Abstract
Streptococcal inhibitor of complement (SIC) was first described in 1996 as a putative inhibitor of the membrane attack complex of complement (MAC). SIC is a 31 000 MW protein secreted in large quantities by the virulent Streptococcus pyogenes strains M1 and M57, and is encoded by a gene which is extremely variable. In order to study further the interactions of SIC with the MAC, we have made a recombinant form of SIC (rSIC) in Escherichia coli and purified native M1 SIC which was used to raise a polyclonal antibody. SIC prevented reactive lysis of guinea pig erythrocytes by the MAC at a stage prior to C5b67 complexes binding to cell membranes, presumably by blocking the transiently expressed membrane insertion site on C7. The ability of SIC and clusterin (another putative fluid phase complement inhibitor) to inhibit complement lysis was compared, and found to be equally efficient. In parallel, by enzyme-linked immunosorbent assay both SIC and rSIC bound strongly to C5b67 and C5b678 complexes and to a lesser extent C5b-9, but only weakly to individual complement components. The implications of these data for virulence of SIC-positive streptococci are discussed, in light of the fact that Gram-positive organisms are already protected against complement lysis by the presence of their peptidoglycan cell walls. We speculate that MAC inhibition may not be the sole function of SIC.
Introduction
Severe Group A streptococcal (GAS) infections have been on the increase world wide in recent years, after a progressive decline prior to the 1970s, and almost complete eradication in developed countries. Alongside the increase in the number of cases of GAS infection reported, several extremely virulent strains have emerged. These include strains of the M1 serotype, which has been the most frequently recorded serotype in surveys of invasive infections in several European countries.1 The factors responsible for the predominance of M1 strains are unknown.
Complement is an important component of the immune response to bacterial infections.2 Complement activation via specific antibody, or directly through the alternative or lectin pathways, results in deposition of C3b and iC3b on bacterial surfaces, a crucial event which targets them for recognition and clearance by phagocytic cells bearing complement receptors. Subsequent cleavage of C5 generates the proteolytic fragment C5a which, together with C3a, is chemotactic for neutrophils and activates them via receptors on their surfaces. The generation of the C5b fragment initiates formation of the complement membrane attack complex (MAC), a lytic assembly of C5b, C6, C7, C8 and multiple molecules of C9. The intermediate complex C5b-7 has a transient membrane binding site; subsequent incorporation of C8 and C9 allows the complex to penetrate lipid bilayers and form a functional pore.2 The MAC is more of a threat to Gram-negative organisms, which have an outer lipid membrane, than to Gram-positive organisms whose membrane is protected by a thick cell wall.3
GAS have developed a number of mechanisms to evade the host defence system and, in particular, exhibit a number of surface virulence proteins involved in the evasion of attack by the complement system. These comprise the M and M-like protein family, some of which bind the host-derived fluid-phase complement inhibitors factor H,4 factor-H-like protein 1 (FHL-1)5 or C4 binding protein (C4bp),6 thus down-regulating complement activation. Another member of this group is C5a peptidase which inactivates C5a, a potent anaphylatoxin, by clipping seven amino acids off the carboxy terminal end.7 The genes for the M-protein family (mrp, emm, enn) and C5a peptidase (scpA) are located in the mga regulon of the GAS genome.
In 1996 Åkesson and co-workers8 identified a new streptococcal protein which they termed streptococcal inhibitor of complement (SIC). The gene for the SIC protein (GenBank Accession No. X92968) is found in very few strains of GAS (namely M1, M12, M55 and M57) and is usually located in the mga regulon,8,9 the exception being M57.9 SIC is a 31 000 MW extracellular protein, secreted in large quantities (up to 10 mg/l), which inhibits lysis by the MAC in haemolytic assays. SIC also binds to clusterin,8 another MAC inhibitor10 and to histidine-rich-glycoprotein8 which has also been reported to interact with complement.11 The exact mechanism by which SIC inhibits complement lysis was not elucidated.
A more recent study addressing the biological significance of SIC implicated a role for SIC in the early stages of establishing infection.12 In that study an M1 isogenic mutant strain was constructed in which the sic gene was inactivated by a novel non-polar mutagenesis strategy. Within 4 days post-infection, mice inoculated with the wild type parental strain had a significantly higher incidence of throat colonization than those inoculated with the sic knock-out mutant strain, demonstrating that SIC promotes the early stages of infection in some way.
The object of the present study was to investigate further the molecular mechanisms by which SIC interferes with the action of the complement MAC.13
Materials and methods
Bacterial strains
Three strains of GAS were obtained from the National Collection of Type Cultures, Central Public Health Laboratory, London, as follows: Type M1, NCTC 8198 (ATCC 12344); Type M12, NCTC 10085; and Type M57, NCTC 10875. All are vaccine strains for production of M-antiserum. The bacteria were grown in Todd Hewitt broth/0·2% yeast extract (Oxoid Ltd, Basingstoke, UK) and maintained on horse blood agar plates. To select for maximum expression of M-proteins and other virulence factors the strains were passaged three times in fresh human plasma as described.14
DNA preparation and polymerase chain reaction
Bacterial genomic DNA was prepared by digestion of the streptococcal cell walls with mutanolysin (Sigma-Aldrich Co. Ltd, Gillingham, UK), followed by lysis with N-lauryl sarcosine, phenol–chloroform extraction and ethanol precipitation. Polymerase chain reaction (PCR) of the entire SIC gene, including the 5′ untranslated region, was performed using the primers S1 (5′ TAAGGAGAGGTCACAAACTA 3′ forward), and S2 (5′ TTACGTTGCTGATGGTGTAT 3′ reverse) as described.15 Cycle conditions were 93° 1 min, 58° 1 min, 72° 1·5 min in a Perkin-Elmer Cetus 4800 thermal cycler (Foster City, CA). Samples were analysed on 1·2% agarose mini-gels in Tris/borate/ethylenediamine tetra-acetic acid (EDTA) buffer containing 0·5 µg/ml ethidium bromide.
Southern blotting
Southern blots16 to detect the presence of the sic gene were performed on HindIII digests of genomic DNA from all three GAS strains, and from strains of Type 2 Streptococcus suis and S. equi (kindly provided by Dr Andrew Allen, Bacterial Infection Group, Centre for Veterinary Science, University of Cambridge) as negative controls. Samples were analysed on 0·8% agarose gels in Tris/acetate/EDTA buffer, blotted onto Hybond N (Amersham Pharmacia Biotech, Little Chalfont, UK), UV cross-linked and probed with an M1 sic PCR amplicon radio-labelled with 32P using the ‘Ready-To-Go’ kit (Amersham Pharmacia Biotech). Filters were washed with increasing stringency down to 0·4 × SSC (standard saline citrate; 60 mm sodium chloride, 6 mm trisodium citrate, pH 7) at 65° and autoradiographed under enhancing conditions.17
Western blotting
Western blot analysis of the binding of factor H, C4bp and C3 to the GAS strains was carried out essentially by the method of Perez-Caballero et al.18 Briefly, aliquots of approximately 108 bacteria, as estimated by OD600, were washed in phosphate-buffered saline (PBS), resuspended in either 50 µl EDTA plasma, 50 µl normal human serum (NHS) or 50 µl NHS/EDTA (NHSE) (each diluted 1/2 in PBS), or 50 µl PBS only as control, and incubated for 2·5 hr at room temperature on a rotator. The samples were then centrifuged, and the pellets washed three times in PBS/0·05% Tween-20 (PBST), resuspended in 50 µl Laemmli sample buffer (2% sodium dodecyl sulphate (SDS), 10% glycerol, 100 mm dithiothreitol (DTT), 60 mm Tris pH 6·8, 0·0001% bromophenol blue) and boiled for 5 min. Samples were run on 15%, 10% or 6% SDS–polyacrylamide gel electrophoresis (PAGE) gels for C3, factor H or C4bp, respectively, in a Mini-PROTEAN II system (Bio-Rad Laboratories Ltd, Hemel Hempstead, UK), then electroblotted onto nitrocellulose. Non-specific protein binding sites were blocked with Tris-buffered saline/5% dried skimmed milk (TBSM), and the blots were probed using either an in-house sheep anti-C3, rabbit anti-C4bp (CN Biosciences UK, Beeston, UK), an in-house sheep anti-factor H, or rabbit anti-factor H (Serotec Ltd, Oxford, UK), followed by alkaline phosphatase-conjugated monoclonal anti-sheep/goat immunoglobulin or goat anti-rabbit immunoglobulin (Sigma-Aldrich Co. Ltd) diluted in TBSM and the nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl-1-phosphate (NBT/BCIP) substrate system (Zymed Laboratories Inc, Cambridge Bioscience, Cambridge, UK).
Production of recombinant SIC
Recombinant SIC (rSIC) was produced using the expression plasmid pBAD TOPO TA (Invitrogen BV, Groningen, The Netherlands) according to the manufacturer's protocol and in the Escherichia coli host TOP 10. The pBAD system adds a V5 epitope and a His-tag to the C-terminal end of the expressed protein to facilitate detection and purification of the recombinant protein. Primers corresponding to nucleotide nos. 97–117 (5′ GAAACGTATACATCACGCAAT 3′ forward) and 895–915 (5′ CGTTGCTGATGGTGTATATGG 3′ reverse) of the published sic gene8 (GenBank Accession No. X92968) were used to produce an amplicon of 818 bp that spanned the entire coding region of the predicted mature protein up to one amino acid before the stop codon. Cycle conditions were 93° 1 min, 62° 1 min, 72° 1 min, 30 cycles in a PTC-100 thermal cycler (MJ Research, Watertown, MA). Confirmation of correct in-phase sequence of positive clones was checked on an ABI Prism 3700 DNA analyser using ‘BigDye’ terminator reaction kits (PE Biosystems, Warrington, UK) at the DNA Sequencing Facility, Department of Biochemistry, University of Cambridge, and the coding sequence was found to be identical to the above published sequence throughout.
Expression in pBAD is positively regulated by the AraC gene product encoded in the plasmid, which is turned on in the presence of l-arabinose. Growth curves and expression conditions were optimized (i.e. titration of the concentration of l-arabinose used to induce expression) according to the manufacturer's protocol. Small scale purifications of rSIC from E. coli TOP 10 lysates were made using the ‘Xpress’ Protein Purification System (Invitrogen BV), and larger-scale preparations from 1-l cultures were purified on a 50-ml nickel-Sepharose column (Chelating Sepharose Fast Flow, Amersham Pharmacia Biotech) eluted with a 50–500 mm imidazole gradient. rSIC-containing fractions were identified by 10% SDS–PAGE and Western blots using horseradish peroxidase-conjugated anti-V5 monoclonal antibody (Invitrogen BV) and chemiluminescence (Super Signal West Pico, Perbio Science UK Ltd, Chester, UK). These fractions were pooled, concentrated and dialysed into PBS/10 mm sodium azide.
Purification of native SIC and antibody production
SIC was purified from 1-l overnight culture supernatants of strain M1 GAS, to which 10 mm EDTA had been added, using the following procedure. A SIC-containing fraction was obtained by precipitation with 30% ammonium sulphate at room temperature. The precipitate was redissolved in, and dialysed against, 50 mm Tris pH 7·5 and then applied to a 10-ml POROS HQ column (PE Biosystems) equilibrated in the same buffer, on an FPLC system (Amersham Pharmacia Biotech). The SIC was eluted with a 0–1·5 m sodium chloride gradient of 30 column volumes which was then stepped to 2 m sodium chloride for two column volumes. SIC-containing fractions were identified by a functional assay to detect complement inhibitory activity (see below) and confirmed by SDS–PAGE and Western blotting using a rabbit anti-SIC (kindly provided by Dr Per Åkesson, Lund University, Lund, Sweden) and alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin (Sigma-Aldrich Co. Ltd). The SIC eluted as a broad trailing peak from approximately 0·9 m sodium chloride. If necessary, the SIC was further purified using hydrophobic interaction chromatography (HIC) on a 2-ml POROS ET column (PE Biosystems). SIC (0·5 mg) was applied to the column in 0·4 m ammonium sulphate/50 mm phosphate buffer pH 7·0, and eluted with a decreasing ammonium sulphate gradient of 10 column volumes down to zero ammonium sulphate in the phosphate buffer. SIC eluted very late in the gradient with a peak around 40 mm ammonium sulphate. SIC-containing fractions were pooled and concentrated, and dialysed into PBS/10 mm sodium azide. The N-terminal sequence of purified SIC was confirmed by the Protein and Nucleic Acid Chemistry Facility, Department of Biochemistry, University of Cambridge.
Purified SIC was used to raise a rabbit antiserum (AbCam Ltd, Cambridge, UK).
Sera and complement reagents
C5, C56 complex, C6 and C7 were ‘in-house’ reagents prepared according to methods outlined in reference 19. These preparations were ‘functionally pure’ in the sense that they were free of other complement components, although they contained other minor protein contaminants. C8 and C9 were purified by affinity chromatography as described20,21 using L6 anti-C8 and C9–8 anti-C9 monoclonal antibodies that were generously provided by Dr Paul Luzio (University of Cambridge) and Professor Paul Morgan (University of Wales College of Medicine), respectively. C7-deficient serum was kindly provided by Dr Menachem Schlesinger (Department of Immunology, Barzilai Medical Center, Ashkelon, Israel). Normal human serum was depleted of C8 or C9 by affinity chromatography as described.20,21 Deficiency/depletion of complement components was confirmed by specific haemolytic assay19 and Western blotting. Partial and complete terminal complement complexes (TCC) were generated by yeast-activation of whole or depleted/deficient sera for 2 hr at 37°. After removal of the zymosan particles by centrifugation, the sera were aliquoted and stored at −80°. The presence of intermediate TCC complexes was confirmed by enzyme-linked immunosorbent assay (ELISA, see below).
Purified clusterin was obtained from Quidel (Qbiogene, Harefield, UK).
Haemolytic assays
To test the effects of SIC upon formation of the complement membrane attack complex (MAC), or to compare its activity with that of clusterin, 25 µl doubling dilutions of SIC, clusterin, or bovine serum albumin (BSA) as control were mixed, in triplicate, with 25 µl of a 5% suspension of guinea pig erythrocytes (GPE) in a 96-well assay plate. Then 25 µl of C56 1/100 and 50 µl NHS (as a source of C7, C8 and C9) 1/40 were added, pre-titrated to give approximately 50% lysis of the GPE. Alternatively, 25 µl C7 1/1000 (pre-titrated, see below) and 25 µl of a mixture of C8 and C9 (each 10 µg/ml) were added instead of whole serum. All dilutions were in PBS/10 mm EDTA. The plate was incubated for 15 min at 37° to promote MAC assembly, then centrifuged to pellet intact cells and cell membrane ‘ghosts’, and 100 µl supernatant from each well was transferred to a fresh plate to measure haemoglobin release. Percentage lysis was calculated by measuring absorbance at 405 nm, and using the following formula:
[(sample − background)/(100% lysis − background)] × 100 where background haemoglobin release was obtained from GPE incubated in buffer only, and 100% lysis was achieved using distilled water.
The above assay was also used to detect the presence of SIC in column fractions during protein purifications. In some experiments, in order to test whether SIC interacted with cell membranes directly, the cells were pre-incubated with dilutions of SIC for 15 min at 37°, then washed and resuspended in 50 µl PBS/10 mm EDTA prior to the addition of complement reagents. In other experiments, SIC was added to the cells after C5b-7 complexes had been formed on their surfaces. In these cases, 2 ml 5% GPE were mixed with 1 ml C56 1/50 and 1 ml C7 1/500 for 15 min at 37°, to allow deposition of C5b-7 complexes. All dilutions were in PBS/10 mm EDTA, and the C7 preparation was pre-titrated to saturate the dose of C56 offered to the cells. The GPE-C5b-7 cells were washed once in PBS/10 mm EDTA, resuspended to 2 ml, and 50 µl was added, in triplicate, to wells of a 96-well assay plate containing 25 µl doubling dilutions of SIC (or BSA as control). Finally, 50 µl NHS 1/40 in PBS/10 mm EDTA was added to each well as a source of C8 and C9, and the plate was incubated at 37° to allow lysis to proceed. Percentage lysis was calculated as described above.
ELISA
To confirm the presence of intermediate TCC complexes in yeast activated sera, ELISA plates (Greiner Labortechnik, Stroud, UK) were coated with a monoclonal anti-C5b neo-epitope (Progen Biotechnik Gmbh, Heidelberg, Germany – obtained through Insight Biotechnology, Wembley, UK) at 1 µg/ml in 50 mm carbonate–bicarbonate buffer pH 9·6 overnight at 4° then blocked with 1% gelatine/PBS. Yeast-activated sera (prepared as above) (and paired non-activated sera as negative controls) diluted 1/200 in PBS/0·05% Tween-20–0·1% gelatine (PBSTG) were added to triplicate wells and incubated for 1 hr at 37°. Complexes were detected with goat anti-C6 (immunoglobulin fraction)22 at 20 µg/ml in PBSTG followed by alkaline phosphatase conjugated monoclonal anti-sheep/goat immunoglobulin (Sigma-Aldrich Co. Ltd) diluted 1/1500 in PBSTG.
To investigate binding of SIC to intermediate forms of the TCC, ELISA plates were coated with SIC or rSIC (in triplicate) at 10 µg/ml total protein (or coating buffer only as negative control). Yeast activated sera diluted 1/200 in PBSTG (or buffer only) were added and incubated for 1 hr at 37°. Complexes were detected with goat anti-C6 (immunoglobulin fraction) followed by alkaline phosphatase-conjugated monoclonal anti-sheep/goat immunoglobulin as above. In another experiment, complexes were detected using the monoclonal anti-C5b neo-epitope at 1 µg/ml followed by alkaline phosphatase-conjugated goat anti-mouse immunoglobulin (Sigma-Aldrich Co. Ltd) diluted 1/1000.
All reagents were diluted in PBSTG, incubations were for 1 hr at 37° and all washes were in PBST except the final wash before substrate addition that was in Tris-buffered saline/0·05% Tween 20 (TBST). The ‘Sigma Fast’ pNPP substrate/buffer system was used throughout (Sigma-Aldrich Co. Ltd). Absorbance was read at 405 nm (reference wavelength 490 nm) in a Bio-Rad 3550 Microplate Reader.
To investigate binding of SIC to individual components of the MAC, triplicate wells were coated with purified C5, C6, C7, C8 or C9 at 2 µg/ml as above or SIC at 1 µg/ml or coating buffer only as positive and negative controls. SIC at 10 µg/ml was added and binding detected with in-house rabbit anti-SIC diluted 1/2000 followed by alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin diluted 1/1500 (Sigma-Aldrich Co. Ltd) and pNPP. All other conditions were as above.
Results
Characterization of bacterial strains
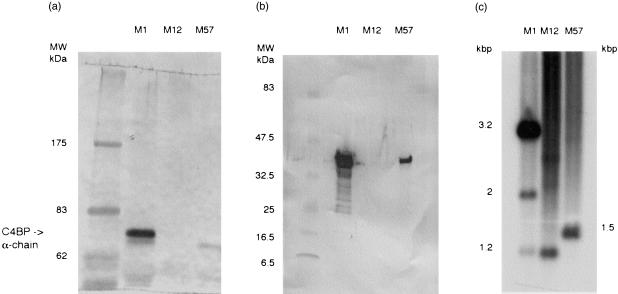
Because only certain strains of GAS express SIC, and only some strains interact with host-derived fluid-phase complement inhibitors, we decided to investigate the particular characteristics of the three strains used in this study. When the bacteria were incubated in whole serum only the M1 strain bound C4bp (Fig. 1a). C3 fragments were deposited equally on all three strains, as detected by Western blot, whereas in the presence of EDTA (which inhibits both classical and alternative pathway activation) no C3 was deposited (data not shown). None of the strains bound Factor H (data not shown). In Western blot analyses, SIC was detectable in culture supernatants of M1 and M57 strains, but not in strain M12 (Fig. 1b). Similarly, the M1 and M57 strains clearly contained the sic gene by PCR, whilst the M12 strain appeared negative (data not shown). However, in Southern blots probed with a PCR amplicon of the M1 sic gene, all three GAS strains contained hybridizing fragments (M1-3·2 and 2 kb, M12-1·5 kb, M57-1·3 kb), indicating that a sic or a sic-related gene was also present in M12 (Fig. 1c). No hybridizing bands were seen in either S. suis or S. equi negative controls.
Figure 1.
Characterization of bacterial strains. (a) Aliquots of strains M1, 12 and 57 GAS were incubated with normal human plasma, and bound proteins were eluted and analysed by SDS–PAGE and Western blotting using polyclonal anti-C4bp. The position of the 70 000 MW α-chain of C4bp is indicated by the arrow. Only strain M1 bound C4bp. (b) Aliquots of overnight culture supernatant from strains M1, M12 and M57 GAS were analysed by SDS-PAGE and Western blotting using polyclonal anti-M1 SIC. This showed that SIC is produced by strains M1 and M57 but not by M12. (c) Southern blot of HindIII digested streptococcal DNA from strains M1, 12 and 57 probed with labelled M1 sic PCR amplicon showing the presence of a sic or sic-like gene in all three GAS strains tested. No hybridizing bands were detected in either of the negative control Streptococci (data not shown).
Purification of native and recombinant SIC
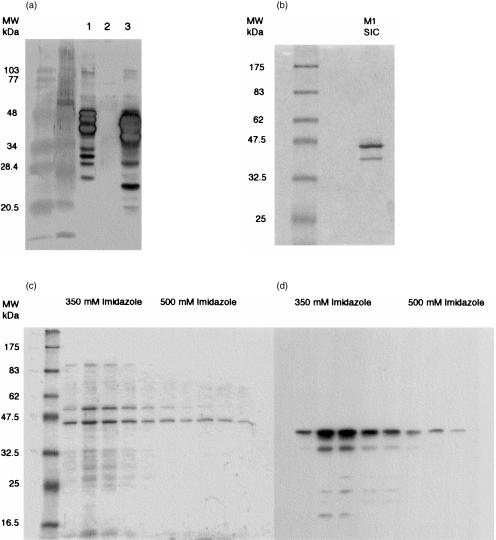
To investigate the interaction of SIC with components of the complement system, we purified native SIC from culture supernatants of the M1 strain, and expressed M1 SIC as a recombinant protein in E. coli. SIC was purified from M1 culture supernatants by a combination of anion-exchange and hydrophobic interaction chromatography, as described in Materials and methods. During preliminary experiments for establishing optimal conditions, it was surprising to find that although SIC is a secreted protein which is theoretically hydrophilic, it bound readily to a relatively weak hydrophobic stationary phase (POROS ET) in low salt concentration. Another interesting observation was that SIC purified from M1 culture supernatant ran at an apparent MW on SDS–PAGE gels of up to 45 000 MW, although its MW is 31 000 MW (Fig. 1b and Fig. 2a,b). Electrophoretic migration of SIC varied according to the voltage used, and the apparent MW of SIC was found to be closer to its calculated MW when gels were run more slowly. Typically, up to 10 mg pure SIC was obtained per litre of overnight culture.
Figure 2.
Western blot analysis of recombinant and native SIC(a) Western blot showing reactivity of rabbit polyclonal antibody to native SIC and recombinant SIC: (1) rSIC; (2) E. coli protein extract negative control; (3) concentrated M1 culture supernatant. Lower MW bands are presumed to represent breakdown products of SIC and rSIC. (b) Coomassie stained 10% SDS–PAGE gel of purified native SIC. The lower band is a breakdown product of SIC. N-terminal sequencing of this band showed that 33 amino acids had been lost from the native protein. (c) Coomassie stained 10% SDS–PAGE gel of rSIC-containing column fractions, and (d) autoradiograph of a Western blot from an identical SDS–PAGE gel demonstrating that rSIC eluted mainly at 350 mm imidazole. rSIC was detected using HRP-conjugated monoclonal anti-V5 epitope and chemiluminescence. Non-reactive bands in panel (c) are residual E. coli proteins.
Pilot experiments revealed that rSIC occurred in the soluble phase of lysed bacteria, and it could be eluted from a nickel–Sepharose column at around 350 mm imidazole using a gradient of 50–500 mm imidazole (Fig. 2c,d). The calculated MW of rSIC was approximately 34 000 MW but migration also varied according to voltage. Typically, 8 mg/l rSIC was obtained in larger scale preparations purified on a 50-ml nickel–Sepharose column, but still contained traces of E. coli proteins. No further purification steps have been carried out to date. The recombinant protein was detectable by SDS–PAGE and Western blots using the rabbit antiserum raised against native SIC (Fig. 2a).
SIC inhibits the MAC by blocking uptake of C5b-7 complexes
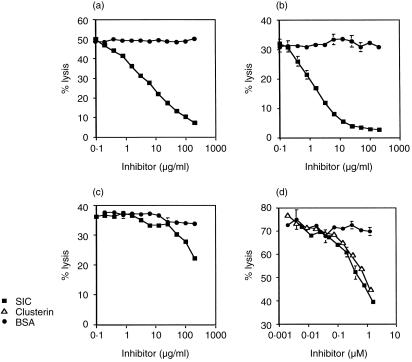
The effect of SIC upon the reactive lysis of GPE in human serum is shown in Fig. 3(a). SIC at a final concentration of 10 µg/ml (0·3 µm) caused 50% inhibition of MAC formation in whole serum. This effect was not observed if cells were pre-incubated with SIC and then washed prior to the addition of serum, indicating that SIC does not inhibit complement through an interaction with the red cell membrane, but works from the fluid-phase (data not shown). To check that SIC inhibition of the MAC was not being mediated by other serum proteins, it was decided to test the effect of SIC upon reactive lysis using purified complement components. SIC inhibited lysis even in these circumstances (Fig. 3b), demonstrating that it is capable of interacting directly with components of the MAC.
Figure 3.
SIC inhibits the binding of C5b-7 complexes to cell membranes. Reactive lysis of guinea pig erythrocytes was initiated by addition of pre-formed C56 complex followed by either (a) 1/40 normal human serum in 10 mm EDTA, or (b) purified C7, C8 and C9. Cells preincubated with SIC were protected against lysis compared to control cells incubated with BSA. When added to cells after C5b-7 sites had been formed on their surfaces (c), SIC had very little effect upon haemolysis, showing that it works by blocking uptake of C5b-7 complexes. A comparison of the activity of SIC and clusterin (d) showed that SIC and clusterin were equally efficient at inhibiting haemolysis of GPE by C56 and 1/40 NHS/EDTA. Data are expressed as mean ± SEM of triplicate results from a typical experiment. Error bars are too small to be seen on some data points.
In order to find out at which stage SIC interfered with MAC assembly, SIC was added to the cells after C5b-7 sites had been pre-formed on their surfaces. SIC then had very little effect upon lysis, indicating that it acts prior to C5b-7 binding to cell membranes (Fig. 3c). SIC resembles clusterin in this respect. A direct comparison of the complement-inhibiting ability of SIC and clusterin showed that they are equally efficient at inhibiting MAC formation in an in vitro reactive lysis assay (Fig. 3d).
SIC and rSIC bind C5b-7, C5b-8 and C5b-9 complexes in ELISA
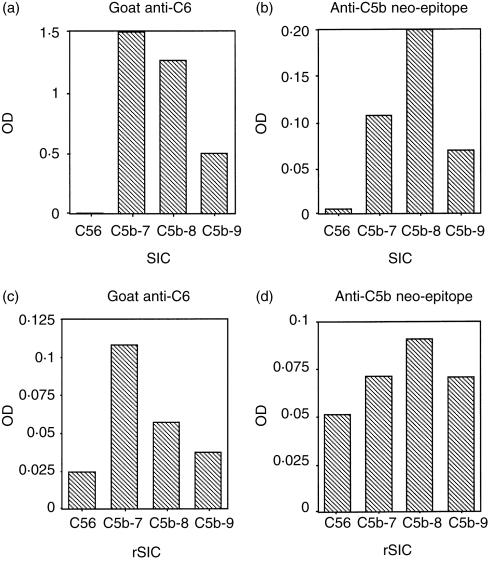
The interaction of SIC with intermediate forms of TCC was measured by ELISA. The partially formed complexes in yeast-activated sera were added to ELISA plates coated with SIC or rSIC, and binding was detected using a polyclonal antibody to C6 or a monoclonal antibody to a C5b neo-epitope which does not react with native C5. C5b-7, C5b-8 and C5b-9 complexes bound to immobilized SIC and rSIC, whereas C5b6 did not appear to do so, whichever antibody was used for detection (Fig. 4a–d). When the anti-C5b neo-epitope was used for detection, the greatest level of binding observed was to C5b-8 complexes (Fig. 4b,d) but when anti-C6 was used, maximum binding occurred with C5b-7 complexes (Fig. 4a,c). The lower levels of binding of intermediate TCC complexes to rSIC may be attributable to there being less rSIC on the plate, since this preparation still contained some E. coli proteins. This probably also accounts for the apparently higher binding of C5b6.
Figure 4.
SIC binds to C5b-7, C5b-8 and C5b-9 complexes in ELISA. ELISA plates were coated with SIC (panels a and b), rSIC (panels c and d) or coating buffer only and intermediate or complete TCC complexes were added. Binding was detected with (i) polyclonal anti-C6 (a and c); or (ii) monoclonal anti-C5b neo-epitope (b and d). SIC and rSIC bind preferentially to C5b-7 complexes and to a lesser extent to C5b-8 and C5b-9 using anti-C6 to detect, but predominantly to C5b-8 using anti-C5b neo-epitope. Results displayed are the net ODs after deduction of background in coated wells plus antibodies but no sera.
SIC binds slightly to C7 and C8 in ELISA
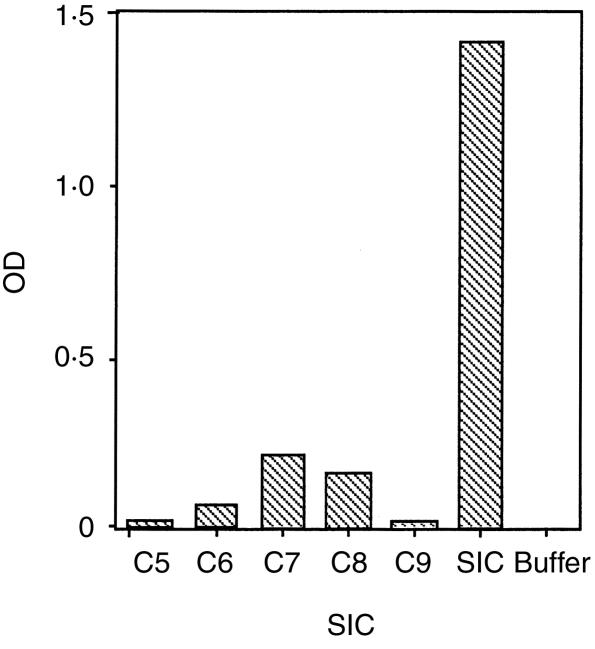
The interaction of SIC with individual components of the TCC was measured by ELISA. SIC was added to an ELISA plate coated with purified complement components and binding was detected using a polyclonal antibody to SIC. SIC bound slightly to C7 and C8 and, to a lesser extent, C6. No binding to C5 or C9 was detected (Fig. 5).
Figure 5.
SIC binds to purified C7 and C8 in ELISA. The ELISA plate was coated with individual purified components of the TCC (or SIC or buffer only as controls). SIC was added and binding detected with polyclonal anti-SIC showing that SIC binds slightly to purified C7 and C8 and to a lesser extent to C6.
Discussion
S. pyogenes type M1 is one of a few predominant strains found in epidemic waves of GAS infection. It is also one of the few strains reported to encode a novel, secreted protein implicated in resistance to complement, and therefore called SIC. The aim of this present study was to investigate further the interaction of SIC with the complement MAC.13
SIC purified from the culture supernatant of M1 GAS exhibited a number of unusual properties. Its behaviour on anion-exchange chromatography was consistent with it carrying a high negative charge, as predicted from the sequence. However, in hydrophobic interaction chromatography using POROS ET, SIC displayed markedly hydrophobic characteristics even though it is a secreted, soluble protein. Its mobility on SDS–PAGE was also variable, depending upon the conditions of electrophoresis – the apparent MW was higher when gels were run faster. This may be due to the unusually high number of prolines in the protein, which together with the lack of cysteines8 results in a non-globular structure that could retard migration of SIC on SDS–PAGE gels.23
Native SIC purified from M1 GAS culture supernatant inhibited lysis by preventing uptake of C5b67 complexes onto cell membranes. No protection against lysis occurred if the cells were washed in between exposure to SIC and addition of complement, showing that SIC did not interact itself with the cells. This may be unsurprising considering that SIC is a secreted protein, but was considered worth investigating in light of the unusual hydrophobic properties of SIC described above.
A number of other fluid phase inhibitors of the terminal complement pathway have been described and, almost exclusively, they act on the C5b67 complex (reviewed in 24). For example, S-protein (or vitronectin), a multifunctional protein whose primary role is as an adhesion molecule, binds to C5b67 and masks the membrane binding site thereby inhibiting insertion into a cell membrane. Clusterin (also known as apolipoprotein J, sulphated glycoprotein 2 or SP-40,40) is another multifunctional protein which has been described as a fluid-phase MAC inhibitor and which acts in the same manner as S-protein. We have shown that SIC and clusterin inhibit C5b67 complex insertion into the cell membrane to approximately the same degree. The physiological significance of any interaction between S-protein or clusterin and the MAC is questionable as C8 is actually the major in vivo inhibitor of C5b-7 complex binding to membranes25 because binding of C8 to the fluid-phase C5b67 complex also prevents membrane insertion. Interestingly, clusterin also binds to SIC8 but the significance of this interaction is not clear. The interaction does not appear to influence the ability of SIC to inhibit MAC lysis of GPE since the level of inhibition by SIC is comparable in the presence or absence of clusterin (Fig. 3a,b). Clusterin appears neither to diminish or augment the MAC inhibitory activity of SIC.
Native SIC and the recombinant version expressed in E. coli bound to C5b-7, C5b-8 and (to a lesser extent) C5b-9 complexes in an ELISA both when these intermediates were generated in depleted sera or using purified individual components. Maximum binding varied depending on the antibody used to detect the complexes and this may be caused by different availability of C6 epitopes due to conformation changes in the developing TCC, whereas availability of the C5b neo-epitope may not vary. Measurement using the monoclonal anti-C5b neo-epitope is possibly the more accurate.
Binding of SIC to C5b-8 and C5b-9 is unlikely to have any functional implications since C5b-8 is already unable to insert into a cell membrane.25 In this respect SIC closely resembles clusterin which has also been shown to bind to C5b-8 and to C5b-910. In other experiments using purified complement components bound to microtitre wells, we found that SIC bound very slightly to C7 and C8 and to a lesser extent to C6, but consider that this is also unlikely to have any significance in vivo.
It is well documented that virulent (M-positive) GAS strains do not activate the alternative pathway, unlike avirulent (M-negative) strains.26 This was attributed to some property of the surface M proteins, subsequently proposed to be their ability to bind the complement regulatory protein factor H.4 However, in a recent study investigating the binding of factor H, FHL-1 and C4bp to 69 clinical isolates of 19 different M-serotypes of GAS, very few strains were found to bind any of these proteins.18 These results are consistent with our findings that only our M1 strain bound C4bp, and none of the three strains we tested (M1, M12 and M57) bound factor H, implying that other complement evasion strategies at must be at work.3
The published sequence of the M12 sic gene,9 which was not available at the beginning of this study, is very different at the 3′ end from the published M1 sequence. The sic gene in the M12 and M55 strains is heavily mutated with deletions scattered throughout its length and has been termed ‘distantly related to SIC’, whereas the M57 sic gene is very similar to that of M1.9 This probably explains why no sic-related gene was detected in M12 by PCR, since it is likely that the 3′ primer used did not hybridize. However, the gene was clearly present in the M12 strain by Southern blotting. The substantial difference in sequence between M1 and M12 SIC also probably explains why no cross-reacting protein was detected in M12 supernatants in Western blots probed with a polyclonal antiserum raised against M1 SIC.
In contrast, the M57 strain contains a sic gene that was readily detectable by PCR and a SIC protein detectable by Western blotting, although in this strain sic is not located within the mga regulon.9 Preliminary pulsed-field gel electrophoresis experiments and Southern blotting of M57 genomic DNA, using a variety of restriction enzymes, failed to reveal a cohybridizing band between the M57 sic gene and the C5a peptidase gene (scpA) (authors' unpublished observations). Similarly, long range PCR experiments using different combinations of primers at each end of the sic and scpA genes did not produce a plausible amplicon. Taken together these results imply that the M57 sic gene is at least 50 kb away from the mga regulon (authors' unpublished observations).
In conclusion, although we have shown that SIC inhibits lysis by blocking uptake of C5b-7 complexes in vitro, it should be borne in mind that GAS are inherently resistant to complement lysis since their cell membranes are inaccessible to the MAC because of the presence of the peptidoglycan cell wall and (in some strains during exponential growth) a hyaluronic acid capsule as well. In addition, the role of fluid-phase inhibitors such as clusterin or SIC may be debatable for reasons we have discussed. In view of the above, we suspect that inhibition of the MAC may not be the only function of SIC and that, like clusterin, it is a multifunctional protein and has other roles important for virulence. Investigations of other possibilities are currently underway.
Acknowledgments
The authors are most grateful to Dr Andrew Allen and Miss Heather Lindsay, Bacterial Infection Group, Centre for Veterinary Science, University of Cambridge for helpful discussions and advice on working with streptococci; to Dr Anne Wilson of our department for advice on ELISA techniques and critical reading of the manuscript; and to Dr Per Åkesson, Department of Cell and Molecular Biology, Section for Molecular Pathogenesis, Lund University, Lund, Sweden for generously providing rabbit anti-SIC which assisted our initial purification procedures.
References
- 1.Efstratiou A. Group A streptococci in the 1990s. J Antimicrob Chemother. 2000;45:3–12. doi: 10.1093/jac/45.suppl_1.3. 10.1093/jac/45.suppl_1.3. [DOI] [PubMed] [Google Scholar]
- 2.Song W-C, Sarrias MR, Lambris JD. Complement and innate immunity. Immunopharmacology. 2000;49:187–98. doi: 10.1016/s0162-3109(00)80303-3. 10.1016/s0162-3109(00)00217-4. [DOI] [PubMed] [Google Scholar]
- 3.Würzner R. Evasion of pathogens by avoiding recognition or eradication by complement, in part via molecular mimicry. Mol Immunol. 1999;36:249–60. doi: 10.1016/s0161-5890(99)00049-8. 10.1016/s0161-5890(99)00049-8. [DOI] [PubMed] [Google Scholar]
- 4.Hortstmann RD, Sievertsen HJ, Knobloch J, Fischetti VA. Antiphagocytic activity of streptococcal M protein: selective binding of complement control protein factor H. Proc Natl Acad Sci USA. 1988;85:1657–61. doi: 10.1073/pnas.85.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnsson E, Berggård K, Kotarsky H, Hellwage J, Zipfel PF, Sjöbring U, Lindahl G. Role of the hypervariable region in streptococcal M proteins: binding of a human complement inhibitor. J Immunol. 1998;161:4894–901. [PubMed] [Google Scholar]
- 6.Thern A, Stenberg L, Dahlbäck D, Lindahl G. Ig-binding surface proteins of Streptococcus pyogenes also bind human C4b-binding protein (C4bp), a regulatory component of the complement system. J Immunol. 1995;154:375–86. [PubMed] [Google Scholar]
- 7.Cleary PP, Prahbu U, Dale JB, Wexler DE, Handley J. Streptococcal C5a peptidase is a highly specific endopeptidase. Infect Immun. 1992;60:5219–23. doi: 10.1128/iai.60.12.5219-5223.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Åkesson P, Sjöholm AG, Björck L. Protein SIC, a novel extracellular protein of Streptococcus pyogenes interfering with complement function. J Biol Chem. 1996;271:1081–8. doi: 10.1074/jbc.271.2.1081. 10.1074/jbc.271.2.1081. [DOI] [PubMed] [Google Scholar]
- 9.Hartas J, Sriprakash KS. Streptococcus pyogenes strains containing emm12 and emm55 possess a novel gene coding for distantly related SIC protein. Microb Pathog. 1999;26:25–33. doi: 10.1006/mpat.1998.0244. 10.1006/mpat.1998.0244. [DOI] [PubMed] [Google Scholar]
- 10.Tschopp J, Chonn A, Hertig S, French LE. Clusterin, the human apolipoprotein and complement inhibitor, binds to complement C7, C8 beta and the b domain of C9. J Immunol. 1993;151:2159–65. [PubMed] [Google Scholar]
- 11.Chang N-S, Leu RW, Rummage JA, Anderson JK, Mole JE. Regulation of complement functional efficiency by histidine-rich glycoprotein. Blood. 1992;79:2973–80. [PubMed] [Google Scholar]
- 12.Lukomski S, Hoe NP, Abdi I, et al. Nonpolar inactivation of the hypervariable streptococcal inhibitor of complement gene (SIC) in serotype M1 Streptococcus pyogenes significantly decreases mouse mucosal colonization. Infect Immun. 2000;68:535–42. doi: 10.1128/iai.68.2.535-542.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernie-King BA, Seilly DJ, Davies A, Lachmann PJ. The membrane attack complex inhibitor of Streptococcus pyogenes (SIC) binds preferentially to C567. Immunopharmacology. 2000;49:74. (Abstract) [Google Scholar]
- 14.Becker CG. Selection of group A streptococci rich in M-protein from populations poor in M-protein. Am J Pathol. 1964;44:51–60. [PMC free article] [PubMed] [Google Scholar]
- 15.Perea Mejia LM, Stockbauer KE, Pan X, Cravioto A, Musser JM. Characterization of Group A Streptococcus strains recovered from Mexican children with pharyngitis by automated DNA sequencing of virulence-related genes: unexpectedly large variation in the gene (SIC) encoding a complement-inhibiting protein. J Clin Microbiol. 1997;35:3220–4. doi: 10.1128/jcm.35.12.3220-3224.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Southern EM. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–17. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 17.Laskey RA, Mills AD. Enhanced autoradiographic detection of 32P and 125I using intensifying screens and hypersensitized film. FEBS Lett. 1977;82:314–6. doi: 10.1016/0014-5793(77)80609-1. [DOI] [PubMed] [Google Scholar]
- 18.Pérez-Caballero D, Alberti S, Vivanco F, Sánchez-Corral P, Rodríguez de Córdoba S. Assessment of the interaction of human complement regulatory proteins with group A Streptococcus. Identification of a high-affinity group A Streptococcus binding site in FHL-1. Eur J Immunol. 2000;30:1243–53. doi: 10.1002/(SICI)1521-4141(200004)30:4<1243::AID-IMMU1243>3.0.CO;2-D. 10.1002/(sici)1521-4141(200004)30:04<1243::aid-immu1243>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 19.Harrison RA, Lachmann PJ. Complement technology. In: Weir DM, Herzenberg LA, Blackwell C, Herzenberg LA, editors. Handbook of Experimental Immunology. 4. Oxford: Blackwell Scientific Publications; 1986. pp. 39.1–39.49. [Google Scholar]
- 20.Abraha A, Morgan BP, Luzio JP. The preparation and characterization of monoclonal antibodies to human complement component C8 and their use in purification of C8 and C8 subunits. Biochem J. 1988;251:285–92. doi: 10.1042/bj2510285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morgan BP, Daw RA, Siddle K, Luzio JP, Campbell AK. Immunoaffinity purification of human complement component C9 using monoclonal antibodies. J Immunol Methods. 1983;64:269–81. doi: 10.1016/0022-1759(83)90434-9. [DOI] [PubMed] [Google Scholar]
- 22.Würzner R, Orren A, Potter P, et al. Functionally active complement proteins C6 and C7 detected in C6- and C7-deficient individuals. Clin Exp Immunol. 1991;83:430–7. doi: 10.1111/j.1365-2249.1991.tb05656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnstone A, Thorpe R. Immunochemistry in Practice. 2. Oxford: Blackwell Scientific Publications; 1987. [Google Scholar]
- 24.Morgan BP, Harris CL. Complement regulatory proteins. London: Academic Press; 1999. [Google Scholar]
- 25.Nemerow GR, Yamamoto KI, Lint TF. Restriction of complement-mediated membrane damage by the eighth component of complement: a dual role for C8 in the complement attack sequence. J Immunol. 1979;123:1245–52. [PubMed] [Google Scholar]
- 26.Peterson PK, Schmeling D, Cleary PP, Wilkinson BJ, Kim Y, Quie PG. Inhibition of alternative complement pathway opsonization by group A streptococcal M protein. J Infect Dis. 1979;139:575–85. doi: 10.1093/infdis/139.5.575. [DOI] [PubMed] [Google Scholar]