Abstract
We have identified a 30-aa peptide that efficiently cleaves single-stranded RNA. The peptide sequence corresponds to a single zinc finger of the human male-associated ZFY protein; a transcription factor belonging to the Cys2His2 family of zinc-finger proteins. RNA cleavage was observed only in the absence of zinc. Coordination with zinc resulted in complete loss of ribonuclease activity. The ribonuclease active structure was determined to be a homodimeric form of the peptide. Dimerization of the peptide occurred through a single intermolecular disulfide between two of the four cystines. The observed hydrolytic activity was single-stranded RNA-specific. Single-stranded DNA, double-stranded RNA and DNA, and 2′-methoxy-modified sequences were not degraded by the peptide. The peptide specifically cleaved pyrimidines within single-stranded RNA and the dinucleotide sequence 5′-pyr-A-3′ was preferred. The RNA cleavage products consisted of a 3′ phosphate and 5′ hydroxyl. The initial rates of cleavage (V0) observed for the finger peptide were comparable to rates observed for human ribonucleases, and the catalytic rate (Kcat) was comparable to rates observed for the group II intron rybozymes. The pH profile exhibited by the peptide is characteristic of general acid–base catalytic mechanisms observed with other ribonucleases. These observations raise interesting questions about the potential biological roles of zinc-finger proteins.
ZFY is a human male-associated transcription factor containing zinc-finger domains and is believed to participate in spermatogenesis and the regulation of male embryogenesis (1–3). A single zinc-finger element, generally 30 aa in length, has been shown to form an independent structural unit through the tetrahedral coordination of the zinc ion with either cysteine thiolates or histidine imidazoles (4–5). The zinc-finger domain of ZFY was shown to consist of finger elements in which the zinc ion was coordinated by two cysteines and two histidines to form the canonical ββα finger structure (4, 6). These finger elements were arranged within the ZFY protein as alternating two-finger repeats. The odd-numbered domains were shown to fit the general zinc-finger consensus sequence, whereas the even numbered domains exhibited a unique zinc-finger sequence (6). The solution structure of two 30-aa peptides representing the odd (ZFY-switch) and even (ZFY-6) numbered zinc fingers showed that these peptides exhibited measurably different higher order structures (7).
Although the metal bound structures of the ZFY finger peptides have been extensively characterized, the structures of the finger peptides in the absence of zinc are less well understood. Weiss et al. (8) showed that a progressive reduction in metal coordination with the ZFY-6 finger peptide resulted in a loss in α-helix formation. A similar loss in α-helix formation was observed for the oxidized form of the peptide in the absence of zinc. We have observed that in the absence of zinc and at concentrations >1 mM, the ZFY-6 peptide formed a structure with a molecular mass consistent with peptide dimerization. Reaction of this higher-molecular-mass species with Ellman reagent [5,5′-dithiobis (2-nitrobenzoic acid)] suggested that the cysteines were oxidized. MS confirmed that in the absence of zinc, the ZFY-6 peptide formed a homodimer through a single intermolecular disulfide bond between two of the four cystines (data not shown). Consistent with the MS data is the observation that the mutant sequence of ZFY-6 in which both cysteines had been substituted with alanines (C5, 8A) as well as the ZFY-6 peptide treated with the thiol blocking agent N-ethlymaleimide did not form dimeric structures (data not shown). In addition, we have observed that the homodimeric forms of the ZFY-6 peptide exhibited highly efficient ribonuclease activity.
MATERIALS AND METHODS
Peptide Synthesis and Characterization. Peptides were sythesized by standard solid phase procedure by using the Applied Biosystems synthesizer and t-BOC amino acids. Synthetic peptides were also purchased from SynPep, Dublin, CA and Chiron. The molecular mass and amino acid content of the peptides were verified by using MS and amino acid analysis. The peptides were determined to be >90% pure by analytical reverse-phase HPLC.
Peptide dimers were prepared by incubating 0.25–0.5 μmol of peptide in 200 mM DTT and 6 M urea for 16 h at 60°C. The reduced peptide was purified by gel filtration in 0.1 M acetic acid. The eluent was lyophilized, and the pellet was resuspended in 50 μl of 20 mM sodium phosphate, pH 8.0. Dimerization of the oxidized peptide was monitored by SDS/PAGE and LC-MS.
The reduced monomeric form of the peptide was prepared as described above, with the exception that the lyophilized pellet was resuspended in 1 ml of 20 mM sodium acetate, pH 4.0. The determination of free sulfhydryl was carried out in 5.2 ml containing 4 μM peptide, 0.4 mg of Ellman reagent, and 100 mM sodium phosphate, pH 8.0. After 15 min, the absorbance was measured at 412 nm and compared with a sulfhydryl standard.
Initial Cleavage Rates.
Initial cleavage rates (V0) for the various peptides and substrates were determined in substrate excess. Cleavage reactions were performed in 100 μl containing 100 nM peptide, 500 nM RNA, 105 cpm 5′-end 32P-labeled RNA, 20 mM sodium phosphate (pH 7.0), 100 mM KCl, 1 mM EDTA, and 0.1% NP40. Digestion reactions were incubated at 37°C and quenched at specific time points in 6 M urea. Digestion products were analyzed by denaturing PAGE. Substrate and product bands were quantitated on a Molecular Dynamics PhosphorImager. The concentration of substrate converted to product was plotted as a function of time. The initial cleavage rate (V0) was obtained from the slope (nM converted substrate per minute) of the best-fit line derived from ≥5 data points within the linear potion (<10% of the total reaction) of the plot. The errors reported were based on three trials.
Single-Turnover Kinetics.
Reactions were performed and analyzed as described for the determination of initial cleavage rates, with the exception that 50 nM RNA and excess peptide ranging in concentration from 50 to 500 nM was used. The reaction rates (kobs) for each peptide concentration were determined by plotting the natural logarithm of the substrate remaining (1 − fraction product) vs. time, and the t1/2 was obtained from the calculated slope. Values for kobs = (ln2)/t1/2. The values for kobs were based on three trials, and the coefficient of variance between individual values for kobs at a single peptide concentration were <20%. The values for kobs were plotted as a function of peptide concentration and fit to a Michaelis–Menten binding curve to obtain the single-turnover Km and kcat.
Multiple-Turnover Kinetics.
Reactions were performed and analyzed as described for the determination of initial cleavage rates with the exception that 100 nM peptide and excess RNA ranging in concentration from 100 to 1,000 nM was used. Reaction rates (kobs) were calculated from the slope of a linear least-squares fit of [product] vs. time over the first 5–10% of the reaction. The reaction rates were plotted as a function of the substrate concentration, and the data were fit to the Michaelis–Menten equation. The multiple-turnover kcat and Km were obtained from a modified Eadie–Hofstee plot of kobs vs. kobs/[S].
Ionic and pH Dependence. Digestion reactions were performed in 100 μl containing 100 nM peptide, 500 nM RNA substrate, 105 cpm 32P-labeled RNA, 20 mM sodium phosphate (pH 7.0), 1 mM EDTA, 0.1% Nonidet P-40, and either 0–500 mM NaCl or 100 mM KCl and 0–10 mM divalent cation. Digestion reactions were incubated at 37°C and quenched at 60 min in 6 M urea. Digestion products were analyzed by denaturing PAGE.
For pH determination, the pH of the reactions was adjusted with 20 mM sodium acetate (pH 4), 20 mM sodium phosphate (pH 5–8), or 20 mM sodium carbonate (pH 9). The reaction rates (kobs) were determined as described for multiple-turnover kinetics. The pH–rate profile was fitted by using the logarithm of the reaction rate.
Cleavage Product Analysis.
The (ACUCCACCAUAGUACACUCC) RNA substrate was internally labeled at the penultimate 3′ phosphate by using [32P]cytidine bisphosphate and T4 RNA ligase. The 3′ terminal phosphate was removed by using calf intestinal phosphatase, and the RNA was digested with the ZFY-6 peptide as described above. The digestion reactions were aliquoted, and one aliquot was subjected to an additional dephosphorylation. The digested products were analyzed by denaturing PAGE.
Kinetic Data Analysis.
Kinetic data were analyzed by using the equations of Cleland (9) and Dixon and Webb (10). Equations were implemented with the program ultrafit (Biosoft, Milltown, NJ) or with the program winnonlin (Scientific Consulting, Mountain View, CA).
RESULTS
The ZFY-6 peptide was observed to hydrolyze RNA in an endonuceolytic manner (Fig. 1). This ribonuclease activity was observed for only the oxidized homodimer in the absence of zinc (Table 1). Neither the zinc-coordinated (Zn⋅ZFY-6) or reduced monomeric forms of the peptide exhibited ribonuclease activity (Fig. 2). Neither was ribonuclease activity observed for the mutant monomeric peptide (C5, 8A) (Table 1, Fig. 2D). In addition, ribonuclease activity was not observed for the ZFY-6 peptide treated with the thiol blocking agent N-ethlymaleimide, suggesting that the formation of intermolecular disulfide bonds was required for ribonuclease activity (data not shown).
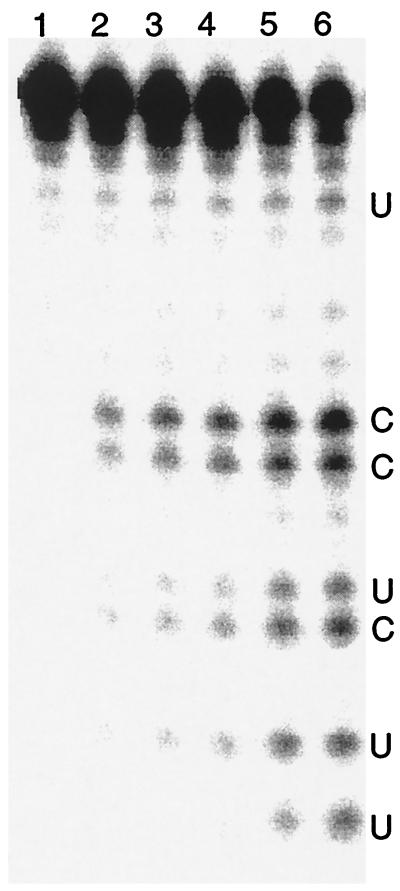
Figure 1.
PAGE of RNA digested by the ZFY-6 peptide dimer. The (UGGUGGGCGCCGUCGGUGUGGGCAA) RNA substrate was incubated in the absence of ZFY-6 peptide (lane 1) for 60 min and with ZFY-6 peptide (lane 2–6) for 5–40 min.
Table 1.
Relationship between peptide structure and RNA cleavage activity for the wild-type and mutant peptides
| Peptide | Initial Rate (V0) pM⋅min−1* | Structure | Sequence |
|---|---|---|---|
| ZN⋅ZFY-6 | Below detection limit | Monomer | KTYQCQYCEYRSADSSNLKTHIKTKHSKEK |
| ZFY-6 | 6000 ± 130 | Dimer | KTYQCQYCEYRSADSSNLKTHIKTKHSKEK |
| E9A | 1800 ± 90 | Dimer | KTYQCQYCAYRSADSSNLKTHIKTKHSKEK |
| H21,26A | 800 ± 30 | Dimer | KTYQCQYCEYRSADSSNLKTAIKTKASKEK |
| C5,8A | Below detection limit | Monomer | KTYQAQYAEYRSADSSNLKTHIKTKHSKEK |
| Reduced ZFY-6 | Below detection limit | Monomer | KTYQAQYAEYRSADSSNLKTAIKTKASKEK |
The initial cleavage rates for the various peptides were determined with the (ACUCCACCAUAGUACACUCC) RNA substrate. Mutant peptides exhibiting ribonuclease activity hydrolyzed the RNA substrate at the same positions observed for the ZFY-6 dimer (see Table 2). Underlined residues indicate the position of the alanine-substituted amino acids within the mutant peptide sequence.
Detection limit = cleavage rates resulting in <1% of the RNA substrate cleaved over 60 min.
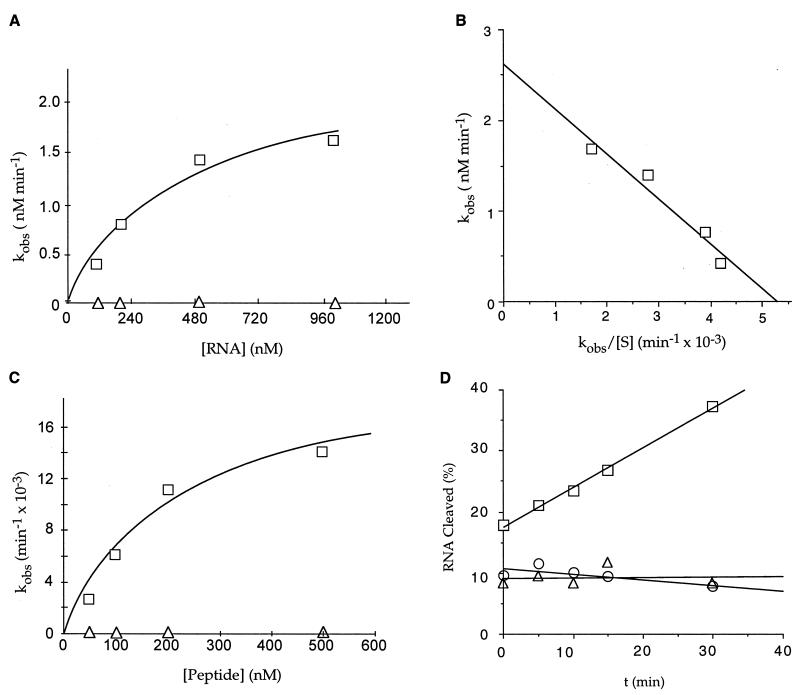
Figure 2.
Kinetics of RNA cleavage by the finger peptide. Multiple- and single-turnover kinetics were performed with the (UGGUGGGCAAUGGGCGUGUU) RNA substrate. (A) Multiple-turnover kinetics for RNA cleavage by the dimeric peptide (□) and zinc-coordinated peptide (▵). The curve represents a nonlinear least-squares fit of the data to the Michaelis–Menten equation. (B) Eadie-Hofstee plot of multiple-turnover kinetics for the dimeric peptide, where [S] is expressed in nM. The slope of the line is equal to −Km, the y intercept is equal to Vmax, and kcat = Vmax/[Et]. The multiple-turnover Km and kcat for the best-fit curve were, respectively, 494 ± 58 nM and 0.026 ± 0.003 min−1. (C) Single-turnover kinetics for the dimeric peptide (□) and the zinc-coordinated peptide (▵). Plotting the kobs as a function of peptide concentration results in a hyperbolic curve with the Km equal to the peptide concentration at half-maximum rate and the kcat equal to the horizontal asymptote. The calculated values for the single-turnover Km and kcat from the best-fit curve were, respectively, 209 ± 64 nM and 0.02 ± .003 min−1. (D) RNA cleavage activity for the reduced (▵) and dimeric (□) forms of the ZFY-6 peptide and the C5,8A peptide mutant (○). The RNA cleavage activity for the reduced form of the peptide was determined by preincubating the peptide and RNA in 20 mM sodium acetate (pH 4) for 30 min and adjusting the pH of the reaction to 7 with 20 mM sodium carbonate (pH 9) for the digestion reaction. The percent RNA cleaved is equal to [product]/[total substrate] × 100.
Various precautions were taken to ensure that the ribonuclease activity observed with the ZFY-6 peptide was not because of single-stranded RNase contamination. The RNase activities reported for the peptide represent activities observed for several syntheses of the ZFY-6 sequence. These peptides were prepared both in our laboratories and through various outside suppliers. Furthermore, the synthesized peptides were subjected to multiple sequential purification procedures that included reverse-phase and size-exclusion HPLC. Allosteric inhibitors of the single-stranded RNase A superfamily, e.g., rRNasin (Promega) and Prime RNase inhibitor (5 Prime → 3 Prime) were included in the digestion reactions. Finally, chelating agents as well as free-radical scavengers, (e.g., EDTA, EGTA, 2-propanol, and isopropyl isocyanate) were also added to eliminate the possibility of contaminating metals that could lead to the degradation of RNA. No effect on the cleavage activity was observed as a result of these treatments.
The ZFY-6 peptide displayed single-stranded endoribonuclease specificity. The ZFY-6 peptide failed to promote the digestion of double-stranded RNA, single-stranded DNA or 2′ methoxy-modified oligonucleotides (Table 2). The sites of cleavage were determined for several oligoribonucleotide substrates. The ZFY-6 peptide cleaved specifically single-stranded RNA at the 3′ side of pyrimidines (Table 2).
Table 2.
Cleavage specificity of the ZFY-6 peptide dimer
| Substrate | Initial rate (V0) pM min−1* | Sequence Specificity |
|---|---|---|
| DNA | Below detection limit | ACTCCACCATAGTACACTCC |
| Double-stranded DNA | Below detection limit | ACTCCACCATAGTACACTCC TGAGGTGGTATCATGTGAGG |
| Double-stranded RNA | Below detection limit | ACUCCACCAUAGUACACUCC UGAGGUGGUAUCAUGUGAGG |
| 2′OMe | Below detection limit | ACUCCACCAUAGUACACUCC |
| ↓↓↓↓ | ||
| RNA | 153 | GGGCGCCGUCGGUGUGG |
| ↓↓↓↓↓↓↓ | ||
| RNA | 422 | UGGUGGGCGCCGUCGGUGUGGGCAA |
| ↓↓↓ | ||
| RNA | 1,200 | UGGUGGGCAAUGGGCGUGUU |
| ↓↓↓↓↓ | ||
| RNA | 6,100 | ACUCCACCAUAGUACACUCC |
Arrows indicate the position of the cleavage site. The length of the arrows indicate the amount of the cleavage product generated for each respective cleavage site.
Detection limit = cleavage rats resulting in <1% of the RNA substrate cleaved over 60 min.
The initial cleavage rates (V0) for the ZFY-6 peptide were determined in oligonucleotide excess. Within the range of oligoribonucleotides studied, V0 varied as a function of sequence but not length of the oligonucleotide (Table 2). Pyrimidine-rich sequences were cleaved more rapidly, and 5′-pyr-A-3′ was the most preferred dinucleotide sequence. The cleavage rate for the RNA sequence containing a single 5′-pyr-A-3′ was ≈3- to 8-fold faster than RNA substrates not containing the preferred dinucleotide sequence. An additional 3-fold increase in the cleavage rate was observed for the RNA substrate containing three 5′-pyr-A-3′ dinucleotide sequences. These cleavage rates are comparable to cleavage rates observed for human RNase H type 1 measured under similar conditions, (11). Comparisons with single-stranded ribonuclease proteins are difficult because these activities were reported under different conditions. Nevertheless, the cleavage rates for the ZFY-6 peptide appear to be several orders of magnitude slower than those observed for single-stranded RNases. Values for kcat of 0.026 and 0.020 min−1 were observed for the dimer under, respectively, multiple- and single-turnover conditions and are comparable to the kcat values observed for the group II intron rybozymes (ref. 12; Fig. 2 B and C).
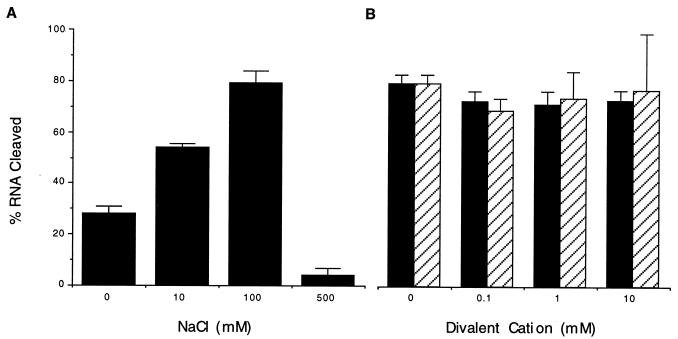
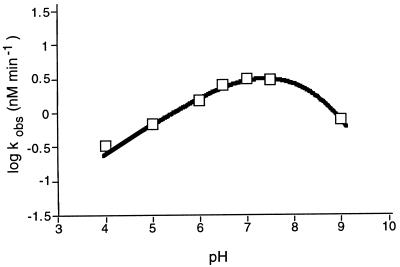
Analysis of the biochemical properties of the cleavage reaction showed that increasing the sodium chloride concentration from 0 to 100 mM resulted in a corresponding increase in the cleavage rate, whereas a marked decrease in the cleavage rate was observed at NaCl concentrations >100 mM (Fig. 3A). A similar profile was observed for potassium chloride. The addition of divalent cations to the reaction did not enhance the cleavage rate of the peptide, suggesting that cleavage is effected via a metal-independent mechanism (Fig. 3B). The pH profile curve was bell-shaped, with the optimal cleavage rate observed at pH 7 (Fig. 4). Finally, analysis of the RNA cleavage products generated by the ZFY-6 peptide revealed that the termini consisted of 3′-phosphate and 5′-hydroxyl (Fig. 5).
Figure 3.
Ionic dependence on RNA cleavage by the dimeric form of the ZFY-6 peptide. Cleavage reactions were prepared with 500 nM (UGGUGGGCAAUGGGCGUGUU) RNA and 100 nM ZFY-6 peptide. The percent RNA cleaved is equal to [product]/[total substrate] × 100 after 60 min. (A) RNA cleavage activity of the ZFY-6 peptide as a function of NaCl concentration. (B) RNA cleavage activity in the presence of either Mg2+ (filled bars) or Mn2+ (hatched bars).
Figure 4.
The pH dependence of RNA cleavage by the ZFY-6 peptide dimer. kobs for the hydrolysis of the (ACUCCACCAUAGUACACUCC) RNA substrate by the ZFY-6 dimer were determined under multiple-turnover conditions in reaction buffers ranging in pH from 4.0 to 9.0. The curve is a nonlinear least-squares fit of the data to equations of Dixon and Webb (25).
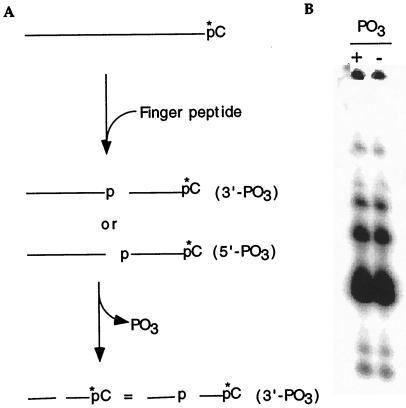
Figure 5.
Cleavage product analysis of the RNA substrate. (A) Schematic illustrating the process used to analyze the RNA cleavage products generated by the ZFY-6 peptide. (B) Comigration of both the untreated (+PO3) and dephosphorylated (−PO3) fractions of the RNA digestion products indicated that the termini consisted of 3′ phosphate and 5′ hydroxyl.
The ZFY-6 peptide sequence contains both histidine and glutamic acid residues and therefore could conceivably follow either an RNase A or T1 mechanism. To differentiate these possibilities, we designed peptide mutants in which both histidines (H21, 26A) or glutamic acid (E9A) were substituted with alanine (Table 1). The ZFY-6 dimer exhibited a cleavage rate 3- to 8-fold faster than the dimeric form of the mutant sequences. The substitution of these amino acids with alanine did not result in complete ablation of the ribonuclease activity, suggesting that these amino acids were not required for catalysis.
DISCUSSION
Compared with other synthetic peptides that have been designed to mimic ribonucleases, the ZFY-6 peptide is extraordinary (13–17). Unlike these other ribonuclease mimics, the ZFY-6 peptide requires no intercalating moieties or other conjugates to enhance binding to the substrate. The ZFY-6 peptide degrades RNA in substrate excess and at peptide concentration orders of magnitude lower than those required for the RNase mimic peptides. In comparison to the conditions required for RNA cleavage by RNase mimics, the cleavage conditions optimized for the ZFY-6 peptide more closely approximate physiological conditions. Most important, the cleavage rates for the ZFY-6 peptide are many orders of magnitude faster than the rates observed for the synthetic RNase peptides. For example, these RNase mimics required >10 hours, and in some cases several days, to achieve complete degradation of the RNA (16–17).
The biochemical properties as well as the cleavage specificity exhibited by the ZFY-6 peptide are consistent with other single-stranded RNases and in particular the enzymatic properties observed with the RNase A superfamily, (18). These enzymes have been shown to (i) produce cleavage products consisting of 3′-phosphate and 5′-hydroxyl termini (19–20); (ii) cleave specifically at pyrimidines; (iii) exhibit a bell-shaped response in the cleavage rate with respect to changes in pH (21–23); and (iv) not require divalent metals for catalysis (19, 23–24). Given these observations, the ZFY-6 peptide and these ribonucleases likely have similar cleavage mechanisms. The cleavage mechanism described for RNase A involves a metal-independent, amino acid-induced general acid–base hydrolysis of the RNA, in which His-12 deprotonates the 2′-hydroxyl, resulting in nucleophilic attack of the phosphorous center by the 2′-oxyanion, subsequent formation of a pentacoordinate phosphate intermediate, and proton transfer from the phosphate to the leaving 5′-oxygen by His-119. A similar mechanism has been proposed for the single-stranded-specific RNase T1. In this case, His-92 is believed to act as the acid and Glu-58 as the base. In addition to the biochemical similarities, the observation that neither DNA nor 2′-methoxy-modified oligonucleotides were cleaved by the peptide further support a single-stranded ribonuclease mechanism that solicits the 2′-hydroxyl of the RNA for catalysis (Table 2).
In contrast to RNases A and T1, the substitution of either histidine (H21, 26A) or glutamic acid (E9A) with alanine did not ablate the ribonuclease activity of the ZFY-6 peptide, suggesting that if a single-stranded ribonuclease mechanism is involved, other residues within the dimeric peptide are acting as the proton donor and acceptor to promote the hydrolysis of the RNA. Clearly, additional mutants must be studied to identify the key residues involved in catalysis. The cleavage rates exhibited by the H21, 26A, and E9A mutants were slower than the rates observed for the wild-type ZFY-6 peptide (Table 1). Given that both mutant sequences were shown to form homodimeric structures, the slower cleavage rates observed for the mutant peptides compared with the wild-type peptide may be the result of differences in hydrophilicity and/or structure between these sequences.
The observations reported here for the ZFY-6 peptide raise a number of interesting biological questions. First, considering that the coordination of zinc with the ZFY-6 peptide inhibits RNase activity, is there evidence of zinc-finger proteins existing in a metal free state and are there cellular mechanisms to regulate the zinc content of finger proteins? The cysteine-rich intestinal protein plays a role in intestinal zinc transport via the zinc-finger domain (LIM) (25). The transport of zinc through the intestine by cysteine-rich intestinal protein appears to be regulated by levels of intestinal metallothionein that competitively inhibit the binding of zinc to the finger domain (26). A similar mechanism may be involved with the ZFY protein. Second, when the zinc content of such finger proteins is reduced, do these proteins cleave RNA? Recently, tristetraprolin (TTP) was shown to regulate tumor necrosis factor α (TNF-α) production via a negative-feedback loop (27). TTP belongs to the Cys3His class of zinc finger proteins. The production of TTP is induced by the same factors that induce TNF-α and by TNF-α itself, and the increased production of TTP appears to coincide with a reduction in the level TNF-α mRNA. TTP was shown to inhibit the production of TNF-α by binding to and destabilizing TNF-α mRNA, and it is conceivable that TTP may directly degrade TNF-α RNA. Third, do peptides derived from other members of the Cys2His2 class of zinc-finger proteins share these properties? Perhaps the ribonuclease activity observed for the ZFY-6 peptide is unique to this zinc-finger sequence, but considering these findings, examination of other zinc-finger peptides for RNase activity is warranted.
Acknowledgments
We thank Steven Hofstadler and Hans Gaus for mass spectral analysis, Fred Hassman for peptide synthesis, and David Ecker, Richard Griffey, Frank Bennett, and Susan Freier for helpful discusions and periodic critical review.
ABBREVIATIONS
- TNF-α
tumor necrosis factor α
- TTP
tristetraprolin
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Page D C, Fisher E M C, McGillivray B, Brown L G. Nature (London) 1990;346:279–281. doi: 10.1038/346279a0. [DOI] [PubMed] [Google Scholar]
- 2.Palmer M S, Sinclair A H, Berta P, Ellis N A, Goodfellow P N, Abbas N E, Fellous M. Nature (London) 1989;342:937–939. doi: 10.1038/342937a0. [DOI] [PubMed] [Google Scholar]
- 3.Koopman P, Gubbay J, Colignon J, Lovell-Badge R. Nature (London) 1989;342:940–942. doi: 10.1038/342940a0. [DOI] [PubMed] [Google Scholar]
- 4.Berg J M. Science. 1987;232:485–487. doi: 10.1126/science.2421409. [DOI] [PubMed] [Google Scholar]
- 5.Frankel A D, Berg J M, Pabo C O. Proc Natl Acad Sci USA. 1987;84:4841–4845. doi: 10.1073/pnas.84.14.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gibson T J, Postama J P M, Brown R S, Argos P. Protein Eng. 1988;2:209–218. doi: 10.1093/protein/2.3.209. [DOI] [PubMed] [Google Scholar]
- 7.Kochayan M, Keutmann H T, Weiss M A. Biochemistry. 1991;30:9396–9402. doi: 10.1021/bi00103a002. [DOI] [PubMed] [Google Scholar]
- 8.Weiss M A, Mason K A, Dahl C E, Keutmann H T. Biochemistry. 1990;29:5660–5664. doi: 10.1021/bi00476a002. [DOI] [PubMed] [Google Scholar]
- 9.Cleland W W. The Enzymes. Vol. 19. New York: Academic; 1990. pp. 99–158. [Google Scholar]
- 10.Dixon M, Webb E C. Enzymes. 3rd Ed. New York: Academic; 1979. pp. 148–153. [Google Scholar]
- 11.Wu, H. J. Lima, W. F. & Crooke, S. T. (1999) J. Biol. Chem., in press. [DOI] [PubMed]
- 12.Michels W J, Pyle A M. Biochemistry. 1995;34:2965–2977. doi: 10.1021/bi00009a028. [DOI] [PubMed] [Google Scholar]
- 13.Brittain I J, Huang X, Long E C. Biochemistry. 1988;37:12113–12120. doi: 10.1021/bi9806605. [DOI] [PubMed] [Google Scholar]
- 14.Barbier B, Brack A. J Am Chem Soc. 1988;110:6880–6882. [Google Scholar]
- 15.Barbier B, Brack A. J Am Chem Soc. 1992;114:3511–3515. [Google Scholar]
- 16.Podyminogin M A, Vlassov V V, Giege R. Nucleic Acids Res. 1993;21:5950–5956. doi: 10.1093/nar/21.25.5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ching-Hsun T, Wei Z, Leibowitz M J, Stein S. Proc Natl Acad Sci USA. 1992;89:7114–7118. doi: 10.1073/pnas.89.15.7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beintema J J, Breukelman H J, Carsana A, Furia A. In: Ribonucleases, Structures and Function. D’Alessio G, Riordan J F, editors. New York: Academic; 1997. pp. 245–265. [Google Scholar]
- 19.Roberts G C K, Dennis E A, Meadows D H, Cohen J S, Jardetzky O. Proc Natl Acad Sci USA. 1969;62:1151–1158. doi: 10.1073/pnas.62.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikehara M, Ohtsuka E, Togunaga T, Nishikawa S, Uesugi S, Tanaka T, Aoyana Y, Kikyodani S, Fugimoto K, Yanase K, et al. Proc Natl Acad Sci USA. 1986;83:4695–4699. doi: 10.1073/pnas.83.13.4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Herries D G, Mathias A P, Rabin B R. Biochem J. 1962;85:127–131. doi: 10.1042/bj0850127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Del Rosario E J, Hammes G G. Biochemistry. 1969;8:1884– 1889. doi: 10.1021/bi00833a017. [DOI] [PubMed] [Google Scholar]
- 23.Blackburn P, Moore S. In: The Enzymes. 3rd Ed. Boyer P D, editor. Vol. 15. New York: Academic; 1982. pp. 317–434. [Google Scholar]
- 24.Takahashi K, Moore S. In: The Enzymes. 3rd Ed. Boyer P D, editor. Vol. 15. New York: Academic; 1982. pp. 435–468. [Google Scholar]
- 25.Hempe J M, Cousins R J. Proc Natl Acad Sci USA. 1991;88:9671–9674. doi: 10.1073/pnas.88.21.9671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Escobar O, Sandoval M, Vargas A, Hempe J M. Pediatr Res. 1995;37:321–327. doi: 10.1203/00006450-199503000-00012. [DOI] [PubMed] [Google Scholar]
- 27.Carballo E, Lai W S, Blackshear P J. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]