Abstract
Immunoglobulin A (IgA) is the major antibody class present in external secretions and is also an important component of serum immunoglobulins. On mucosal surfaces, IgA represents a first line of defence by neutralizing invading pathogens. The number of IgA constant-region genes (Cα) present in different mammalian species is variable. Immunoglobulin Cα genes differ mainly in the sequences located in the hinge region. IgA molecules, whose hinge regions are remarkably similar to those of the respective human molecules, are present in hominoid primates. In this report, we show that two alleles of a single immunoglobulin Cα are present in rhesus macaques (Macaca mulatta). In addition, we show that intraspecies immunoglobulin Cα allelic polymorphism is very high in this non-human primate species. Specifically, five different hinge regions, some of which are proline-rich, were identified from a total of eight rhesus macaque immunoglobulin Cα-chains. The five hinge regions were different from those present in hominoid primates, both in length and in sequence. These results represent the first example of high levels of intraspecies immunoglobulin constant-region variability and suggest that IgAs of variable structure and function may be present in rhesus macaques. As rhesus macaques are widely used as animal models for the development of vaccines for acquired immune deficiency syndrome (AIDS), the possible presence of structurally and functionally variable IgA molecules in different animals should be taken into account when designing experimental strategies to induce mucosal antibody responses to human immunodeficiency virus (HIV).
Introduction
Immunoglobulin A (IgA) is the major antibody class present in external secretions and is also an important component of serum immunoglobulins.1 The secretory forms of IgA include a J-chain and a secretory component, and are therefore polymeric, whereas serum IgA is present predominantly in monomeric form in primates.2 The rate of synthesis of IgA is extremely high. Indeed, the total daily production of IgA outpaces that of all other immunoglobulin isotypes combined. On mucosal surfaces, IgA prevents adsorption of bacterial pathogens, thus representing a first line of defence.3 Although IgA responses are usually T-cell dependent, IgA directed against cell-wall antigens and proteins of commensal bacteria is produced independently of T cells or germinal centre formation.4
The IgA isotype was generated during evolution before segregation of birds and mammals5 and has been found in virtually all mammalian species that have been examined.6 However, the number of IgA subclasses present in different mammalian species is variable. There is only one IgA subclass in mice and swine.7,8 Conversely, 13 IgA isotypes have been identified in rabbits.9 Two IgA subclasses – IgA1 and IgA2 – are present in humans.10 IgA1 molecules include an elongated hinge region rich in proline residues. The IgA2 hinge region differs from the IgA1 hinge region over a 13-residue stretch of amino acids, presumably as the result of an evolutionary response by bacterial IgA1-specific proteases.1 Indeed, the hinge region of the IgA molecules is characterized by evolutionary instability.10 It is subject to rapid evolutionary changes in length that may result from adaptations to conflicting functional demands.10,11 Two allelic IgA2 variants, A2m(1) and A2m(2), have been identified.10 A third allelic variant seems to be the product of recombination or gene conversion between two IgA2 alleles.12 IgA subclasses whose hinge regions are remarkably similar to those of the respective human molecules are present in chimpanzees (Pan troglodytes),13 gorillas (Gorilla gorilla)14 and gibbons (Hylobates lar).15 The orang-utan (Pongo pygmaeus) possesses only one IgA subclass, which resembles the human IgA1.15 A single IgA isotype has been identified in cynomolgus monkeys (Macaca fascicularis).15 In this report, we show that two alleles of a single immunoglobulin-α constant-region gene (Cα) are present in rhesus macaques (Macaca mulatta). In addition, we show that intraspecies immunoglobulin Cα allelic polymorphism is very high in rhesus macaques.
Materials and methods
Genomic DNA extraction
Peripheral blood mononuclear cells (PBMCs) were purified from heparinized blood by Ficoll–Hypaque gradient centrifugation. DNA was extracted from the pelleted PBMCs using the QIAamp Blood Kit (Qiagen, Valencia, CA).
Primers for polymerase chain reaction amplification and sequencing
The nucleotide sequences of the polymerase chain reaction (PCR) primers and internal sequencing primers that were used to amplify the human and rhesus macaque Cα genes, as well as their relative positions, are shown in Table 1. Primer sequences were derived from regions conserved among the Cα genes of humans, chimpanzees and gorillas (GenBank acc. nos: J00220, M60192, X53702, X53706, X15045 and X53707). The primer IgA5F is located at the intron–exon junction of CH1; the primer IgA4R is located at the 3′ end of Cα3. The primers IgA1F, IgA2R and IgA6R are located in Cα2. The primer IgA3F is located in Cα3. To amplify the Cα gene, ≈ 1 µg of genomic DNA was used. The amplification reaction was run in a total volume of 100 µl using the HotWaxOptiStart Kit (Invitrogen, Carlsbad, CA) and the high-fidelity expanded polymerase (Roche Molecular Biochemicals, Indianapolis, IN). The reaction contained a final concentration of 60 mm Tris-HCl, pH 9·0, 15 mm (NH4)2SO4, 2·5 mm MgCl2 and 50 pmol each of the primers IgA5F and IgA4R. After an initial denaturation step at 94° for 4 min, the reaction was run for 35 cycles with each cycle consisting of 1 min at 94°, 1 min at 58° and 2 min at 72°. A final incubation at 72° for 10 min was employed.
Table 1.
Oligonucleotide primers used for the polymerase chain reaction (PCR) and sequencing
| Name | Sequence | Nt loc |
|---|---|---|
| IgA1F | TCGAGGACCTGCTCTTAGGTTCAG | 755–778 |
| IgA2R | TTTTGAGAGGGTGGCGGTTAG | 997–1017 |
| IgA3F | CGAGAAGTACCTGACTTGGGCATC | 1395–1418 |
| IgA4R | CAACAGACACATTGACATGGGTG | 1584–1606 |
| IgA5F | CGCATTCTGTGTTCCAGCATCC | 127–146 |
| IgA6R | CTTCTGAACCTAAGAGCAGGTCCTC | 757–781 |
The nucleotide (Nt) locations of the primers are based on the corresponding site in the human immunoglobulin Cα1 GenBank acc. no.: J00220. Nucleotides of exon I in the IgA5F primer are in bold.
Molecular cloning, DNA sequencing and computer-assisted analysis
For cloning, 100 µl of a PCR reaction was run on a 1% agarose gel. The specific band of interest was excised from the gel, purified (Qiagen) and ligated into either the pCRII or the TopoTA vector (Invitrogen). After transformation into the appropriate strain of Escherichia coli, the plasmid DNA from at least 10 colonies in each sample was amplified. Plasmid DNA was screened on a 1% agarose gel after digestion with EcoRI, PstI and BamHI to confirm the correct size of the DNA fragments and to identify different polymorphic patterns. All DNA sequences were determined using the ABI Prism DNA sequencing kit dye-terminator cycle sequencing ready reaction (Perkin Elmer, Branchburg, NJ) on a ABI model 373 automated sequencer. The universal and reverse M13 primers, as well as the IgA1, IgA2, IgA3, IgA4, IgA5 and IgA6 primers, were used for sequencing. Plasmid DNA from at least three clones in each sample was sequenced completely; plasmid DNA from the remaining clones was sequenced with the IgA6 primers to confirm the sequences of the hinge region.
Analysis of DNA sequences
Overlapping regions were identified and sequences were edited by using the MacVector software program (Accelrys, Burlington, MA). Sequences were aligned with the known Cα-chains by using the Clustal W program, part of the MacVector software package.
Genomic Southern blot analysis
PBMC genomic DNA (10 µg) from the four rhesus macaques was digested with EcoRI. In addition, 10 µg of PBMC genomic DNA from two (RhB and RhC) of the four macaques was digested with StuI. The digested DNA was electrophoresed through a 0·7% agarose gel and then blotted onto a positively charged nylon membrane (Roche Molecular Biochemicals), according to the manufacturer's instructions. Membranes were incubated overnight at 65° with a 32P-labelled full-length genomic human Cα-chain or with a 32P-labelled full-length genomic rhesus-specific Cα-chain. Filters were washed in 0·1 × saline sodium citrate (SSC) and 0·1% sodium dodecyl sulphate (SDS) at 65° and subsequently exposed to Kodal XAR-5 film.
Results
Cloning and sequence analysis: two immunoglobulin Cα genes are present in each rhesus macaque
Rhesus macaque PBMCs were selected as the tissue source for identification of the Cα genes. The PCR was used to amplify and clone rhesus macaque Cα genes of almost full length. The amino acid sequences of IgA from humans, mice and rabbits exhibit ≈ 50% homology. As IgAs from humans and hominoid primates exhibit higher homology, we designed several primer pairs based upon the conserved sequences shared by the human, gorilla and chimpanzee Cα genes. The primer IgA5 is located in the intron–exon junction of the Cα1 domain, whereas the primer IgA4 is located at the 3′ end of the Cα3 exon. Therefore, amplification with these primers yielded germline Cα genes of almost full length (the last 10 amino acids of the Cα3 exon were not present in the final product). The primers IgA1 and IgA6 are located downstream the hinge region in the Cα2 exon. The Ig2 primer is located in the Cα2 exon. The IgA3 and the IgA4 primers are located in the Cα3 exon. All primers were designed to allow amplification of both human IgA subclasses (IgA1 and IgA2).
First, we determined whether or not amplification with the primers described above would lead to successful cloning of the human Cα genes. Therefore, the IgA5 and IgA4 primer pair was used to amplify human PBMC DNA. A fragment of ≈ 1480 bp was amplified and cloned into the pCRII vector, which was then transformed into E. coli. Ten E. coli colonies containing the recombinant pCRII vector were selected for plasmid DNA amplification. Plasmid DNAs were analysed by restriction analyses using the EcoRI, BamHI and PstI restriction enzymes. An EcoRI restriction enzyme site is known to be present in the CH1 exon of the Cα2 A2 m(2) and Cα2A2 (n).12 This restriction site is not present in the other IgA2 allotype m(1) nor in IgA1. A conserved PstI restriction enzyme site is present in the Cα3 domain of all human IgA subtypes and a unique PstI site is located at the 5′ end of the CH1 of IgA1. By restriction analysis, we were able to exclude the presence of the allotypes Cα2 A2 m(2) and Cα2A2(n). Two clones, one for the IgA1 gene and one for the IgA2 gene, were fully sequenced, allowing identification of the IgA1 gene as well as the IgA2m(1) gene (data not shown).
Having proved the validity of our strategy for the cloning and identification of the human Cα genes, we used the IgA5 and IgA4 primer pair to amplify PBMC DNA from four rhesus macaques, designated RhA, RhB, RhC and RhD. A fragment of ≈ 1500 bp was amplified from each monkey sample and separated by gel electrophoresis. The fragments were gel excised and cloned into either the pCRII or the TopoTA vectors. The vectors were transformed into E. coli and 10 colonies were selected from each sample for plasmid DNA isolation. By restriction analysis of the plasmid DNA with EcoRI, BamHI and PstI, two patterns were identified (data not shown) in all four samples. The corresponding plasmid DNAs were sequenced. The nucleotide sequences of the two genes for each rhesus macaque are shown in Fig. 1. The intron–exon organization of the genes was inferred from the published sequence of the Cα H-chains. The necessary splicing signals were present at the intron boundaries. The appropriate intronic sequences were also present. Each Cα gene had the typical immunoglobulin gene structure in which the protein domains – Cα1, Cα2, and Cα3 – are encoded by separate exons. The hinge region was identified by comparison of the gene sequence with that of humans and hominoid primates. Interestingly, our results showed that two different immunoglobulin Cα genes were present in each individual rhesus macaque. The two genes from a single animal have been designated I and II. Therefore, the various immunoglobulin Cα sequences (of a total of eight) were designated RhA.I and II, RhB.I and II, RhC.I and II, and RhD.I and II. Several clones from at least two different PCR products were sequenced for each individual gene.
Figure 1.
Nucleotide sequences of rhesus macaque immunoglobulin Cα genes (GenBank accession numbers AY039245-AY039252).
Comparison of the nucleotide and derived amino acid sequences: heterogeneity of immunoglobulin Cα genes
IgA molecules consist of three constant domains encoded by three exons – Cα1, Cα2 and Cα3 – with a hinge region encoded at the start of the Cα2 exon and located between the Cα1 and Cα2 domains. The human IgA1 differs from the IgA2 at 22 of 365 amino acids in the C region, with the majority of substitutions located in the Cα1 domain.10 In addition, a 13-amino acid sequence of the IgA1 hinge region is absent in IgA2. The amino acid differences existing in the rhesus macaque Cα regions are shown in Table 2. Comparison of the RhA.I and RhA.II sequences identified three amino acid differences in the Cα1 domain, five amino acid differences in the Cα2 domain outside the hinge region and only one amino acid difference in the Cα3 domain. The RhB.I and RhB.II sequences exhibited three amino acid differences in the Cα1 domain, nine amino acid differences in the Cα2 domain outside the hinge region and no amino acid differences in the Cα3 domain. The RhC.I and RhC.II sequences differed for six amino acids in the Cα1 domain and four amino acids in the Cα2 domain outside the hinge region. There were no amino acid differences in the Cα3 domain. The RhD.I and RhD.II sequences differed for four amino acids in the Cα1 domain, eight amino acids in the Cα2 domain outside the hinge region and only one amino acid in the Cα3 domain. Therefore, in general, the majority of amino acid differences existing between the two Cα-chains from the same rhesus macaque are located in the Cα2 domain and not in the Cα1 domain, as observed in humans.10 Together, these results show that two different Cα genes are present in each animal and that a high level of Cα gene intraspecies heterogeneity can be recognized within rhesus macaques.
Table 2.
Amino acid differences in rhesus macaque immunoglobulin Cα regions
| Residue | HuA1 | A.I | A.II | B.I | B.II | C.I | C.II | D.I | D.II | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cα1 domain | 124 | Ser | Lys* | Arg | Arg | Arg | Arg | Arg | Arg | Arg | |
| 131 | Ser | Ser | Ser | Asn | Ser | Asn | Ser | Ser | Ser | ||
| 133 | Cys | Glu | Glu | Cys | Cys | Cys | Glu | Glu | Glu | ||
| 134 | Ser | Gly | Gly | Ser | Ser | Ser | Gly | Gly | Gly | ||
| 139 | Gly | — | — | — | — | — | — | — | — | ||
| 143 | Ile | Val | Val | Val | Ile | Val | Val | Val | Val | ||
| 154 | Glu | Lys | Glu | Glu | Glu | Glu | Glu | Glu | Glu | ||
| 167 | Gly | Gly | Gly | Gly | Gly | Asp | Gly | Gly | Gly | ||
| 175 | Ser | Arg | Arg | Ser | Arg | Ser | Ser | Arg | Ser | ||
| 212 | Pro | Pro | Pro | Pro | Pro | Pro | Ala | Pro | Ala | ||
| 213 | Ser | Arg | Arg | Ser | Ser | Ser | Arg | Arg | Arg | ||
| 217 | Thr | Ala | Gly | Ala | Ala | Ala | Ala | Gly | Ala | ||
| 221 | Pro | Arg | Arg | — | — | — | — | Arg | — | ||
| Hinge region | Pro | Pro | Pro | Pro | Pro | Ser | Pro | Ser | |||
| Pro | Pro | Pro | Gln | Pro | Gln | Pro | Gln | ||||
| Pro | Pro | Ile | — | Ile | — | Pro | — | ||||
| Lys | Asn | Thr | Pro | Thr | Thr | Asn | Thr | ||||
| Cys | Cys | Pro | Lys | Pro | Lys | Cys | Lys | ||||
| Pro | Arg | Pro | Pro | Pro | Pro | Arg | Pro | ||||
| Ser | Leu | Cys | Cys | Cys | Cys | Leu | Cys | ||||
| Leu | — | Pro | Leu | Pro | Leu | — | Leu | ||||
| Lys | — | Ser | — | Ser | — | — | — | ||||
| Cα2 domain | 242 | Cys | Asp | Asp | Cys | Asp | Cys | Asp | Asp | Asp | |
| 243 | His | Lys | Lys | Glu | Lys | Glu | Glu | Lys | Glu | ||
| 272 | Arg | Arg | Arg | Lys | Arg | Arg | Arg | Arg | Arg | ||
| 280 | Thr | Thr | Ala | Thr | Thr | Thr | Thr | Ala | Thr | ||
| 295 | Glu | Glu | Glu | Lys | Glu | Lys | Lys | Glu | Lys | ||
| 296 | Arg | His | His | Arg | His | Arg | Arg | His | Arg | ||
| 319 | Lys | Val | Val | Glu | Val | Glu | Val | Val | Val | ||
| 322 | Thr | Thr | Asn | Thr | Thr | Thr | Thr | Asn | Thr | ||
| 324 | Thr | Thr | Thr | Thr | Thr | Thr | Lys | Thr | Lys | ||
| 326 | Ala | Glu | Asn | Asn | Glu | Asn | Glu | Asn | Glu | ||
| 329 | Glu | Glu | Glu | Glu | Gln | Glu | Glu | Glu | Glu | ||
| 331 | Lys | Glu | Lys | Glu | Lys | Glu | Glu | Lys | Glu | ||
| 333 | Pro | Gln | Pro | Pro | Pro | Pro | Pro | Pro | Pro | ||
| Cα3 domain | 390 | Leu | Leu | Val | Leu | Leu | Leu | Leu | Val | Leu |
Amino acid differences existing between the two Cα genes from the same rhesus macaque are indicated in bold.
Comparison of the nucleotide and derived amino acid sequences: heterogeneity of the immunoglobulin Cα hinge region
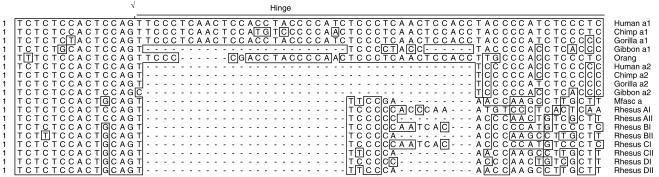
In humans, as well as in hominoid primates, the Cα1 gene is characterized by a hinge-coding region consisting of two 30-bp repeats with a 6-bp overlap.10 The 30-bp repeats, in turn, consist of two tandem 15-bp units. The Cα2 hinge region includes only one 15-bp unit. Because of its involvement in recombinatorial events, a reiterated structure is thought to facilitate rapid evolutionary changes in the length of the hinge region.15 Results from our study show that a hinge structure consisting of repeated units is present in the rhesus macaque Cα genes. However, the number of bases included in each unit are different from those present in humans and hominoid primates. Figure 2 shows a comparison of the Cα hinge-coding region of various primate species, including rhesus macaques (as assessed by analysis of the results obtained from this study).
Figure 2.
Aligned nucleotide sequences of the Cα hinge region from humans, chimpanzees, gorillas, orang-utans, Macaca fascicularis (Mfasc) and rhesus macaques (including the 5′ intron sequences).
The hinge regions from the RhA.I, RhB.I and RhC.I genes consist of 27 bp represented by two repeats of a 15-bp unit with a deletion of three nucleotides in the second tandem. Conversely, the hinge region of the RhD.I gene consists of 21 bp represented by a 15-bp unit and a second unit of 6 bp. Therefore, the RhA.I, RhB.I and RhC.I hinge regions comprise nine amino acids, whereas the RhD.I hinge region comprises seven amino acids. The hinge regions of the RhA.II, RhB.II, RhC.II and RhD.II genes consist of 21 bp with a repeat of 15 bp that shares identity with 14 nucleotides of the first unit of the RhA.I, RhB.I, RhC.I and RhD.I genes, respectively, as well as a second tandem of 6 bp. Consequently, the hinge regions of the RhA.II, RhB.II, RhC.II and RhD.II genes comprise seven amino acids. Although the hinge region of the two Cα genes from the rhesus monkey RhD exhibits the same amino acid length, the sequences are very different, with only one (RhD.I) rich in proline residues. Indeed, Table 2 and Fig. 3 show that a total of five different hinge regions can be identified in the four rhesus macaques included in this study. The RhA.II hinge region is identical to the corresponding RhD.I region, the RhB.I to the RhC.I region, and the RhB.II to the RhC.II region. Therefore, the two Cα hinge regions present in rhesus macaque RhB are also present in rhesus macaque RhC, and one hinge region present in RhA is also present in RhD. However, the rhesus macaque RhA exhibits a unique hinge that is not shared by any other macaque included in our study. A hinge not shared by any other macaque included in our study is also present in RhD.
Figure 3.
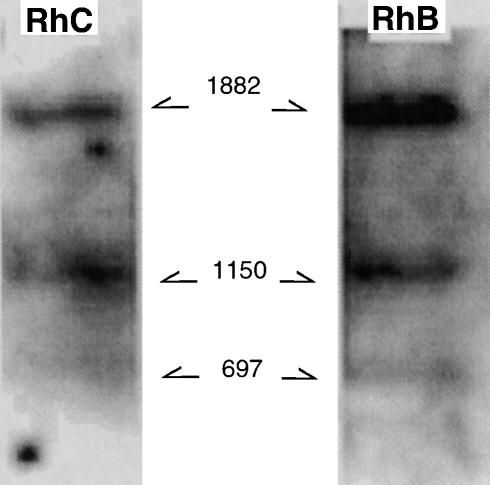
Southern blot analysis of rhesus macaque genomic DNA. DNA purified from peripheral blood mononuclear cells (PBMC) was digested with StuI and hybridized with a rhesus macaque immunoglobulin A (IgA) Cα genomic DNA probe.
Southern blot analysis: two alleles of a single Cα gene are present in each rhesus macaque
Southern blot analysis of the genomic PBMC DNA cleaved with EcoRI yielded only a strong hybridizing band of > 23 000 bp when probed with the human Cα1 gene (data not shown). Therefore, we performed additional Southern blot analyses. A StuI restriction enzyme site (at nucleotide position 649) was present in one of the two amplified Cα-chains (BI and CI) from two macaques, whereas no StuI restriction site was present in the BII and CII clones. As shown in Fig. 3, Southern blot analysis carried out using BI probes yielded three bands. Specifically, the following bands were visualized: a strong band of ≈ 1882 bp corresponding to the full-length Cα gene (1432 bp) and two additional bands derived from the Cα gene containing the StuI restriction site (≈1150 bp for the 3′ end of the StuI-digested clone and ≈ 697 bp for the 5′ end of the StuI-digested clone). The combined size of the two latter bands corresponded approximately to the size of the larger band. Because the outer StuI restriction sites are probably present at the same nucleotide position in the genomic DNA outside amplified and cloned Cα-chain genes, it was reasonable to conclude that an IgA gene existing in two allelic forms is present in each animal.
Discussion
By Southern blot analysis and sequencing of the immunoglobulin Cα regions from four animals, we have shown that two alleles of a single Cα gene can be identified in each rhesus macaque. In addition, we have shown that a highly heterogenous intraspecies population of Cα genes exists in this non-human primate species. Indeed, a total of six different Cα genes were identified within the four rhesus macaques that we examined. Two of these genes (RhB.I and RhC.I) differed for only two amino acid substitutions, at positions 167 (Gly → Asp) and 272 (Lys → Arg), respectively (Table 2). Considering that so many sequences have been found in a total of four animals, it is logical to conclude that the level of rhesus macaque immunoglobulin Cα-gene heterogeneity is apparently higher than the one currently assumed to be present in humans, where two IgA subclasses with a total of three allotypes have been identified to date.1,12 However, it is thought that additional human immunoglobulin Cα-gene diversity might be found as a greater number of individual genomes are characterized.16
Five different hinge regions consisting of separated units were identified within the eight rhesus macaque immunoglobulin Cα-chains. The number of bases included in each unit was different from that present in humans and hominoid primates, thus confirming the evolutionary instability of the IgA hinge region.14 Both human IgA1 and IgA2 hinge regions are proline-rich.1 Proline residues are present in all macaque hinge regions, which contain a minimum of one and a maximum of five proline residues. Such heterogeneity in amino acid sequences suggests that IgAs of variable structure and function may be present in rhesus macaques.
These findings are particularly interesting, especially in view of the important role played by the IgA hinge region. Not only is the IgA hinge responsible for providing a flexible region that facilitates effective antigen binding, but it also provides a structure that may confer protection from IgA-specific proteases. Indeed, it is well established that the human IgA hinge region is the target of bacterial IgA proteases produced by pathogens of the genera Neisseria, Haemophilus and Streptococcus.17 The majority of IgA proteases cleave only IgA1 molecules because of the presence of a susceptible site. Such a site is represented by either a prolyl-seryl (for proteases type I) or prolyl-threonyl (for proteases type II) peptide bond in a heavily glycosylated duplicated sequence (Pro-Ser-Thr-Pro-Pro-Thr-Pro-Ser). This sequence is present in the hinge region of IgA1 molecules but absent in the hinge region of IgA2 molecules.17,18 The Clostridium ramosum protease represents an exception. It cleaves not only human IgA1 molecules, but also the IgA2m(1) allotype, at a prolyl-valyl peptide bond located outside the hinge region.19 Additional substrates for bacterial proteases have been identified in the Cα1 hinge region of gorillas and chimpanzees, as well as in the orang-utan Cα-chain.20 The information currently available on the susceptibility of rhesus macaque IgA molecules to bacterial proteases is contradictory. According to the results of a study performed using 20 primate species, including rhesus macaques, IgA substrates for bacterial proteases are present only in humans, chimpanzees and gorillas.21 However, results from a subsequent study22 carried out using IgA proteases (protease type 2) produced by a human isolate (ATCC 27336), as well as animal isolates of S. pneumoniae, indicate that IgA molecules from a rhesus monkey were susceptible to cleavage. A latter study performed using the IgA protease from the same S. pneumoniae human isolate (ATCC 27336) failed to demonstrate cleavage of two IgA preparations obtained from individual rhesus monkeys.23 These contradictory results might be explained taking into account our findings related to the heterogeneity of IgA molecules present in the rhesus macaque population. This heterogeneity is of particular interest, as no other examples of such extensive intraspecies immunoglobulin constant-region variability have been reported. It is intriguing to speculate that the existence of multiple Cα genes results in the expression of distinct IgA molecular forms that differentially participate in the protective mechanisms operating within the mucosal immune system of rhesus macaques. The characterization of the physiological properties of these molecular forms may lead to the recognition of additional protective IgA mechanisms. The practical importance of our findings is related to the major role that rhesus macaques play as animal models for acquired immune deficiency syndrome (AIDS) vaccine development.24 The ability to induce mucosal immune responses is considered to be one of the most desirable features of many human immunodeficiency virus (HIV) vaccine candidates. Clearly, when designing AIDS vaccine strategies to be tested on the rhesus macaque animal model, it will be necessary to take into account the individual differences in IgA structure that may potentially result in distinct levels of protection.
Acknowledgments
The authors thank Dr Harold McClure for providing rhesus macaque blood samples. This work was supported by NIH Grant RR10755.
References
- 1.Kerr MA. The structure and function of human IgA. Biochem J. 1990;271:285. doi: 10.1042/bj2710285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mestecky J, McGhee JR. Immunoglobulin A (IgA): molecular and cellular interactions involved in IgA biosynthesis and immune response. Adv Immunol. 1987;40:153. doi: 10.1016/s0065-2776(08)60240-0. [DOI] [PubMed] [Google Scholar]
- 3.Childers NK, Bruce MG, McGhee JR. Molecular mechanisms of immunoglobulin A defense. Annu Rev Microbiol. 1989;43:503. doi: 10.1146/annurev.mi.43.100189.002443. [DOI] [PubMed] [Google Scholar]
- 4.Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222. doi: 10.1126/science.288.5474.2222. 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 5.Nezlin R. Structure and Function. New York: Academic Press; 1998. The Immunoglobulins. [Google Scholar]
- 6.Peppard JV, Russell MW. Phylogenetic development and comparative physiology of IgA. In: Ogra PL, Mestecky J, Lamm ME, Strober W, Bienenstock J, McGhee JR, editors. Mucosal Immunology. New York: Academic Press; 1999. pp. 63–79. [Google Scholar]
- 7.Shimizu A, Takahashi N, Yaoita Y, Honjo T. Organization of the constant region gene family of the mouse immunoglobulin heavy chain. Cell. 1982;28:499. doi: 10.1016/0092-8674(82)90204-5. [DOI] [PubMed] [Google Scholar]
- 8.Brown WR, Butler JE. Characterization of a Cα gene of swine. Mol Immunol. 1994;31:633. doi: 10.1016/0161-5890(94)90171-6. [DOI] [PubMed] [Google Scholar]
- 9.Burnett RC, Hanly WC, Zhai SK, Knight KL. The IgA heavy-chain gene family in rabbit: cloning and sequence analysis of 13 Cα genes. EMBO J. 1989;8:4042. doi: 10.1002/j.1460-2075.1989.tb08587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flanagan JG, Lefranc M-P, Rabbitts TH. Mechanisms of divergence and convergence of the human immunoglobulin α1 and α2-constant region gene sequences. Cell. 1984;36:681. doi: 10.1016/0092-8674(84)90348-9. [DOI] [PubMed] [Google Scholar]
- 11.Tucker PW, Slightom JL, Blattner FR. Mouse IgA heavy chain gene sequence: implication for evolution of immunoglobulin hinge exons. Proc Natl Acad Sci USA. 1981;78:7684. doi: 10.1073/pnas.78.12.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chintalacharuvu KR, Raines M, Morrison SL. Divergence of human α-chain constant region gene sequences: a novel recombinant α2 gene. J Immunol. 1994;152:5299. [PubMed] [Google Scholar]
- 13.Cole MF, Hale CA, Sturzenegger S. Identification of two subclasses of IgA in the chimpanzee (Pan troglodytes) J Med Primatol. 1992;21:275. [PubMed] [Google Scholar]
- 14.Ueda S, Matsuda F, Honjo T. Multiple recombinatorial events in primate immunoglobulin epsilon and alpha genes suggest a closer relationship of human to chimpanzee than to gorilla. J Mol Evol. 1988;27:77. doi: 10.1007/BF02099732. [DOI] [PubMed] [Google Scholar]
- 15.Kawamura S, Omoto K, Ueda S. Evolutionary hypervariability in the hinge region of the immunoglobulin alpha gene. J Mol Biol. 1990;215:201. doi: 10.1016/S0022-2836(05)80336-5. [DOI] [PubMed] [Google Scholar]
- 16.Knight KL, Winstead CR. Organization and expression of genes encoding IgA heavy chain, polymeric immunoglobulin receptor, and J chain. In: Ogra PL, Mestecky J, Lamm ME, Strober W, Bienenstock J, McGhee JR, editors. Mucosal Immunology. New York: Academic Press; 1999. pp. 153–62. [Google Scholar]
- 17.Kilian M, Mestecky J, Russell M. Defense mechanisms involving Fc-dependent functions of immunoglobulin A and their subversion by bacterial immunoglobulin A proteases. Microbiol Rev. 1988;52:296. doi: 10.1128/mr.52.2.296-303.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plaut AG. The IgA1 proteases of pathogenic bacteria. Annu Rev Microbiol. 1983;37:603. doi: 10.1146/annurev.mi.37.100183.003131. [DOI] [PubMed] [Google Scholar]
- 19.Fujiyama Y, Kobayashi K, Senda S, Benno Y, Bamba T, Hosoda S. A novel IgA protease from Clostridium sp. capable of cleaving IgA1 and IgA2 : a2m(1) allotype, but not IgA2 : a2m(2) allotype paraproteins. J Immunol. 1985;134:573. [PubMed] [Google Scholar]
- 20.Qiu J, Brackee GP, Plaut AG. Analysis of the specificity of bacterial immunoglobulin A (IgA) proteases by a comparative study of ape serum IgAs as substrates. Infect Immun. 1966;64:933. doi: 10.1128/iai.64.3.933-937.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornefeld SJ, Plaut AG. Secretory immunity and the bacterial IgA proteases. Rev Infect Dis. 1981;3:521. doi: 10.1093/clinids/3.3.521. [DOI] [PubMed] [Google Scholar]
- 22.Proctor M, Manning PJ. Production of immunoglobulin A protease by Streptococcus pneumoniae from animals. Infect Immun. 1990;58:2733. doi: 10.1128/iai.58.9.2733-2737.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reinholdt J, Kilian M. Lack of cleavage of immunoglobulin A (IgA) from rhesus monkeys by bacterial IgA1 proteases. Infect Immun. 1991;59:2219. doi: 10.1128/iai.59.6.2219-2221.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nathanson N, Mathieson BJ. Towards an AIDS vaccine: the role of nonhuman primates. J Med Primatol. 1999;28:146. doi: 10.1111/j.1600-0684.1999.tb00263.x. [DOI] [PubMed] [Google Scholar]