. There was no detectable hSR-A transgene expression in the liver cDNA sample. In the spleen cDNA sample, the type III hSR-A-specific band was of higher intensity than that for macrosialin, suggesting that the transgene mRNA was present at higher levels than the endogenous gene. Analysis of type III hSR-A mRNA expression in a similar panel of tissues from III.11 Tg+ mice revealed a similar pattern of transgene expression (data not shown). Hence, CD68 gene sequences can direct transgene expression to Mφ populations in vivo, in a pattern that is suggestive of Mφ-specific targeting.
Type III hSR-A protein is not readily detected in Mφs from transgenic mice as a result of its rapid degradation
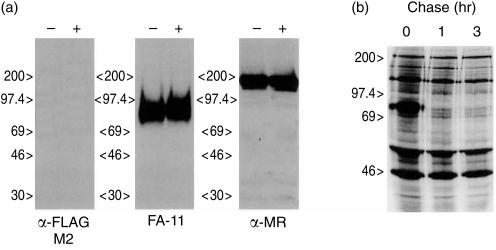
To examine further the cell type specificity of transgene expression, immunohistochemistry was performed on a panel of tissues from III.4 and III.11 Tg+ mice. Tissue sections were probed with the anti-FLAG monoclonal antibody (mAb) and anti-hSR-A polyclonal antibodies, but no staining pattern unique to Tg+ mice was observed for either transgenic line (data not shown). Appropriate staining patterns were observed with anti-macrosialin and anti-F4/80 antibodies, and were identical in tissue sections from Tg+ and Tg– mice (data not shown), indicating normal Mφ distributions in these mice. In an effort to optimize staining protocols for the detection of type III hSR-A protein using anti-FLAG and anti-hSR-A antibodies, we performed immunofluorescence staining of BMDMs from both transgenic lines. Despite numerous attempts using many different experimental conditions, no type III hSR-A protein could be detected (data not shown). As an alternative approach to detect type III hSR-A protein expression, Western blotting of protein lysates from BMDMs derived from III.11 mice was performed as shown in Fig. 5(a). Under conditions in which we could readily detect type III hSR-A protein expressed by transfected RAW-264 cells, no expression could be detected in Tg+ BMDMs. Identical blots probed for macrosialin (FA-11) or mannose receptor (α-MR) showed that other Mφ antigens were readily detectable in these cells (Fig. 5a). Hence, despite the expression of type III hSR-A mRNA in isolated Mφs and tissues from both lines of transgenic mice, no type III hSR-A protein could be detected.
Figure 5.
Analysis of human class A scavenger receptor (hSR-A) III protein expression in bone marrow-derived macrophages (BMDMs) isolated from line III.11 transgenic mice. (a) Western blotting. Protein lysates were generated from adherent BMDMs cultured for 6 days in medium containing macrophage colony-stimulating factor (M-CSF). Total protein lysate (30 µg) from non-transgenic (–) and transgenic (+) BMDMs was separated by 6% non-reducing sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE). After transfer to nitrocellulose, blots were probed with anti-FLAG antibody, M2 (10 µg/ml), anti-mouse macrosialin antibody, FA-11 (10 µg/ml), or anti-mouse mannose receptor polyclonal antibody (1 : 5000) and the appropriate horseradish peroxidase (HRP)-conjugated secondary antibody prior to development with enhanced chemiluminescence (ECL) reagent. (b) Pulse-chase. BMDMs cultured for 6 days in medium containing M-CSF were pulse-labelled for 30 min with 35S-labelled cysteine and -methionine and chased for the indicated times prior to lysis and immunoprecipitation using anti-FLAG antibody, M2, and protein G–sepharose beads. Samples were separated by SDS–PAGE, on an 8% gel, under reducing conditions prior to visualization by autoradiography.
Type III hSR-A protein is retained in the endoplasmic reticulum of cells where it is able to exert dominant negative properties, presumably by inducing the degradation of type I and type II SR-A.10 The protein can be readily detected at steady-state levels when expressed in RAW-264 cells (Fig. 1b). However, it is possible that a combination of lower rates of synthesis and higher rates of degradation in the endoplasmic reticulum of Mφs of CD68-type III hSR-A transgenic mice mean that steady-state levels of type III hSR-A protein do not accumulate. To test this hypothesis, BMDMs from Tg+ hSR-A III.11 mice were pulse labelled for 30 min with 35S-methionine and cysteine, and chased for the times indicated in Fig. 5(b). Type III hSR-A protein was immunoprecipitated with an anti-FLAG antibody, and samples were separated under reducing conditions by SDS–PAGE prior to visualization by autoradiography. A band of the correct size for monomeric type III hSR-A protein (Mr ≈ 80 kDa) was clearly evident after the initial pulse, but this disappeared after 1 or 3 hr of chase. The other bands present at constant intensity through the chase period probably represent Fc receptors synthesized by Mφs and immunoprecipitated because of their interaction with the murine anti-FLAG antibody. Attempts to alter the kinetics of type III hSR-A protein degradation through use of the protease inhibitors lactacystin or NH4Cl were unsuccessful owing to a significant inhibition in the basal levels of protein synthesis under the experimental conditions used (data not shown). Hence, type III hSR-A protein is synthesized by Tg+ Mφs from hSR-A III.11 mice, but is rapidly degraded, consistent with the finding that no steady-state protein could be detected in isolated cells or tissues from Tg+ mice.
Discussion
Mφs are cells with unique functional properties, being able to efficiently recognize and phagocytose pathogenic particles and subsequently present antigens to T cells. Many of these characteristics are the result of expression of Mφ-restricted cell-surface receptors that generate unique cell biological properties. The absence of tools to overexpress novel Mφ genes in Mφs, allowing them to be studied in their correct cellular environment, has hampered elucidation of their function. The data presented in this report show that CD68 gene sequences are able to generate higher levels of stable gene expression in transiently transfected RAW-264 murine Mφs than standard expression plasmids based on the human CMV promoter. Indeed, the overexpression of type I hSR-A in RAW-264 cells leads to a striking change in cell morphology (Fig. 1b), which is consistent with the documented role of SR-A in Mφ adhesion.3
The use of CD68 gene sequences as the basis for a stable expression system allowed us to produce large quantities of soluble hSR-AI protein. The yield of 6 mg of protein from 1 l of cell supernatant compares favourably with a yield of ≈ 0·75 mg/l of a similar shSR-AI protein from supernatant produced using a CHO cell-expression system.46 In addition, the stable RAW cell line (RAW-1F7), used here to produce shSR-AI, was generated within 6 weeks of transfection, unlike the CHO cell line which required over 8 months of selection using increasing concentrations of methotrexate (M. Krieger, personal communication). The generation of shSR-AI protein using a Mφ cell line may also prove advantageous as ‘Mφ-specific’ post-translational modifications may influence shSR-AI-binding properties, helping to mimic the authentic cell-surface receptor. Hence, the combination of the RAW cell line and CD68 promoter-driven expression constructs appears to offer a useful approach for generating large quantities of recombinant proteins, particularly those expressed by Mφs.
The ability of the purified shSR-AI protein to block SR-A-mediated endocytosis is consistent with previous studies showing that adenoviral expression of this form of hSR-A is able to block the endocytosis of AcLDL and oxidized LDL (OxLDL) by RAW-264 cells and inhibit Mφ foam cell formation.44 The inhibition of endocytosis of modified forms of LDL by shSR-AI highlights the ability of shSR-AI to act as a ‘decoy receptor’ and block the function of cellular SR-A. The generation of large quantities of purified shSR-AI protein will enable these inhibitory properties of shSR-AI to be explored in in vivo experiments in an attempt to further define the role of SR-A in atherosclerotic lesion development and host defence. Purified shSR-AI protein will be a useful tool for use in the molecular characterization of the epitopes on known SR-A ligands, such as OxLDL and bacterial endotoxin, required for SR-A binding, and for the identification of novel SR-A ligands, especially for the SRCR domain of type I SR-A.
The analysis of the expression pattern of the type III hSR-A transgene mRNA in the two lines of transgenic mice showed a pattern that is consistent with Mφ-restricted expression. Transgene mRNA was readily detectable in both thioglycollate-elicited and bone marrow-derived Mφ populations, and showed the highest levels of expression in spleen and lymph node, organs that contain large numbers of Mφs. However, the rapid degradation of type III hSR-A protein meant that it was not possible to examine fully the cell-type specificity of gene expression given by the CD68 gene sequences. To address this issue we are currently generating more transgenic lines expressing type I hSR-A under the control of the CD68 promoter, as this protein has previously been shown to be stable in murine Mφs in transgenic mice.51 As well as answering questions regarding the cell type specificity of the −2·9 kb + IVS-1 CD68 sequences, these mice should prove useful in further defining the in vivo function of SR-A.
The pattern and levels of gene expression provided by CD68 gene sequences in the transgenic mice described here compare favourably with other promoter fragments that have been described for the expression of transgenes in Mφs in vivo. The majority of these promoters, including human CD11b, human lysozyme and human c-fes, are from genes that show myeloid-restricted expression, and so at best the transgene is expressed by both neutrophils and Mφs.23,25,26,28,52 Human SR-A promoter elements direct transgene expression in Mφs in mouse atherosclerotic lesions, spleen and testes, but not in other organs that contain significant numbers of Mφs.24 However, it is difficult to compare levels of transgene expression between transgenic lines owing to differences in transgene copy number and the reporter gene used.
The CD68 promoter and intron sequences described in this article provide a novel tool for sustained, high-level expression of genes in Mφs in vitro and in vivo. The future application of CD68 gene sequences for Mφ-specific gene targeting should allow many questions regarding Mφ cell biology, and the in vivo functions of this unique cell type, to be addressed.
Acknowledgments
We are very grateful to Colin Hetherington for technical assistance. P.J.G. was the recipient of an Arthritis Research Campaign studentship and a Goodger Scholarship from the University of Oxford. D.R.G. is a British Heart Foundation Basic Science Lecturer. Work in the laboratory of S.G. is funded by the UK Medical Research Council and an external research grant from Glaxo Wellcome.
Abbreviations
- bGHpA
bovine growth hormone polyadenylation signal
- BMDM
bone marrow-derived macrophage
- CMV
human cytomegalovirus
- hSR-A
human class A scavenger receptor
- IVS-1
first intron
- Mφ
macrophage
- poly C
polycytidylic acid
- poly I
polyinosinic acid
- shSR-AI
soluble secreted form of type I human class A scavenger receptor
- SR-A
class A scavenger receptor
- Tg+
transgenic
- Tg–
non-transgenic.
References
- 1.Gordon S. The mononuclear phagocyte system. In: McGee JO, editor. Oxford Textbook of Pathology. Oxford: Oxford University Press; 1992. pp. 236–58. [Google Scholar]
- 2.Raines EW, Rosenfeld ME, Ross R. The role of macrophages. In: Fuster V, Ross R, Topol EJ, editors. Atherosclerosis and Coronary Artery Disease. Philadelphia: Lippincott-Raven Publishers; 1996. pp. 539–55. [Google Scholar]
- 3.Krieger M, Herz J. Structures and functions of multiligand lipoprotein receptors – macrophage scavenger receptors and LDL receptor-related protein (LRP) Annu Rev Biochem. 1994;63:601–37. doi: 10.1146/annurev.bi.63.070194.003125. [DOI] [PubMed] [Google Scholar]
- 4.Brown MS, Goldstein JL. Lipoprotein metabolism in the macrophage – implications for cholesterol deposition in atherosclerosis. Annu Rev Biochem. 1983;52:223–61. doi: 10.1146/annurev.bi.52.070183.001255. [DOI] [PubMed] [Google Scholar]
- 5.Fraser I, Hughes D, Gordon S. Divalent cation-independent macrophage adhesion inhibited by monoclonal-antibody to murine scavenger receptor. Nature. 1993;364:343–5. doi: 10.1038/364343a0. [DOI] [PubMed] [Google Scholar]
- 6.Platt N, Suzuki H, Kurihara Y, Kodama T, Gordon S. Role for the class-A macrophage scavenger receptor in the phagocytosis of apoptotic thymocytes invitro. Proc Natl Acad Sci USA. 1996;93:12456–60. doi: 10.1073/pnas.93.22.12456. 10.1073/pnas.93.22.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miki S, Tsukada S, Nakamura Y, et al. Functional and possible physical association of scavenger receptor with cytoplasmic tyrosine kinase Lyn in monocytic THP-1-derived macrophages. FEBS Lett. 1996;399:241–4. doi: 10.1016/s0014-5793(96)01332-4. 10.1016/s0014-5793(96)01332-4. [DOI] [PubMed] [Google Scholar]
- 8.Freeman M, Ashkenas J, Rees DJG, Kingsley DM, Copeland NG, Jenkins NA, Krieger M. An ancient, highly conserved family of cysteine-rich protein domains revealed by cloning type-I and type-II murine macrophage scavenger receptors. Proc Natl Acad Sci USA. 1990;87:8810–4. doi: 10.1073/pnas.87.22.8810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emi M, Asaoka H, Matsumoto A, et al. Structure, organization, and chromosomal mapping of the human macrophage scavenger receptor gene. J Biol Chem. 1993;268:2120–5. [PubMed] [Google Scholar]
- 10.Gough PJ, Greaves DR, Gordon S. A naturally occurring isoform of the human macrophage scavenger receptor (SR-A) gene generated by alternative splicing blocks modified LDL uptake. J Lipid Res. 1998;39:531–43. [PubMed] [Google Scholar]
- 11.Rohrer L, Freeman M, Kodama T, Penman M, Krieger M. Coiled-coil fibrous domains mediate ligand-binding by macrophage scavenger receptor type-II. Nature. 1990;343:570–2. doi: 10.1038/343570a0. [DOI] [PubMed] [Google Scholar]
- 12.Kodama T, Freeman M, Rohrer L, Zabrecky J, Matsudaira P, Krieger M. Type-I macrophage scavenger receptor contains alpha-helical and collagen-like coiled coils. Nature. 1990;343:531–5. doi: 10.1038/343531a0. [DOI] [PubMed] [Google Scholar]
- 13.Penman M, Lux A, Freedman NJ, et al. The type-I and type-II bovine scavenger receptors expressed in Chinese-Hamster Ovary cells are trimeric proteins with collagenous triple helical domains comprising non-covalently associated monomers and Cys83-disulfide-linked dimers. J Biol Chem. 1991;266:23985–93. [PubMed] [Google Scholar]
- 14.Araki N, Higashi T, Mori T, et al. Macrophage scavenger receptor mediates the endocytic uptake and degradation of advanced glycation end-products of the Maillard reaction. Eur J Biochem. 1995;230:408–15. doi: 10.1111/j.1432-1033.1995.0408h.x. [DOI] [PubMed] [Google Scholar]
- 15.El-Khoury J, Thomas CA, Loike JD, Hickman SE, Cao L, Silverstein SC. Macrophages adhere to glucose-modified basement membrane collagen IV via their scavenger receptors. J Biol Chem. 1994;269:10197–200. [PubMed] [Google Scholar]
- 16.El-Khoury J, Hickman SE, Thomas CA, Cao L, Silverstein SC, Loike JD. Scavenger receptor-mediated adhesion of microglia to beta-amyloid fibrils. Nature. 1996;382:716–9. doi: 10.1038/382716a0. [DOI] [PubMed] [Google Scholar]
- 17.Paresce DM, Ghosh RN, Maxfield FR. Microglial cells internalize aggregates of the Alzheimer's-disease amyloid beta-protein via a scavenger receptor. Neuron. 1996;17:553–65. doi: 10.1016/s0896-6273(00)80187-7. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki H, Kurihara Y, Takeya M, et al. A role for macrophage scavenger receptors in atherosclerosis and susceptibility to infection. Nature. 1997;386:292–6. doi: 10.1038/386292a0. [DOI] [PubMed] [Google Scholar]
- 19.Gough PJ, Gordon S. The role of scavenger receptors in the innate immune system. Microbes Infect. 2000;2:305–11. doi: 10.1016/s1286-4579(00)00297-5. 10.1016/s1286-4579(00)00297-5. [DOI] [PubMed] [Google Scholar]
- 20.Haworth R, Platt N, Keshav S, et al. The macrophage scavenger receptor type A is expressed by activated macrophages and protects the host against lethal endotoxic shock. J Exp Med. 1997;186:1431–9. doi: 10.1084/jem.186.9.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sinclair JH, Baillie J, Bryant LA, Taylor-Wiedeman JA, Sissons JG. Repression of human cytomegalovirus major immediate early gene expression in a monocytic cell line. J Gen Virol. 1992;73:433–5. doi: 10.1099/0022-1317-73-2-433. [DOI] [PubMed] [Google Scholar]
- 22.Clarke S, Gordon S. Myeloid-specific gene expression. J Leukoc Biol. 1998;63:153–68. doi: 10.1002/jlb.63.2.153. [DOI] [PubMed] [Google Scholar]
- 23.Dziennis S, Van Etten RA, Pahl HL, et al. The CD11b promoter directs high-level expression of reporter genes in macrophages in transgenic mice. Blood. 1995;85:319–29. [PubMed] [Google Scholar]
- 24.Horvai A, Palinski W, Wu H, Moulton KS, Kalla K, Glass CK. Scavenger receptor A gene regulatory elements target gene expression to macrophages and to foam cells of atherosclerotic lesions. Proc Natl Acad Sci USA. 1995;92:5391–5. doi: 10.1073/pnas.92.12.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clarke S, Greaves DR, Chung LP, Tree P, Gordon S. The human lysozyme promoter directs reporter gene expression to activated myelomonocytic cells in transgenic mice. Proc Natl Acad Sci USA. 1996;93:1434–8. doi: 10.1073/pnas.93.4.1434. 10.1073/pnas.93.4.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dighe AS, Campbell D, Hsieh CS, et al. Tissue-specific targeting of cytokine unresponsiveness in transgenic mice. Immunity. 1995;3:657–66. doi: 10.1016/1074-7613(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 27.Jin DI, Jameson SB, Reddy MA, Schenkman D, Ostrowski MC. Alterations in differentiation and behaviour of monocytic phagocytes in transgenic mice that express dominant suppressors of ras signalling. Mol Cell Biol. 1995;15:693–703. doi: 10.1128/mcb.15.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Back A, East K, Hickson D. Leukocyte integrin CD11b promoter directs expression in lymphocytes and granulocytes in transgenic mice. Blood. 1995;85:1017–24. [PubMed] [Google Scholar]
- 29.Chen JW, Cha Y, Yuksel KU, Gracy RW, August JT. Isolation and sequencing of a cDNA clone encoding lysosomal membrane glycoprotein mouse lamp-1 – sequence similarity to proteins bearing onco-differentiation antigens. J Biol Chem. 1988;263:8754–8. [PubMed] [Google Scholar]
- 30.Fambrough DM, Takeyasu K, Lippincottschwarz J, Siegel NR, Somerville D. Structure of lep100, a glycoprotein that shuttles between lysosomes and the plasma-membrane, deduced from the nucleotide-sequence of the encoding cDNA. J Cell Biol. 1988;106:61–7. doi: 10.1083/jcb.106.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fukuda M, Viitala J, Matteson J, Carlsson SR. Cloning of cDNAs encoding human lysosomal membrane-glycoproteins, h-lamp-1 and h-lamp-2 – comparison of their deduced amino-acid sequences. J Biol Chem. 1988;263:18920–8. [PubMed] [Google Scholar]
- 32.Holness CL, Simmons DL. Molecular-cloning of CD68, a human macrophage marker related to lysosomal glycoproteins. Blood. 1993;81:1607–13. [PubMed] [Google Scholar]
- 33.Holness CL, Dasilva RP, Fawcett J, Gordon S, Simmons DL. Macrosialin, a mouse macrophage-restricted glycoprotein, is a member of the lamp/lgp family. J Biol Chem. 1993;268:9661–6. [PubMed] [Google Scholar]
- 34.Strobl H, Scheinecker C, Csmarits B, Majdic O, Knapp W. Flow cytometric analysis of intracellular CD68 molecule expression in normal and malignant hematopoiesis. Br J Haematol. 1995;90:774–82. doi: 10.1111/j.1365-2141.1995.tb05195.x. [DOI] [PubMed] [Google Scholar]
- 35.Smith MJ, Koch GLE. Differential expression of murine macrophage surface glycoprotein antigens in intracellular membranes. J Cell Sci. 1987;87:113–9. doi: 10.1242/jcs.87.1.113. [DOI] [PubMed] [Google Scholar]
- 36.Pulford KAF, Sipos A, Cordell JL, Stross WP, Mason DY. Distribution of the CD68 macrophage/myeloid associated antigen. Int Immunol. 1990;2:973–80. doi: 10.1093/intimm/2.10.973. [DOI] [PubMed] [Google Scholar]
- 37.Rabinowitz S, Horstmann H, Gordon S, Griffiths G. Immunocytochemical characterisation of the endocytic and phagolysosomal compartments in peritoneal macrophages. J Cell Biol. 1992;116:95–112. doi: 10.1083/jcb.116.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabinowitz SS, Gordon S. Macrosialin, a macrophage-restricted membrane sialoprotein differentially glycosylated in response to inflammatory stimuli. J Exp Med. 1991;174:827–36. doi: 10.1084/jem.174.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warnke RA, Pulford KAF, Pallesen G, Ralfkiaer E, Brown DC, Gatter KC, Mason DY. Diagnosis of myelomonocytic and macrophage neoplasms in routinely processed tissue biopsies with monoclonal-antibody kp1. Am J Pathol. 1989;135:1089–95. [PMC free article] [PubMed] [Google Scholar]
- 40.Athanasou NA, Heryet A, Quinn J, Gatter KC, Mason DY, McGee JO. Osteoclasts contain macrophage and megakaryocyte antigens. J Pathol. 1986;150:239–46. doi: 10.1002/path.1711500403. [DOI] [PubMed] [Google Scholar]
- 41.Jones E, Quinn CM, See CG, et al. The linked human elongation initiation factor 4A1 (EIF4A1) and CD68 genes map to chromosome 17p13. Genomics. 1998;53:248–50. doi: 10.1006/geno.1998.5515. 10.1006/geno.1998.5515. [DOI] [PubMed] [Google Scholar]
- 42.Greaves DR, Quinn CM, Seldin MF, Gordon S. Functional comparison of the murine macrosialin and human CD68 promoters in macrophage and nonmacrophage cell lines. Genomics. 1998;54:165–8. doi: 10.1006/geno.1998.5546. 10.1006/geno.1998.5546. [DOI] [PubMed] [Google Scholar]
- 43.Quinn CM, Wiles AP, El-Shanawany T, et al. The human eukaryotic initiation factor 4AI gene (EIF4A1) contains multiple regulatory elements that direct high-level reporter gene expression in mammalian cell lines. Genomics. 1999;62:468–76. doi: 10.1006/geno.1999.6031. 10.1006/geno.1999.6031. [DOI] [PubMed] [Google Scholar]
- 44.Laukkanen J, Lehtolainen P, Gough PJ, Greaves DR, Gordon S, Yla-Herttuala S. Adenovirus-mediated gene transfer of a secreted form of human macrophage scavenger receptor inhibits modified low-density lipoprotein degradation and foam-cell formation in macrophages. Circulation. 2000;101:1091–6. doi: 10.1161/01.cir.101.10.1091. [DOI] [PubMed] [Google Scholar]
- 45.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–1. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 46.Resnick D, Chatterton JE, Schwartz K, Slayter H, Krieger M. Structure of class-A macrophage scavenger receptors – electron-microscopic study of flexible, multidomain, fibrous proteins and determination of the disulfide bond pattern of the scavenger receptor cysteine-rich domain. J Biol Chem. 1996;271:26924–30. doi: 10.1074/jbc.271.43.26924. 10.1074/jbc.271.43.26924. [DOI] [PubMed] [Google Scholar]
- 47.Resnick D, Freedman NJ, Xu SZ, Krieger M. Secreted extracellular domains of macrophage scavenger receptors form elongated trimers which specifically bind crocidolite asbestos. J Biol Chem. 1993;268:3538–45. [PubMed] [Google Scholar]
- 48.Hogan B, Beddington R, Costantini F, Lacey E. Manipulating the Mouse Embryo. New York: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 49.Hume DA, Gordon S. Optimal conditions for proliferation of bone marrow-derived mouse macrophages in culture – the roles of CSF-1, serum, Ca-2+, and adherence. J Cell Physiol. 1983;117:189–94. doi: 10.1002/jcp.1041170209. [DOI] [PubMed] [Google Scholar]
- 50.Gough PJ, Greaves DR, Suzuki H, et al. Analysis of macrophage scavenger receptor (SR-A) expression in human aortic atherosclerotic lesions. Arterioscler Thromb Vasc Biol. 1999;19:461–71. doi: 10.1161/01.atv.19.3.461. [DOI] [PubMed] [Google Scholar]
- 51.de Winther MP, van Dijk KW, van Vlijmen BJ, et al. Macrophage specific overexpression of the human macrophage scavenger receptor in transgenic mice, using a 180-kb yeast artificial chromosome, leads to enhanced foam cell formation of isolated peritoneal macrophages. Atherosclerosis. 1999;147:339–47. doi: 10.1016/s0021-9150(99)00204-x. 10.1016/s0021-9150(99)00204-x. [DOI] [PubMed] [Google Scholar]
- 52.Heydemann A, Warming S, Clendenin C, Sigrist K, Hjorth JP, Simon MC. A minimal c-fes cassette directs myeloid-specific expression in transgenic mice. Blood. 2000;96:3040–8. [PubMed] [Google Scholar]