Abstract
We previously reported that expression of the T-cell receptor (TCR) α and lck genes is extinguished in hybrids between mouse T-lymphoma EL4 cells and mouse fibroblast B82 cells. In the present study, we found that the activities of the TCRα minimum enhancer and the lck promoter monitored by the luciferase or chloramphenicol acetyltransferase (CAT) assays were markedly inhibited in the hybrids. Expression of the TCF-1, LEF-1, GATA-3, Ikaros, c-myb and Fli-1 genes, which encode the haematopoietic cell-restricted transcription factors that appear to be responsible for the activities of the enhancer and the promoter, was fully extinguished or markedly suppressed in the hybrids. On the other hand, expression of the transcription factor genes observed in both parental cells, such as the AML1 and c-ets-1 genes, and that of the genes encoding ubiquitously expressed transcription factors, such as the E2A, CREB and c-ets-2 genes, was not significantly suppressed in the hybrids. These results suggest that the genes encoding haematopoietic cell-restricted transcription factors are targets for negative regulation in fibroblastic background and that the repression of these genes may consequently lead to suppression of the promoter and/or enhancer activities of several T-cell-specific structural genes in T-lymphoma × fibroblast cell hybrids.
Introduction
Cell differentiation is the process in which cells acquire defining phenotypes as a result of a co-ordinated programme of cell type-specific gene expression. It is tightly linked to hierarchical networks of activation of key transcriptional regulators.1,2 Determination and maintenance of specific cell types are believed to be mediated through a combination of expression of appropriate genes and repression of inappropriate genes in cells.
Somatic cell hybridization may be a useful approach to understanding the molecular mechanisms of the repression of inappropriate genes because differentiated properties in expressing cells are generally extinguished by cell fusion with non-expressing cells, referred to as hybrid cell extinction.3 Well-known examples of extinction are the shut-off of immunoglobulin gene expression in myeloma × fibroblast cell hybrids4 and of liver-specific gene expression in hepatoma × fibroblast cell hybrids.3,5 In the myeloma × fibroblast cell hybrids, extinction of immunoglobulin gene expression is accompanied by repression of the oct-2 gene encoding a B-cell-specific transcription factor responsible for the expression of the immunoglobulin genes.4,6 In the hepatoma × fibroblast cell hybrids, acquisition of repressor molecules derived from tissue-specific extinguisher-1 (Tse-1) loci in the fibroblasts is involved in the extinction of liver-specific gene expression,5,7 as well as in the repression of the HNF-1 and HNF-4 genes encoding liver cell-restricted transcription factors responsible for the expression of the genes.8,9 In T-cell-specific gene expression, however, the molecular basis of extinction is not yet clearly understood. Furthermore, it is still not evident whether extinction of expression of the genes encoding tissue-specific transcription factors is a general phenomenon in cell hybrid extinction.
In our previous studies, we reported that expression of the T-cell receptor (TCR) α-chain gene and the lck proto-oncogene was extinguished or markedly suppressed in T-lymphoma × fibroblast cell hybrids despite the existence of the genes.10,11 T-cell-specific expression of the TCR and lck genes is believed to be controlled by a combination of ubiquitously expressed transcription factors with several groups of haematopoietic cell-restricted transcription factors bound to the enhancers and promoters.12,13 In the present study, we examined whether several haematopoietic cell-restricted transcription factor genes that appear to be critical for T-cell-specific gene expression are targets for transcriptional repression.
Materials and methods
Cell culture
Five hybrid clones previously isolated by two independent cell fusions between hypoxanthine guanine phosphoribosyl transferase (HGPRT)-deficient mouse T-lymphoma EL4 cells and thymidine kinase (TK)-deficient mouse fibroblast B82 cells were used.10 The hybrid cells exhibited typical fibroblastic morphology. They were cultured for the shortest possible period of time to minimize chromosome segregation.11 Chromosome preparations and isolation of DNA and RNA were performed in the same passage generations.
Chloramphenicol acetyltransferase (CAT) and luciferase assay
The TCRα-TK-luciferase and TK100-luciferase constructs14 were kindly donated by Dr K. A. Jones, the Salk Institute for Biological Studies, La Jolla, CA. To generate the lck-CAT construct, a 3·5-kilobase (kb) AccI–BamHI fragment of the lck distal promoter kindly presented by Dr R. M. Perlmutter, Howard Hughes Medical Institute Research Laboratories, University of Washington, WA, was subcloned into pSV00CAT. Five micrograms of the CAT or luciferase reporter plasmids was transiently transfected into the parental and hybrid cells with 3 µg of the plasmid containing the bacterial β-galactosidase gene driven by the cytomegalovirus promoter by using the diethylaminoethyl–dextran method.15 Cells were harvested 36 hr after transfection for protein extraction. CAT and luciferase activities were normalized for transfection efficiency by the β-galactosidase assay. For CAT assay, cellular lysates were incubated with [14C]chloramphenicol and acetyl-coenzyme A for 12 hr at 37°. The CAT activity of the reporter constructs was assayed by measuring the amount of acetylated [14C]chloramphenicol by thin-layer chromatography. For luciferase assay, luciferase reagent buffer (Boehringer Mannheim, Germany) was mixed with cellular lysates and the activities were measured using a Lumat LB9507 luminometer (Berthold, Bad Wildbad, Germany).
Northern and Southern blot analysis
Twenty micrograms of total RNA samples was fractionated through 2·2 m formaldehyde–1% agarose gel. Ten-microgram samples of DNA were digested with 50 units of restriction enzymes, separated in 0·7% agarose gel and denatured with 0·2 m NaOH in 0·6 m NaCl. RNA and DNA were transferred onto a nylon membrane (Biodyne A, Pall Co, Port Washington, NY) The membranes were then hybridized with 32P-labelled probes in hybridization solution, washed in 0·5 × saline sodium citrate–0·1% sodium dodecyl sulphate at 65° and were then exposed to X-ray films according to our routine method.10
Polymerase chain reaction (PCR) analysis
Mouse chromosome-specific polymorphic minisatelite DNA fragments were amplified by PCR from genomic DNA. The primers for relevant chromosomes, D2Mit43 (chromosome #2), D3Mit73 (chromosome #3), D5Mit 138 (chromosome #5) and D10Mit10 (chromosome #10) (Murine MapPairs™) were purchased from Research Genetics, Inc. (Huntsville, AL). PCR was performed using 30 cycles, each consisting of 94° for 1 min, 55° for 2 min and 72° for 2 min. The amplified products were separated through 6 or 8% polyacrylamide gel, stained with ethidium bromide, and then photographed under ultraviolet illumination.
Results
Activities of the TCRα enhancer and lck promoter are suppressed in intraspecific hybrids between murine T-lymphoma cells and fibroblasts
We previously reported that expression of the TCRα gene and that of the lck proto-oncogene is extinguished or markedly suppressed in intraspecific somatic cell hybrid clones between mouse T-lymphoma EL4 cells and mouse fibroblast B82 cells, designated BEL hybrid clones, which maintained most chromosomes from both parental cells.10,11
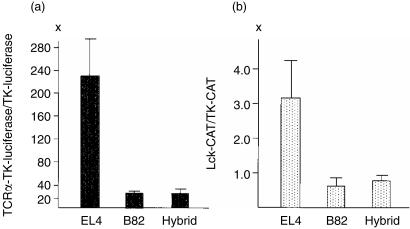
In the present study, to analyse the molecular basis of extinction of gene expression, we first examined the activities of the minimal TCRα enhancer and the lck promoter in the parental cells and their hybrids. As shown in Fig. 1(a), five independent luciferase reporter gene assays reproducibly revealed that the TCRα minimum enhancer activity was increased approximately 230-fold in EL4 cells when the relative enhancer activity was calculated on the basis of the activity of the TK promoter used as a standard in the same cells. In contrast, significantly less luciferase activity was observed in B82 cells and BEL hybrid cells; the relative enhancer activities were 27-fold in B82 cells and 28-fold in BEL hybrid cells. These results indicate that the activation of TCRα minimum enhancer was more than eight-fold greater in EL4 cells than in B82 cells or in BEL hybrid cells. Similarly, the activation of the lck distal promoter, the mainly used promoter in EL4 cells,11 was 3–4-fold greater in EL4 cells than in B82 fibroblast or BEL hybrid cells in six independent experiments of CAT assay (Fig. 1b). These results suggest that the TCRα minimum enhancer and the lck promoter are targets for extinction in BEL hybrids and that extinction of TCRα and lck gene expression in the hybrids is partly due to suppression of the enhancer and promoter activities.
Figure 1.
(a) Relative luciferase activities of the TCRα enhancer in parental EL4 T-cell lymphoma and B82 fibroblastic cells and their hybrid. (b) Relative CAT activities of the lck distal promoter in EL4, B82 and their hybrid. Error bars indicate SD for five to six independent experiments.
Expression of several haematopoietic cell-restricted transcription factor genes is extinguished in the hybrids
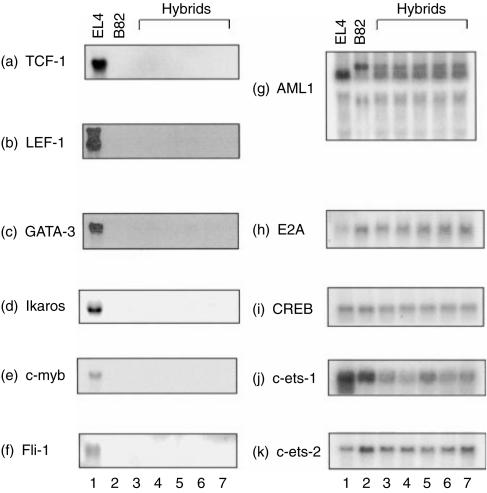
The activities of the TCRα/β enhancers and the lck promoters are controlled by the several haematopoietic cell-restricted transcription factors as well as by ubiquitously expressed ones.12,13 Therefore, we investigated the expression of several haematopoietic transcription factor genes that appear to be responsible for T-cell-specific expression of the genes in the hybrids. The genes examined were the TCF-1,16 LEF-1,17 Sox-4,18 GATA-3,19 Ikaros,20 c-myb,21 c-ets-1,22 c-ets-2,23 Fli-1,24 AML1/PEBP2αB,25 E2A26 and CREB27 genes.
The TCF-1, LEF-1 and Sox-4 genes encode HMG box proteins of the transcription factors in lymphoid cells. The predominant levels of TCF-1 and LEF-1 transcripts were observed in EL4 cells (Fig. 2a,b). No Sox-4 mRNA was observed in our EL4 cells (data not shown), although original EL4 cells are reported to express Sox-4 mRNA to a high level.16 Transcripts of the TCF-1, LEF-1 and Sox-4 genes were not detected in B82 cells. Expression of the TCF-1 and LEF-1 genes was not detected in any of BEL hybrid clones, although they maintained most chromosomes from both parental cells, suggesting that expression of the genes was extinguished in the hybrids.
Figure 2.
Suppression of expression of the haematopoietic cell-restricted transcription factor genes in hybrid clones between EL4 and B82 cells. Expression of the genes were monitored by Northern blot analysis.
Expression of the GATA-3 and Ikaros genes encoding zinc-finger transcription factors was high in EL4 cells and was not detected in B82 cells (Fig. 2c,d). Expression of the GATA-3 gene was markedly suppressed and that of the Ikaros gene was completely extinguished in the hybrids.
The c-myb mRNA was abundantly expressed in EL4 cells but no c-myb mRNA was observed in B82 cells and BEL hybrids (Fig. 2e), suggesting that expression of c-myb is also extinguished in hybrid cells between T lymphoma cells and fibroblasts.
The Fli-1 gene, a member of the ets family of oncogenes, was highly expressed in EL4 cells but also completely extinguished in the hybrids (Fig. 2f).
In contrast, expression of the AML1, E2A, CREB, c-ets-1 and c-ets-2 genes, which was observed in both of the parental cells, was maintained and not remarkably suppressed in the hybrids (Fig. 2g–k), suggesting that expression of the genes encoding transcription factors expressed ubiquitously or expressed in both parental cells is not suppressed at all after cell fusion.
The genes encoding the haematopoietic cell-restricted transcription factors are maintained in the hybrids
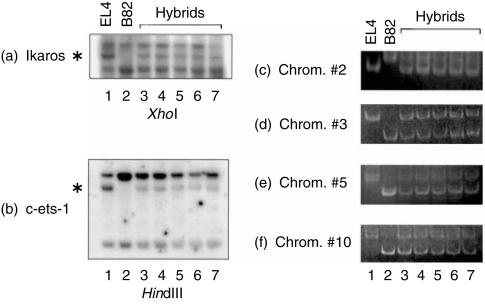
The BEL hybrid clones we used in this study were previously confirmed to maintain almost the expected sum of total chromosome numbers from both parental EL4 and B82 cells.10 It is therefore unlikely that the suppression of the expression of haematopoietic cell-restricted transcription factor genes we examined in this study is merely attributed to segregation of chromosomes for these genes in five independent hybrid clones. XhoI digest showed restriction fragment length polymorphism (RFLP) in the Ikaros gene on mouse chromosome 11 and an EL4-specific fragment was proven to be retained in all BEL hybrids (Fig. 3a), suggesting that EL4-derived Ikaros gene was maintained in the hybrid cells. Although no RFLP was found for the Fli-1 gene, the existence of EL4-derived c-ets-1 gene in the hybrids detected by RFLP with HindIII digest (Fig. 3b) probably supports the concomitant existence of the EL4-derived Fli-1 gene in the cells, since the Fli-1 gene is just adjacent to the c-ets-1 gene on mouse chromosome 9. Since no available RFLP was found for the GATA-3, LEF-1, TCF-1 and c-myb genes in EL4 and B82 cells using several restriction endonucleases, instead, polymorphic minisatellite DNA fragments were detected by PCR analysis using the primers for mouse chromosomes 2, 3, 5 and 10 on which the genes are located, respectively. Amplification of genomic DNA revealed that the hybrid clones maintained the polymorphic fragments from both of the parental cells for the chromosomes (Fig. 3c–f). All these results support the notion that the haematopoietic cell-restricted transcription factor genes we examined were maintained in the hybrids.
Figure 3.
Maintenance of the haematopoietic cell-restricted transcription factor genes in hybrid clones between EL4 and B82 cells. RFLP analyses were carried out for the
Ikaros and
-ets-1 genes. The asterisks indicate EL4-specific fragments. Polymorphic minisatellite DNA fragments were detected by PCR analyses (c–f) using the primers for mouse chromosomes 2, 3, 5 and 10 on which the GATA-3, LEF-1, TCF-1 and c-myb genes are located, respectively.
Discussion
We have shown that the activities of the TCRα minimum enhancer and the lck distal promoter were markedly reduced when mouse T-lymphoma EL4 cells were fused with mouse fibroblast B82 cells. Reduction of their activities appeared to be associated with extinction or marked suppression of expression of several haematopoietic cell-restricted transcription factor genes that appear to be critical for TCRα and lck gene expression.13 Extinction was not specifically observed in a certain family of transcription factor genes but rather appeared to be dependent on their expression patterns in tissues: expression of the genes encoding haematopoietic cell-restricted transcription factors was extinguished by fusing with fibroblast but no suppression was observed in expression of the genes encoding ubiquitously expressed transcription factors in general.
The TCRα minimum enhancer contains the binding motifs for CREB, TCF-1/LEF-1, AML1 and Ets transcription factors which are critical for TCRα gene expression.14 All these transcription factors were available in EL4 cells and the reporter assay showed that the TCRα minimum enhancer was highly activated in the cells, whereas no expression of TCF-1/LEF-1 and Fli-1 was found in BEL cell hybrids where the TCRα minimum enhancer activity was significantly reduced. Expression of GATA-3 and c-Myb, which is also critical for T cell-specific expression of the lck and the TCRβ genes as well as the TCRα gene, was very high in EL4 cells but it was also reduced in the hybrids. Expression of the TCRβ and Thy-1 genes was also extinguished in the hybrids (data not shown).
Our results suggest that extinction of expression of tissue-specific transcription factor genes may be a general role in hybrid cells between haematopoietic cells and fibroblasts. Our results also provided evidence that tissue-specific promoters and/or enhancers are targets in hybrid cell extinction. The haematopoietic cell-restricted transcription factors we examined are known to be co-operatively involved in the control of expression of many T-cell-specific genes.1,12 Since one haematopoietic cell-restricted transcription factor controls the expression of several sets of haematopoietic cell-specific structural genes, it may be efficient to shut off these genes by shutting off a few sets of haematopoietic cell-restricted transcription factors.
There are several mechanisms to shut off inappropriate genes which include DNA methylation,28 histone deacetylation,29 acquisition of direct repressors,30 extinction of necessary transcription factors6 and post-transcriptional regulation.31,32 Our previous study on extinction of the lck gene in the hybrids showed that DNA methylation status around the lck promoter and coding regions was not changed as far as examined.11 The results in the present study suggest that extinction of positive regulators for keeping haematopoietic characters in T cells is at least one of the mechanisms in the shut off of T-cell-specific structural gene expression in non-haematopoietic cells like fibroblasts. Extinction of expression of tissue-specific transcription factor genes may be one of the biologically significant ways for commitment and determination of cell fate and lineage specificity, since only a small fraction of the genome is expressed in each type of differentiated cells. The putative repressors in fibroblast may be a part of such a developmental programme to prevent haematopoietic cell-restricted transcription factor genes from being expressed in non-haematopoietic cells. In this regard, we previously reported that expression of PU.1, an ets family oncogene encoding a B-cell- and macrophage-specific transcription factor, is extinguished in hybrids between myeloma and embryonic carcinoma cells.33 At present, however, we do not know the precise molecular mechanisms of the extinction of the transcription factor genes and whether the extinction mechanism is common for the LEF-1, TCF-1, GATA-3, Ikaros, c-myb and Fli-1 genes. The most recent study on T-cell-specificity of GATA-3 gene expression has revealed that an upstream regulatory region from the GATA-3 transcriptional initiation site acts as a silencer in non-T cells and that competition between the basic helix-loop-helix (bHLH) proteins E2A/HEB and the repressor protein ZEB is involved in the silencer activity.34 The ZEB protein has also been reported to be involved in silencer activity in the immunoglobulin gene in non-B cells.35 Similarly, the Id and Hes-1 HLH repressor proteins have been reported to be implicated in suppression of the function of master positive regulators such as MyoD, NeuroD and SCL, probably through sequestration of E2A/HEB proteins.36,37 Whether the ZEB, Id and Hes-1 proteins are involved in the repression of haematopoietic cell-restricted transcription factor genes examined in this study remains to be determined. Since it is likely that the regulatory regions of the transcriptional factor genes are also targets for extinction, expression of only a very few sets of putative master negative regulators could inhibit the expression of several haematopoietic cell-restricted transcription factor genes and lead subsequently to extinction of many haematopoietic cell-specific structural genes in fibroblasts. A system in which many genes can be shut off by only a small fraction of repressor proteins has been reported in liver cells and in neural cells, where Tse-13–5 and REST/NRSF38 act as direct negative repressors in non-hepatic cells and non-neural cells, respectively, to control the cells' fate.
Our results do not exclude the possibility that direct negative repressors are also involved in the extinction of expression of the haematopoietic-specific genes in a non-haematopoietic background. Reverse transcription-PCR analysis revealed that temporal exogenous expression of TCF-1 and GATA-3 did not recover the endogenous TCRα and lck gene expression in BEL hybrids (data not shown), suggesting that additional extinction mechanisms may exist. This is similar to the results reported by Bulla39 in which ectopic co-expression of the two major liver-specific transcriptional activators of the α1-anti-trypsin gene, HNF1α and HNF4, failed to prevent extinction of liver-specific expression of the gene in hepatoma × fibroblast cell hybrids. On the other hand, forced Oct-2 expression has been shown to prevent extinction of the immunoglobulin gene expression in B × T-cell hybrids.40 Both loss of tissue-specific transcriptional activators and acquisition of direct negative regulators have been proposed to be responsible for extinction of liver-specific gene expression in hepatoma × fibroblast hybrids.41 Thus, multiple mechanisms, including loss of haematopoietic cell-restricted transcription factors and acquisition of direct negative factors, may also contribute to the extinction of T-cell-specific gene expression in cell hybrids.
Somatic cells appear to have much more plasticity than ever thought and the cell fate might be changed when the genes for master positive and negative regulators are introduced or reprogrammed.42–45 For example, introduction of the myoD gene converts fibroblasts into myogenic cells43 and introduction of the v-raf oncogene converts Eµ-myc transgenic B cells into macrophages.45 Recent reports that a variety of blood cell types were produced from neural stem cells after transplantation into irradiated hosts46 may also be such an example. It has also been reported that globin gene expression is reprogrammed in chimeras generated by injecting adult haematopoietic stem cells into mouse blastocysts.47 Thus, cell fate seems to be determined by the balance between expression of master positive and negative regulators. In this sense, it is very important to identify in future studies the genes encoding such putative master negative regulators that could shut off the expression of several haematopoietic cell-specific genes.
Acknowledgments
This work was supported by Grants-in-Aid for Scientific Research (B)(2) from the Ministry of Education, Science and Culture, Japan, and partly from the Uehara Memorial Foundation, Tokyo, Japan. We thank Drs K. Jones and R. Wildin for the plasmids of the mouse TCRα enhancer and the mouse lck distal promoter. We also thank Drs H. Clevers, S. Fujimoto, M. Yamamoto, S. Ishii, T. S. Papas, J. M. Leiden, A. Bernstein, M. Satake, R. H. Goodman and H. Yagita for the plasmids of the mouse TCF-1, LEF-1, GATA-3, c-myb, c-ets-1, c-ets-2, Fli-1, AML-1/PEBP2αB, CREB, E2A and Thy1 genes. We are also grateful for the continuous encouragement from Dr J. Akiyama, OB-GYN Akiyama Memorial Hospital, Hakodate, Japan.
References
- 1.Clevers HC, Oosterwegel MA, Georgopoulos K. Transcription factors in early T cell development. Immunol Today. 1991;14:591–6. doi: 10.1016/0167-5699(93)90198-T. [DOI] [PubMed] [Google Scholar]
- 2.Orkin SH. Transcription factors and hematopoietic development. J Biol Chem. 1995;270:4955–8. doi: 10.1074/jbc.270.10.4955. [DOI] [PubMed] [Google Scholar]
- 3.Gourdeau H, Fournier REK. Genetic analysis of mammalian cell differentiation. Annu Rev Cell Biol. 1990;6:69–94. doi: 10.1146/annurev.cb.06.110190.000441. [DOI] [PubMed] [Google Scholar]
- 4.Junker S, Pedersen S, Schreiber E, Mathias P. Extinction of an immunoglobulin promoter in cell hybrids is mediated by the octamer motif and correlates with suppression of oct-2 expression. Cell. 1990;61:467–74. doi: 10.1016/0092-8674(90)90528-m. [DOI] [PubMed] [Google Scholar]
- 5.Boshart M, Weih F, Schmidt A, Fournier REK, Shütz G. A cyclic AMP response element mediates repression of tyrosine aminotransferase gene transcription by the tissue-specific extinguisher locus Tse-1. Cell. 1990;61:905–16. doi: 10.1016/0092-8674(90)90201-o. [DOI] [PubMed] [Google Scholar]
- 6.Yu H, Porton B, Shen L, Eckhardt A. Role of the octamer motif in hybrid cells extinction of immunoglobulin gene expression: Extinction is dominant in two enhancer system. Cell. 1989;58:441–8. doi: 10.1016/0092-8674(89)90425-x. [DOI] [PubMed] [Google Scholar]
- 7.Jones KW, Shapero MH, Chevrette M, Fournier REK. Subtractive hybridization cloning of a tissue-specific extinguisher: Tse1 encodes a regulatory subunit of protein kinase A. Cell. 1991;66:861–72. doi: 10.1016/0092-8674(91)90433-y. [DOI] [PubMed] [Google Scholar]
- 8.Nitsch D, Boshart M, Shütz G. Extinction of tyrosine amino-transferase gene activity in somatic cell hybrids involves modification and loss of several essential transcriptional activators. Genes Dev. 1993;7:308–19. doi: 10.1101/gad.7.2.308. [DOI] [PubMed] [Google Scholar]
- 9.Griffo G, Hamon-Benais C, Angrand P-O, et al. HNF-4 and HNF-1 as well as a panel of hepatic function are extinguished and reexpressed in parallel in chromosomally reduced rat hepatoma-human fibroblast hybrids. J Cell Biol. 1993;121:887–98. doi: 10.1083/jcb.121.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamada T, Hitomi Y, Shimizu K, Ohki M, Oikawa T. Extinction of T cell receptor alpha-chain gene expression accompanied by loss of the lymphoid enhancer-binding factor 1 (LEF-1) in murine somatic cell hybrids. Mol Cell Biol. 1993;13:1943–50. doi: 10.1128/mcb.13.3.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oikawa T, Yamada T, Kubota Y, Kondoh N, Hitomi Y, Uchiumi F, Yamamoto T. Suppression of lck proto-oncogene expression in murine somatic cell hybrids between T lymphoma cells and fibroblasts. Cytogenet Cell Genet. 1996;72:12–19. doi: 10.1159/000134152. [DOI] [PubMed] [Google Scholar]
- 12.Clevers HC, Grosschedl R. Transcriptional control of lymphoid development: lessons from gene targeting. Immunol Today. 1996;17:336–43. doi: 10.1016/0167-5699(96)10019-0. 10.1016/0167-5699(96)10019-0. [DOI] [PubMed] [Google Scholar]
- 13.Leiden JM. Transcriptional regulation of T cell receptor genes. Annu Rev Immunol. 1993;11:539–70. doi: 10.1146/annurev.iy.11.040193.002543. [DOI] [PubMed] [Google Scholar]
- 14.Mayall TP, Sheriden PL, Montminy MR, Jones KA. Distinct role of P-CREB and LEF-1 in TCRα enhancer assembly and activation on chromatin templates in vitro. Genes Dev. 1997;11:887–99. doi: 10.1101/gad.11.7.887. [DOI] [PubMed] [Google Scholar]
- 15.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Introduction of DNA into mammalian cells. Transfection using DEAE-dextran. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current Protocols in Molecular Biology. New York: John Wiley & Sons,; 1987. p. 921. [Google Scholar]
- 16.Oosterwegel M, van de Wetering M, Dooijes D, Klomp L, Winoto A, Georgopoulos K, Meijlink F, Clevers H. Cloning of murine TCF-1, a T cell-specific transcription factor interacting with functional motif in CD3-ε and T cell receptor α enhancers. J Exp Med. 1991;173:1133–42. doi: 10.1084/jem.173.5.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Travis A, Amsterdam A, Belanger C, Grossschedl R. LEF-1, a gene encoding a lymphoid-specific protein with an HMG domain, regulates T-cell receptor α enhancer function. Genes Dev. 1991;5:880–94. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 18.van de Wetering M, Oosterwegel M, van Norren K, Clevers H. Sox-4, an Sry-like HMG box protein, is a transcriptional activator in lymphocytes. EMBO J. 1993;12:3847–54. doi: 10.1002/j.1460-2075.1993.tb06063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pandolfi PP, Roth ME, Karis A, Leonard MW, Dziezak E, Grosveld FG, Engel JD, Lindenbaum MH. Targeted disruption of the GATA-3 gene causes severe abnormalities in the nervous system and in fetal liver hematopoiesis. Nature Genet. 1995;11:40–4. doi: 10.1038/ng0995-40. [DOI] [PubMed] [Google Scholar]
- 20.Georgopoulos K, Winandy S, Avitahl N. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu Rev Immunol. 1997;15:155–76. doi: 10.1146/annurev.immunol.15.1.155. [DOI] [PubMed] [Google Scholar]
- 21.Hernandez-Munain C, Krangel MS. c-Myb and core-binding factor/PEBP2 display functional synergy but bind independently to adjacent sites in the T-cell receptor δ enhancer. Mol Cell Biol. 1995;15:3090–9. doi: 10.1128/mcb.15.6.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho I-C, Bhat NK, Gottschalk LR, Lindsten T, Thompson CB, Papas TS, Leiden JM. Sequence-specific binding of human Ets-1 to the T cell receptor α gene enhancer. Science. 1990;250:814. doi: 10.1126/science.2237431. [DOI] [PubMed] [Google Scholar]
- 23.Bhat NK, Thompson CB, Lindstein T, June CH, Fujiwara S, Koizumi S, Fisher BJ, Papas TS. Reciprocal expression of human ETS1 and ETS2 genes during T-cell activation: Regulatory role for protooncogene ETS1. Proc Natl Acad Sci USA. 1990;87:3723–7. doi: 10.1073/pnas.87.10.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mélet F, Motro B, Rossi DJ, Zhang L, Bernstein A. Generation of a novel Fli-1 protein by gene targeting leads to a defect in thymus development and a delay in Friend virus-induced erythroleukemia. Mol Cell Biol. 1996;16:2708–18. doi: 10.1128/mcb.16.6.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Satake M, Nomura S, Yamaguchi-Iwai Y, Takahama Y, Hashimoto Y, Niki M, Kitamura Y, Ito Y. Expression of the Runt domain-encoding PEBP2α genes in T cells during thymic development. Mol Cell Biol. 1995;15:1662–70. doi: 10.1128/mcb.15.3.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bain G, Engel I, Maandag ECR, et al. E2A deficiency leads to abnormalities in αβ T-cell development and to rapid development of T-cell lymphomas. Mol Cell Biol. 1997;17:4782–91. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Georgopoulos K, Morgan BA, Moore DD. Functionally distinct isoforms of the CRE-BP DNA-binding protein mediate activity of a T-cell-specific enhancer. Mol Cell Biol. 1992;12:747–57. doi: 10.1128/mcb.12.2.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones PA. The DNA methylation paradox. Trends Genet. 1999;15:34–7. doi: 10.1016/s0168-9525(98)01636-9. [DOI] [PubMed] [Google Scholar]
- 29.Ng HH, Bird A. Histone deacetylases: silencers for hire. Trends Biol Sci. 2000;25:121–6. doi: 10.1016/s0968-0004(00)01551-6. [DOI] [PubMed] [Google Scholar]
- 30.Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–34. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- 31.Verma IM, Stevenson JK, Schwarz EM, Antwerp DV, Miyamoto S. Rel/NF-κB/IκB family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–35. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 32.Graef IA, Crabtree GR. The transcriptional paradox: Octamer factors and B and T cells. Science. 1997;277:193–4. doi: 10.1126/science.277.5323.193. 10.1126/science.277.5323.193. [DOI] [PubMed] [Google Scholar]
- 33.Hitomi Y, Yamada T, Oikawa T. Extinction of expression of the PU.1/Sfpi-1 putative oncogene encoding a B-cell- and macrophage-specific transcription factor in somatic cell hybrids. Cancer Res. 1993;53:5759–65. [PubMed] [Google Scholar]
- 34.Gregoire JM, Romeo PH. T-cell expression of the human GATA-3 gene is regulated by a non-lineage-specific silencer. J Biol Chem. 1999;274:6567–78. doi: 10.1074/jbc.274.10.6567. 10.1074/jbc.274.10.6567. [DOI] [PubMed] [Google Scholar]
- 35.Genetta T, Ruezinsky D, Kadesch T. Displacement of an E-box-binding repressor by basic helix-loop-helix proteins. implications for B-cell specificity of the immunoglobulin heavy-chain enhancer. Mol Cell Biol. 1994;14:6153–63. doi: 10.1128/mcb.14.9.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Norton JD, Deed RW, Craggs G, Sablitzky F. Id helix-loop-helix proteins in cell growth and differentiation. Trends Cell Biol. 1998;8:58–65. 10.1016/s0962-8924(97)01183-5. [PubMed] [Google Scholar]
- 37.Ishibashi M, Moriyoshi K, Sasaki Y, Shiota K, Nakanishi S, Kageyama R. Persistent expression of helix-loop-helix factor HES-1 prevents mammalian neural differentiation in the central nervous system. EMBO J. 1994;13:1799–805. doi: 10.1002/j.1460-2075.1994.tb06448.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schenherr CJ, Anderson DJ. The neuron-restrictive silencer factor (NRSF): a coordinate repressor of multiple neuron-specific genes. Science. 1993;267:1360–3. doi: 10.1126/science.7871435. [DOI] [PubMed] [Google Scholar]
- 39.Bulla GA. Extinction of α1-antitrypsin expression in cell hybrids is independent of HNF1α and HNF4 and involves both promoter and internal DNA sequences. Nucl Acid Res. 1999;27:1190–7. doi: 10.1093/nar/27.4.1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Radomska HS, Shen C-P, Kadesch T, Erkhardt LA. Constitutively expressed October-2 prevents immunoglobulin gene silencing in myeloma x T cell hybrids. Immunity. 1994;1:623–34. doi: 10.1016/1074-7613(94)90034-5. [DOI] [PubMed] [Google Scholar]
- 41.Boshart M, Nitsch D, Schütz G. Extinction of gene expression in somatic cell hybrids–a reflection of important regulatory mechanisms? Trends Genet. 1993;9:240–5. doi: 10.1016/0168-9525(93)90088-y. [DOI] [PubMed] [Google Scholar]
- 42.Olson EN, Klein WH. bHLH factors in muscle development: dead lines and commitments, what to leave in and what to leave out. Genes Dev. 1994;8:1–8. doi: 10.1101/gad.8.1.1. [DOI] [PubMed] [Google Scholar]
- 43.Nerlov C, McNagny KMD, Döderlein G, Kowenz-Leutz E, Graf T. Distinct C/EBP functions are required for eosinophil lineage commitment and maturation. Genes Dev. 1998;12:2413–23. doi: 10.1101/gad.12.15.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamaguchi Y, Zon LI, Ackerman SJ, Yamamoto M, Suda T. Forced GATA-1 expression in the murine myeloid cell line M1: Induction of c-Mpl expression and megakaryocytic/erythroid differentiation. Blood. 1998;91:450–7. [PubMed] [Google Scholar]
- 45.Klinken SP, Alexander WS, Adams JM. Hematopoietic lineage switch: v-raf oncogene converts Eµ-myc transgenic B cells into macrophages. Cell. 1988;53:857–67. doi: 10.1016/s0092-8674(88)90309-1. [DOI] [PubMed] [Google Scholar]
- 46.Bjornson CRR, Reitze RL, Reynolds BA, Magli MC, Vescovi AL. Turning brain into blood: a hematopoietic fate adopted by adult neural stem cells in vivo. Science. 1999;283:534–7. doi: 10.1126/science.283.5401.534. 10.1126/science.283.5401.534. [DOI] [PubMed] [Google Scholar]
- 47.Geiger H, Sick S, Bonifer C, Müller AM. Globin gene expression is reprogrammed in chimeras generated by injecting adult hematopoietic stem cells into mouse blastocysts. Cell. 1998;93:1055–65. doi: 10.1016/s0092-8674(00)81210-6. [DOI] [PubMed] [Google Scholar]