Abstract
Members of the CD1 family of antigen-presenting molecules bind and present a variety of mammalian and microbial glycolipids for specific recognition by T cells. CD1 proteins accomplish their antigen-presenting function by binding the alkyl chains of the antigens within a deep, hydrophobic groove on the membrane distal surface of CD1, making the hydrophilic elements of the antigen available for contact with the variable regions of antigen-specific T-cell receptors. Most models of CD1-restricted T cells function in infectious, neoplastic, or autoimmune diseases and are based on the premise that CD1-restricted T-cell responses are initiated by alterations in cellular glycolipid content. Although a growing number of self, altered self and foreign glycolipid antigens have been identified, the cellular mechanisms that could lead to the generation of antigenic glycolipids within cells, or control the presentation of particular classes of altered self or microbial glycolipids in disease states have only recently come under investigation. Here we review the structures of known glycolipid antigens for T cells and discuss how the chemical nature of these antigens, which is quite different from that of peptides, influences their recognition by T cells.
Introduction
Research over the past two decades has firmly established the mechanisms of peptide antigen presentation by the major histocompatibility complex (MHC) class I and class II proteins.1–3 The information gained from these studies has provided important insights into the pathogenesis of human diseases involving tissue transplantation, infectious diseases and autoimmunity, and now forms the basis for most targeted approaches to the development of vaccines and specific immunotherapies. However, it is now clear that MHC/peptide complexes are not the only targets of T-cell recognition. The CD1 family of antigen-presenting molecules mediates presentation of a variety of cellular and microbial glycolipids to antigen-specific T cells.
Glycolipids are not gene products, but their biosynthesis is rigorously controlled by enzymatic pathways, resulting in a myriad of structures that contain information about their origin that may be useful to the immune system in the detection of infected, stressed, or transformed cells. Yet, fundamental differences between peptides and glycolipids in their mode of synthesis, cellular trafficking and biophysical properties require the application of certain basic principles of lipid chemistry to classical immunological paradigms of antigen recognition. This review will summarize the structures of the known glycolipid antigens for CD1-restricted T cells, and discuss how the chemical nature of glycolipids impacts on the cellular mechanisms by which they are loaded onto CD1 proteins and activate T cells.
CD1–glycolipid antigen complexes
CD1 antigen-presenting molecules are transmembrane glycoproteins that are related in structure to MHC class I and class II proteins, both in their primary amino acid sequences, and in their overall domain organization (Fig. 1).4,5 The human CD1 complex is encoded outside the MHC and consists of five genes, CD1A, CD1B, CD1C, CD1D and CD1E.6 Four of the proteins encoded in this locus, CD1a, CD1b, CD1c, CD1d are known to mediate T-cell activation by glycolipid antigens.4 CD1E is translated, but little is known concerning the function of CD1e proteins.7,8 Shortly after the discovery of the CD1 locus by Calabi and Milstein, they classified members of the human CD1 gene family as belonging to either group 1 (CD1A, CD1B, CD1C) or group 2 (CD1D) based on their predicted primary amino acid sequences.9 Homologues of human group 2 proteins are conserved in all mammals studied to date, but group 1 proteins are not found in rats or mice.
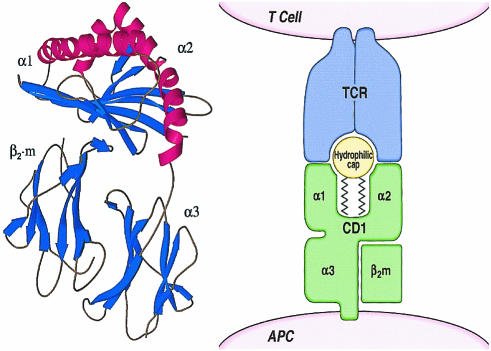
Figure 1.
CD1–glycolipid complexes. The murine CD1d protein crystal structure demonstrates that the α1 and α2 domains of the CD1 heavy chain form a groove structure. The groove is lined by non-polar amino acids, which render it able to bind the aliphatic hydrocarbon chains of antigens, positioning the hydrophilic elements of the antigen for direct contact with antigen-specific TCRs. Figures are reprinted with permission from Blackwell Science Ltd and the American Advancement of Science.5,15
The structure of murine CD1, a homologue of human CD1d, is known from crystallographic studies completed by Wilson and colleagues.5 The membrane distal α1 and α2 domains are organized into a β-sheet floor that supports α-helical domains, forming a groove structure that is at least superficially similar to the grooves found in MHC-encoded antigen-presenting proteins (Fig. 1). However, the CD1 groove is comprised of two large and deep pockets, A′ and F′, rather than a shallow one, so that the CD1 groove is much larger in its overall volume, having a surface area of 1390 Å2 compared to approximately 850 Å2 for a typical MHC class I groove. In addition, the mCD1d groove is partially closed at the top and at both ends, so that solvent molecules are thought to access the interior of the groove only over a relatively narrow portal above the F′ pocket. Importantly, the inner surface of the CD1 groove is lined almost exclusively by non-polar amino acids, providing a hydrophobic surface for interaction with the aliphatic hydrocarbon chains of the small amphipathic glycolipids that it presents. Although human CD1 isoforms have not yet been crystallized, they also contain a high percentage of non-polar amino acids in the α1 and α2 domains, particularly at locations that are predicted to line the interior of the groove based on homology to murine CD1.4,5
The depth, enclosed nature and hydrophobicity of the interior of the CD1 groove make it well adapted to carry out its proposed function in antigen presentation by sequestering the hydrocarbon chains of amphipathic glycolipids within the globular head of the CD1 protein. Therefore, the free energy of desolvation derived from the insertion of lipid tails into the hydrophobic groove contributes to CD1 binding to glycolipids. This contrast with peptides, which rely mainly on electrostatic and hydrogen bonds for interactions with the shallow pockets on the surface of MHC-encoded antigen-presenting molecules. The ability of CD1 proteins to bind directly the glycolipids that activate T cells has been demonstrated by eluting glycolipids from cellular CD1 proteins and by in vitro assays of lipid binding to recombinant CD1b and CD1d proteins.10–12 In addition, CD1d-multimers loaded with glycolipid antigens can directly bind to the surface of CD1d-restricted T cells, providing strong evidence that CD1–glycolipid complexes are the molecular targets of the T-cell response.13,14
Insertion of the aliphatic hydrocarbon chains of the antigen into the hydrophobic CD1 groove is likely to position the rigid and hydrophilic elements of the antigen on the α-helical surface of the groove, so that they are available to interact with T-cell antigen receptors.15,16 This mode of binding suggests that the hydrophilic cap of the antigen, including the carbohydrate, phosphate, or polar substitution of the lipid, are likely to determine the specificity of interactions of the antigen with T-cell receptors (TCR). Consistent with this model, studies of the fine specificity of CD1b-, CD1d- and CD1c-presented glycolipids confirm that T cells can discriminate among glycolipids that vary only in the orientation of a single hydroxyl on the carbohydrate portion of the antigen.17–19
Presentation of synthetic α-galactosylceramides by CD1d
Perhaps the most extensively studied CD1-presented antigens are α-galactosyl ceramide, a glycolipid that is naturally produced by marine sponges.19 This antigen was discovered by Taniguchi and colleagues using a screen for compounds with anti-tumour effects, and it was subsequently demonstrated that it potently activated CD1d-restricted natural killer (NK) T cells.19,20 The α-galactosyl-ceramide-reactive NK T-cell population is of great interest because these T cells secrete large amounts of interleukin-4 (IL-4) and interferon-γ (IFN-γ) in the absence of prior priming, and are thought to regulate early events in the generation of peptide antigen-specific immune responses.21,22 Indeed, administration of α-galactosyl ceramide as a pharmacological agent has potent effects in models of tumour immunity, autoimmunity and infectious diseases.20,23–28
Mammalian cells produce glycosyl ceramides (cerebrosides and related compounds) that are related in structure to synthetic α-galactosyl ceramide antigens (Fig. 2). However, these natural mammalian glycolipids are not known to activate NK T cells, a finding that contributes to the central biological mystery of this system: what, if any, relationship does this marine-sponge-derived glycolipid have with natural glycolipids that may control NK T-cell responses in vivo?19 One hypothesis is that α-galactosyl ceramide is structurally homologous to an as yet undiscovered mammalian glycolipid that serves as an intercellular signalling molecule for CD1d-expressing antigen-presenting cells (APCs), allowing them to communicate with immunoregulatory NK T cells. Therefore, it is useful to consider the precise molecular features of synthetic α-galactosyl ceramides that distinguish them from non-antigenic monoglycosyl ceramides naturally produced by mammalian cells.
Figure 2.
Natural and synthetic glycosyl sphingolipids (GSL). Synthetic antigens for NK T cells have an α-anomeric linkage and are comprised of phytosphingosines, whereas natural GSLs found in mammalian cells typically have a β-anomeric linkage and are usually made from sphingosines.
Natural mammalian monoglycosyl ceramides differ from synthetic NK T-cell antigens in that the former are typically composed of a sphingosine base, whereas the latter are produced from phytosphingosines. That is, non-antigen mammalian glycolipids have a double bond between C4 and C5 rather than a hydroxyl group at C4 (Fig. 2). NK T cells show little or no activation by synthetic analogues that lack the hydroxyl groups at C3 and C4, demonstrating the functional importance of this subtle chemical alteration on controlling the T-cell response.19,29 A second chemical feature that distinguishes non-antigenic mammalian ceramides from NK T-cell antigens is the stereochemistry of the anomeric linkage of the carbohydrate to the lipid (Fig. 2). Most mammalian glycosyl ceramides have carbohydrates that are β-anomerically linked to the sphingosine base, whereas only α-linked synthetic compounds have been shown to be stimulatory for T cells.19 Therefore, although the chemical structures of endogenous ceramides for NK T cells are not known, these observations suggest the possibility that cellular events which alter the enzymatic pathways which control the oxidation and glycosylation state of glycosyl ceramides could serve as the initial stimulus for NK T-cell activation in vivo.
TCR and antigen variability
Early studies of T-cell specificity for glycolipid antigen demonstrated that various CD1-restricted T-cell populations recognized individual glycolipid antigens without cross-reactivity.17,30,31 This implied that a clonally distributed receptor on T cells mediates antigen recognition. More recent studies have shown that T-cell recognition of glycolipid antigens can be conferred by insertion of certain TCR chains into the germ-line of mice or by transfecting CD3low T lymphoblastoma cells with TCRα and β chains.16,19,32 These studies prove that clonally variable regions of the TCR function in antigen recognition. However, the extent to which the TCR repertoire of CD1-restricted T cells varies has not been established and is a current topic of interest because it has implications for the role of CD1-restricted T cells as effectors of innate required immunity.
Many studies have shown that human and murine NK T cells circulate in relatively large numbers and populate certain somatic tissues, particularly the liver and other gastrointestinal organs.33–37 Many of these T cells express what have been called canonical, invariant, or semi-invariant TCRs, which in mice are Vα14Jα281 typically paired with Vβ2, Bβ7, or Vβ8, and humans use Vα24JαQ genes.37,38 The widely used but oxymoronic term ‘semi-invariant’ is meant to convey that the TCRs of NK T cells are not identical and are therefore not derived from a single parental clone, but they do show marked skewing of TCR gene usage, which has now been shown to result from the positive selection of these cells on the non-polymorphic CD1d protein.39–41 NK T cells expressing the semi-invariant TCRs can be activated in bulk by synthetic α-galactosyl ceramides and structurally related synthetic glycolipids. Semi-invariant NK T cells are normally present in fairly large numbers and have a markedly limited receptor and antigen repertoire, semi-invariant TCRs have been compared to receptors that mediate innate immune responses.42 This T-cell population is thought to perform a stereotyped or immunoregulatory function early in immune responses, though the precise function of these T cells in innate immunity remains to be defined.
In addition to α-galactosyl ceramide reactive NK T-cell populations, there is now clear evidence for the existence of many T-cell populations that are restricted by CD1a, CD1b, CD1c and CD1d that express varied TCRs and recognize many glycolipid antigens that are not structurally related to α-glycosyl ceramides (Fig. 3). Several laboratories have described CD1d-restricted T cells with diverse TCR gene usage, including rearranged TCRα chains with apparently random incorporation variable region gene segments.43,44 Whether these varied TCRs mediate recognition of a wide array of glycolipids remains to be determined.45 However, murine T-cell hybridomas can be activated by phosphatidylinositol and phosphatidylethanolamine in vitro.46 Furthermore, there is no evidence for conserved TCR gene usage in T cells that recognize group 1 CD1 proteins. CD1a-, CD1b- and CD1c-restricted T-cell clones express receptors composed of varied Vα and Vβ gene segments and N-region additions.16 These TCR genes differed from the semi-invariant TCRs of human NK T cells (Va24) and also differ from one another.
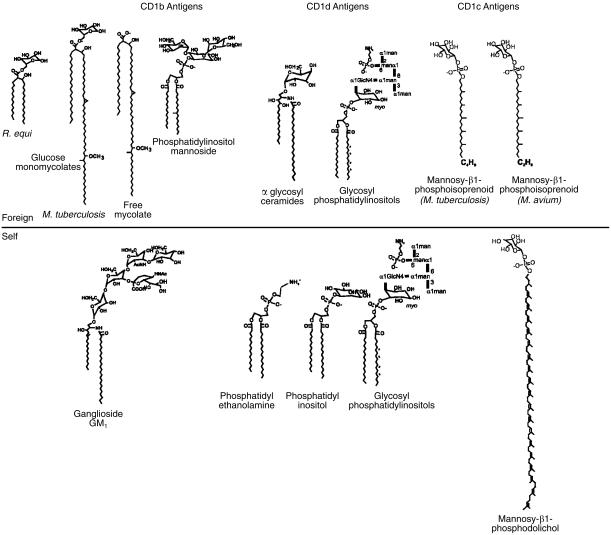
Figure 3.
CD1-presented glycolipid antigens. The CD1-presented glycolipid antigens of known structure are listed according to whether they were derived from microbial or mammalian sources.10,17–19,30,31,46,55 PIM containing two mannosyl residues is depicted, but antigenic PIMs typically have additional mannosyl residues.
In addition, the list of known glycolipid targets of CD1-mediated immune responses continues to grow (Fig. 3).15 Four of the human CD1 isoforms have now been shown to present glycolipid antigens to T cells, and some CD1 isoforms have been shown to present more than one class of glycolipids. For example, CD1b proteins can bind and present glycolipids containing mycolate, diacylglycerol or sphingolipid moieties, demonstrating that despite the lack of polymorphism in the structure of antigen-binding regions of CD1, a single isoform can present at least three major classes of cellular glycolipids (Fig. 3).17,30,31,47 Moreover, given the large variety of known glycosylation patterns of glycolipids within each of these classes, it is possible that the number of glycolipid antigens recognized by CD1-restricted T cells is much greater than is currently known. Therefore, considered as a whole, the CD1-restricted T-cell repertoire represents multiple populations of cells that respond without cross-reactivity to a variety of glycolipid ligands. These glycolipid antigens differ in their expression among different organisms, vary in their pathways of biosynthesis and accumulate in different cell types and disease states.48–51 Thus, it is unlikely that CD1-restricted T cells have a single housekeeping function, but instead they probably have diverse functions in immune response that have only recently come to be appreciated.15,52
Foreign microbial glycolipid antigens
All three of the group 1 CD1 proteins are now known to mediate T-cell recognition of glycolipid components from the mycobacterial cell wall. Porcelli, Brenner and colleagues discovered the first-known CD1-presented antigen by study of CD1b-restricted T cells that were activated by Mycobacterium tuberculosis-derived antigens.53 The stimulatory cell wall component was found to be an α-branched, β-hydroxy fatty acid with very long alkyl chains (C70–90) known as mycolic acid, so named because it is produced only by mycobacteria and closely related actinomyces species.31,49 This CD1b-presented antigen, and two others discovered shortly thereafter, lipoarabinomannan (LAM) and glucose monomycolate (GMM), do not have readily identifiable structural homologues in mammalian cells (Fig. 3).17,30 Thus, they may be considered intrinsically foreign to the mammalian immune system. Likewise the recently identified CD1c-presented antigens are mycobacterial mannosyl-β-1-phosphoisoprenoids (MPI), which possess an unusual polymethylated alkyl chain that is only known to occur in mycobacteria (Fig. 3).18 These mycobacterial polyisoprenoid antigens are homologous to mammalian mannosyl phosphodolichols. However, the antigenic mycobacterial lipids have a shorter prenyl moiety that is C30, rather than C95 dolichol which is found in mammalian cells. In fact, many microbes, including pathogenic protozoa and fungi, produce acyclic polyisoprenols, which are shorter than those found in mammalian cells, suggesting that naturally occurring differences in prenyl length that distinguish self from foreign polyprenols could control CD1c-mediated T-cell responses (Fig. 3).51 In addition to these antigens of well-defined structure, functional evidence suggests that CD1a-restricted T cells can also respond to lipid components of the mycobacterial cell wall.54
The role of group 2 CD1 proteins in the presentation of microbial glycolipids during infection is less clear. Natural and synthetic glycosyl phosphatidylinositols (GPI), including those from pathogenic protozoa, can activate CD1d-restricted T cells.55 Although the immunizing effect of GPIs on murine antibody production in vivo has been challenged, synthetic GPIs do appear to activate CD1d-restricted T cells in vitro, and natural phospholipids containing phosphatidylinositol units have been eluted directly from cellular CD1d proteins.10,56,57 In addition, purified phosphatidylinositol (PI), which is the core structure of GPI, can bind to recombinant CD1d proteins and activate CD1d-restricted T-cell hybridomas.46 Taken together, these studies indicate that GPI and other glycolipids, which contain a PI core structure, can bind to CD1d proteins and activate T cells in vitro. However, both GPI and PI are abundantly present in mammalian cells, and the structural basis by which T cells could discriminate these ubiquitous self lipids from altered self or microbial glycolipids has not been established.
In addition to these studies, many studies have shown that in vivo activation of NK T cells in bulk by the systemic administration of large amounts α-galactosyl ceramides alters the outcome of bacterial, protozoal and viral infections. These strong effects are probably due to the massive release of IFN-γ and other cytokines by NK T cells.27,28,58 These studies provide useful information about the potential therapeutic applications of α-galactosyl ceramides. However, since the natural homologue of α-galactosyl ceramide is unlikely to be released in such a rapid and systemic manner, these studies do not constitute evidence for a natural function of CD1d in microbial infections, a question that can be investigated by experimental infections of animals lacking CD1d proteins.59
Mammalian glycolipids as self-antigens for T cells
Recently, several studies have indicated that CD1-restricted T cells can be activated by treating APCs with large doses of mammalian glycolipids of normal structure. For example, DeLibero and colleagues have identified human T-cell clones that recognize CD1b-expressing APCs only when they have been previously treated with high concentrations of mammalian gangliosides, including GM1 ganglioside.47 In addition, CD1d-restricted murine T-cell hybridomas can be activated by APCs treated with phosphatidylinositol or by recombinant CD1d proteins complexed to PI or phosphatidylethanolamine.46 Furthermore, CD1c-restricted T cells that are activated by mycobacterial MPI also cross-react with certain di-trans poly-cis mannosyl phosphodolichols that are typical of eukaryotic cells.18 Thus, at least under certain experimental circumstances, T-cell activation may occur in response to APCs whose lipid content has been altered by adding exogenous mammalian glycolipids of normal structure.
In addition, several groups have described ‘CD1-autoreactive’ T cells that recognize murine CD1d and human CD1a, CD1b, CD1c and CD1d.60–63 Activation of such autoreactive T cells requires that APCs express CD1 proteins, but does not require that an exogenous antigen be added to APC cultures. These CD1 autoreactive cell populations do not generally cross-react with more than one CD1 isoform, demonstrating the specificity of the response for the structure of the CD1 protein or possibly the structure of an endogenous glycolipid bound to CD1. It remains formally possible that the molecular targets of some autoreactive T cells are unliganded CD1 proteins or CD1–glycolipid complexes in which the glycolipid does not contribute to the specificity of the response. However, evidence suggests that autoreactive T cells recognize CD1 proteins that are complexed to endogenous self antigens and that the structure of the antigen determines the T-cell response. This has been most clearly demonstrated for invariant NK T cells, which were originally described as being autoreactive to CD1d proteins, but are now known to be strongly activated by CD1d-α-galactosyl ceramide complexes and are indeed quite specific for the structure of the α-galactosyl ceramide unit.12,19,21,29
Glycolipid processing by cleavage of covalent bonds
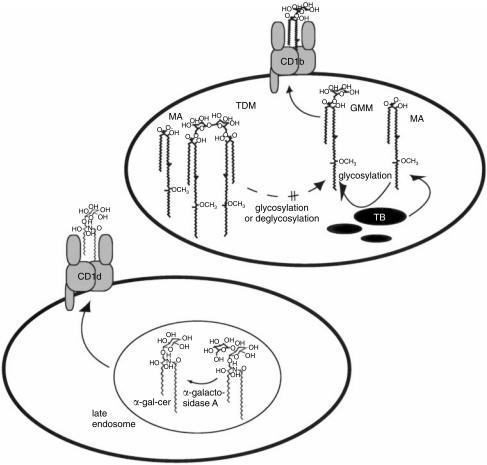
The role of alterations in glycolipid structure that occur during antigen-processing reactions is not yet well understood. However, two recent reports have clearly demonstrated that APCs can alter the structure of glycolipids by glycosylation or glycan trimming in ways that affect their recognition by T cells.32,64 GMM is a CD1b-presented antigen that is composed of long-chain mycobacterial mycolates esterified via the sixth carbon to glucose (Fig. 3 and Fig. 4).17 Mycobacteria cannot produce this glycolipid antigen when growing in the absence of an exogenous source of glucose, but they can acquire glucose from host tissues and couple it to their own mycolates to produce antigenic GMM during infection of glucose-rich mammalian tissues.32,65 This glycosylation reaction is presumably completed by a mycobacterial enzyme, since human dendritic cells cannot glucosylate free mycolic acid to generate GMM.17 In addition, dendritic cells cannot efficiently cleave larger mycolyl glycolipids, such as cord factor and trehalose monomycolate to generate antigenic GMM within activated human dendritic cells, even though these larger glycolipids contain antigenic GMM as a substructure (Fig. 4).17,66 Taken together, these studies support the hypothesis that GMM is a hybrid antigen produced from glycosylation of mycobacterial mycolates with host-derived glucose using a mycobacterial mycolyltransferase, which remains to be identified. Since this reaction requires precursors of both host and pathogen origin, GMM may be thought of as an antigen that is selectively expressed during mycobacterial infection and not in saprophytic mycobacteria growing in the environment (Fig. 4).
Figure 4.
Glycolipid antigen-processing reactions in cells. Treatment of activated macrophages with trehalose monomycolate, or trehalose dimycolate (TDM, cord factor) does not lead to the activation of glucose monomycolate (GMM) -specific T cells, even though these larger glycolipids contain antigenic GMM as a substructure.17,66 In addition, activated macrophages cannot efficiently glucosylate free mycolic acid (MA) to yield GMM in the absence of mycobacteria. However, mycobacteria growing within cells or tissues can capture glucose from host cells to generate antigenic GMM using a mycolyltransferase that remains to be identified.32 Dendritic cells can use α-galactosidase A to cleave the terminal galactosyl residue of synthetic galactosyl (α1 → α2) α-galactosyl ceramide to generate antigenic α-galacosyl ceramide within the late endosomes or lysosomes of antigen-presenting cells.19,64
In contrast to GMM antigens, that could not be generated from larger natural precursors, a recent study by Prigozy and colleagues has demonstrated that covalent cleavage of terminal glycosyl residues of α-glycosyl ceramides within APCs can reveal antigenic T-cell epitopes.64 The non-antigenic, synthetic disaccharide Gal(α1–2)GalCer required the removal of the terminal, two-linked galactosyl residue to generate antigenic α-galactosyl ceramides. This process was mediated by a lysosomal enzyme, α-galactosidase A, allowing the generation of the antigenic monosaccharide epitope within late endosomal or lysosomal compartments of APCs (Fig. 4). In addition, lysosomes are rich in acid-hydrolases that could carry out analogous processing reactions on neutral glycolipids in this compartment, which is thought to be the sight of loading of antigens onto CD1b and CD1d proteins.45,63 These studies suggest that APCs may influence T-cell responses through enzymatically controlled glycosylation and de-glycosylation reactions.
For protein antigens, a key part of cellular processing reactions involves the cleavage of very large proteins with hundreds of amino acids into small peptides of 8–20 amino acids, so that they can fit within the groove of MHC class I or class II proteins.67 Glycolipid processing reactions may also lead to the creation of smaller antigens from larger ones, as is the case for peptides, retaining a lipid moiety that corresponds to the size of the CD1 groove.5 For example, mammalian, microbial diacylglycerols and ceramides have two hydrocarbon chains with an overall length of C30 to C42, a size that corresponds to the combined length of the two alkyl chains in glycosylphosphatidylinositols eluted from CD1d proteins and the predicted volume of the CD1 groove based on computerized analysis of the murine CD1 crystal structure.5,10 However, two classes of antigens, CD1b-presented mycolyl glycolipids and CD1c-presented dolichyl glycolipids, have alkyl chain lengths in the range of C70–80 and C90–100, respectively, which greatly exceed the predicted size of the CD1 groove.17,18,31 If these antigens are presented by insertion of the lipid into the groove, then this apparent problem could be solved either by trimming the length of the lipid chains via possibly a free-radical process or by allowing the lipid to protrude from the groove.
Immune discrimination of self from foreign glycolipids
Activation of CD1-restricted T cells by microbial glycolipids of intrinsically foreign structure suggests that CD1-restricted T cells, particularly those that recognize antigens presented by group 1 CD1 proteins, function in host defence. Consistent with this hypothesis, it has been demonstrated that human CD1-restricted T cells possess effector mechanisms, such as IFN-γ secretion, cytolysis and granulysin production, which are known to have antimicrobial effects in vivo.54,68,69 Furthermore, we have recently reported that CD1c-restricted lipid-specific T-cell responses have been detected at higher levels in the peripheral blood of M. tuberculosis patients compared to control subjects, indicating that CD1c-restricted T cells are activated during the natural course of M. tuberculosis infections in humans.18 However, CD1b, and to some extent CD1c, can only present microbial glycolipids after they have been taken up into mammalian APCs and loaded onto CD1c in endosomal compartments.31,61 Therefore, these foreign lipids are probably intermixed with the much larger pool of self glycolipids prior to loading onto CD1 proteins.
Since T-cell activation by abundant self glycolipids of normal structure would lead to autoimmunity, it is widely assumed that immune mechanisms exist which promote the selective presentation and recognition of foreign or altered self glycolipids in preference to the larger pool of self glycolipids. This premise underlies most models of CD1 function in infection, autoimmunity, tumour immunity and immunosurviellance. Although a strong theoretical rationale underlies this speculation, there is remarkably little direct information regarding the particular chemical features of altered self or foreign glycolipids that allow them to be distinguished from self glycolipids. The regulation of T-cell activation by particular classes of glycolipids may occur by controlling the production of glycolipids, antigen-processing reactions, glycolipid loading onto CD1 proteins, co-stimulatory pathways or shaping of the T-cell repertoire by selection events.
Since peptide antigen presentation pathways by MHC-encoded proteins are understood in such remarkable detail, they provide a framework for studies of glycolipid antigen presentation pathways. However, certain basic principles of peptide antigen presentation are unlikely to be applicable to glycolipids due to the important differences in the biophysical properties of these molecules. Peptide presentation pathways concern antigens that move within aqueous compartments separated by membranes, and lipid antigen presentation is about understanding how antigens move within membranes that are separated by aqueous compartments. Since the concentrations of glycolipid antigens required for T-cell activation generally exceed their critical micellar concentrations, glycolipid antigens aggregate into lipid–protein complexes and membranes within cells. CD1 antigen presentation is not controlled by TAP transporters, but instead is likely to be regulated by lipid-transport proteins, flippases and the like.59
Peptide antigens are soluble as monomers at high concentrations in aqueous solutions, so their binding to MHC-encoded antigen-presenting molecules can be accurately measured and interpreted, both in vivo and in vitro, with relatively simple kinetic models.70 Although in vitro binding studies of glycolipid antigens with CD1 proteins conducted in aqueous buffers have been extremely useful for demonstrating the existence of CD1–glycolipid complexes, these methods are unlikely to recapitulate the substantially more complex phase behaviour of glycolipid interactions in membranes within cells.11,12 Therefore, the chemical features that control the hierarchy of presentation of particular classes of glycolipid antigens and other more subtle questions related to antigen loading are likely to come from cell-based studies in which potentially relevant protein cofactors and membranes are present.
The discovery of lipid antigens for T cells prompted one prominent editorialist to lament, ‘Now, just when… [immunologists] are becoming comfortable with high-performance liquid chromatography, mass spectrometry and protein crystallography…to ensure a place on the fast track, the modern immunologist needs to fret about fat.’.71 Coming to grips with the complex, multiphase behaviour of lipids in cells represents a technical challenge, but also a remarkable opportunity to understand the immune recognition of a whole new class of antigens that are tucked into the membranes of mammalian cells.
Acknowledgments
D. B. Moody is supported by grants from the NIH-NIAMS (AR01988), NIH-NIAID (AI49313) and an American College of Rheumatology Research and Education Foundation Investigator Award. G. S. Besra, a Lister Institute–Jenner Research Fellow, acknowledges support from The Medical Research Council (Co-operative Group Grant Number G9901077 (ID 49343) and Component Project G0000895), The Wellcome Trust (060750) and NIH (AI-48933).
References
- 1.Cresswell P, Howard JC. Antigen recognition. Curr Opin Immunol. 1997;9:71–4. doi: 10.1016/s0952-7915(97)80161-6. [DOI] [PubMed] [Google Scholar]
- 2.Garcia KC, Degano M, Stanfield RL, Brunmark A, Jackson MR, Peterson PA, Teyton L, Wilson IA. An alpha beta T cell receptor structure at 2.5 Å and its orientation in the TCR-MHC complex. Science. 1996;274:209–19. doi: 10.1126/science.274.5285.209. [DOI] [PubMed] [Google Scholar]
- 3.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–41. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 4.Porcelli SA. The CD1 family: a third lineage of antigen-presenting molecules. Adv Immunol. 1995;59:1–98. doi: 10.1016/s0065-2776(08)60629-x. [DOI] [PubMed] [Google Scholar]
- 5.Zeng Z, Castaño AR, Segelke BW, Stura EA, Peterson PA, Wilson IA. Crystal Structure of Mouse CD1: an MHC-Like Fold with a Large Hydrophobic Binding Groove. Science. 1997;277:339–45. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 6.Calabi F, Milstein C. A novel family of human major histocompatibility complex-related genes not mapping to chromosome 6. Nature. 1986;323:540–3. doi: 10.1038/323540a0. [DOI] [PubMed] [Google Scholar]
- 7.Angenieux C, Salamero J, Fricker D, Cazenave JP, Goud B, Hanau D, de la Salle H. Characterization of CD1e, a third type of CD1 molecule expressed in dendritic cells. J Biol Chem. 2000;275(48):37757–64. doi: 10.1074/jbc.M007082200. 10.1074/jbc.m007082200. [DOI] [PubMed] [Google Scholar]
- 8.Mirones I, Oteo M, Parra-Cuadrado JF, Martinez-Naves E. Identification of two novel human CD1E alleles. Tissue Antigens. 2000;56(2):159–61. doi: 10.1034/j.1399-0039.2000.560208.x. 10.1034/j.1399-0039.2000.560208.x. [DOI] [PubMed] [Google Scholar]
- 9.Calabi F, Jarvis JM, Martin L, Milstein C. Two classes of CD1 genes. Eur J Immunol. 1989;19:285–92. doi: 10.1002/eji.1830190211. [DOI] [PubMed] [Google Scholar]
- 10.Joyce S, Woods AS, Yewdell JW, De Bennink JRSA, Boesteanu A, Balk SP, Cotter RJ, Brutkiewicz RR. Natural ligand of mouse CD1d: cellular glycosylphosphatidylinositol. Science. 1998;279:1541–4. doi: 10.1126/science.279.5356.1541. [DOI] [PubMed] [Google Scholar]
- 11.Ernst WA, Maher J, Cho S, et al. Molecular interaction of CD1b with lipoglycan antigens. Immunity. 1998;8:331–40. doi: 10.1016/s1074-7613(00)80538-5. [DOI] [PubMed] [Google Scholar]
- 12.Naidenko O, Maher J, Ernst DN, Sakai T, Modlin R, Kronenberg M. Binding and antigen presentation of ceramide-containing glycolipids by soluble mouse and human CD1d molecules. J Exp Med. 1999;190:1069–80. doi: 10.1084/jem.190.8.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med. 2000;191(11):1895–903. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang CR, Koezuka Y, Kronenberg M. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192(5):741–54. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moody DB, Besra GS, Wilson IA, Porcelli SA. The molecular basis of CD1-mediated presentation of lipid antigens. Immunol Rev. 1999;172:285–96. doi: 10.1111/j.1600-065x.1999.tb01373.x. [DOI] [PubMed] [Google Scholar]
- 16.Grant EP, Degano M, Rosat JP, Stenger S, Modlin RL, Wilson IA, Porcelli SA, Brenner MB. Molecular recognition of lipid antigens by T cell receptors. J Exp Med. 1999;189:195–205. doi: 10.1084/jem.189.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moody DB, Reinhold BB, Guy MR, et al. Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science. 1997;278:283–6. doi: 10.1126/science.278.5336.283. [DOI] [PubMed] [Google Scholar]
- 18.Moody DB, Ulrichs T, Muhlecker W, et al. CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature. 2000;404:884–8. doi: 10.1038/35009119. [DOI] [PubMed] [Google Scholar]
- 19.Kawano T, Cui J, Koezuka Y, et al. CD1d-restricted and TCR-mediated activation of Valpha14 NKT cells by glycosylceramides. Science. 1997;278(5343):1626–9. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 20.Kawano T, Nakayama T, Kamada N, et al. Antitumor cytotoxicity mediated by ligand-activated human Valpha24 NKT cells. Cancer Res. 1999;59:5102–5. [PubMed] [Google Scholar]
- 21.Bendelac A, Lantz O, Quimby ME, Yewdell JW, Bennink JR, Brutkiewicz RR. CD1 recognition by mouse NK1+ T lymphocytes. Science. 1995;268:863–5. doi: 10.1126/science.7538697. [DOI] [PubMed] [Google Scholar]
- 22.Yoshimoto T, Bendelac A, Watson C, Hu-Li J, Paul WE. Role of NK1.1+ T cells in a TH2 response and in immunoglobulin E production. Science. 1995;270:1845–7. doi: 10.1126/science.270.5243.1845. [DOI] [PubMed] [Google Scholar]
- 23.Shin T, Nakayama T, Akutsu Y, et al. Inhibition of tumor metastasis by adoptive transfer of IL-12-activated Valpha14 NKT cells. Int J Cancer. 2001;91:523–8. doi: 10.1002/1097-0215(20010215)91:4<523::aid-ijc1087>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 24.Moodycliffe AM, Nghiem D, Clydesdale G, Ullrich SE. Immune suppression and skin cancer development: regulation by NKT cells. Nat Immunol. 2000;1(6):521–5. doi: 10.1038/82782. 10.1038/82782. [DOI] [PubMed] [Google Scholar]
- 25.Terabe M, Matsui S, Noben-Trauth N, et al. NKT cell-mediated repression of tumor immunosurveillance by IL-13 and the IL-4R-STAT6 pathway. Nat Immunol. 2000;1(6):515–20. doi: 10.1038/82771. [DOI] [PubMed] [Google Scholar]
- 26.Oishi Y, Sumida T, Sakamoto A, et al. Selective reduction and recovery of invariant Valpha24JalphaQT, cell receptor T cells in correlation with disease activity in patients with systemic lupus erythematosus. J Rheumatol. 2001;28:275–83. [PubMed] [Google Scholar]
- 27.Gonzalez-Aseguinolaza G, de Oliveira C, Tomaska M, et al. Alpha-galactosylceramide-activated Valpha14 natural killer T cells mediate protection against murine malaria. Proc Natl Acad Sci USA. 2000;97(15):8461–6. doi: 10.1073/pnas.97.15.8461. 10.1073/pnas.97.15.8461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kawakami K, Kinjo Y, Yara S, Koguchi Y, Uezu K, Nakayama T, Taniguchi M, Saito A. Activation of Valpha14 (+) natural killer T cells by alpha-galactosylceramide results in development of Th1 response and local host resistance in mice infected with Cryptococcus neoformans. Infect Immun. 2001. pp. 213–20. [DOI] [PMC free article] [PubMed]
- 29.Brossay L, Naidenko O, Burdin N, Matsuda J, Sakai T, Kronenberg M. Structural requirements for galactosylceramide recognition by CD1-restricted NK T cells. J Immunol. 1998;161:5124–8. [PubMed] [Google Scholar]
- 30.Sieling PA, Chatterjee D, Porcelli SA, et al. CD1-restricted T cell recognition of microbial lipoglycan antigens. Science. 1995;269:227–30. doi: 10.1126/science.7542404. [DOI] [PubMed] [Google Scholar]
- 31.Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1-restricted alpha-beta+ T cells. Nature. 1994;372:691–4. doi: 10.1038/372691a0. [DOI] [PubMed] [Google Scholar]
- 32.Moody DB, Guy MR, Grant EP, Brenner MB, Besra GS, Porcelli SA. CD1b-mediated T cell recognition of a glycolipid antigen generated from mycobacterial lipid and host carbohydrate during infection. J Exp Med. 2000;192:965–76. doi: 10.1084/jem.192.7.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bleicher PA, Balk SP, Hagen SJ, Blumberg RS, Flotte TJ, Terhorst C. Expression of murine CD1 on gastrointestinal epithelium. Science. 1990;250:679–82. doi: 10.1126/science.1700477. [DOI] [PubMed] [Google Scholar]
- 34.Lantz O, Bendelac A. An invariant T cell receptor alpha chain is used by a unique subset of major histocompatibility complex class I-specific CD4+ and CD4−8− T cells in mice and humans. J Exp Med. 1994;180:1097–106. doi: 10.1084/jem.180.3.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacDonald HR. NK1.1+ T cell receptor-alpha-beta+ cells: new clues to their origin, specificity, and function. J Exp Med. 1995;182:633–8. doi: 10.1084/jem.182.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blumberg RS, Gerdes D, Chott A, Porcelli SA, Balk SP. Structure and function of the CD1 family of MHC-like cell surface proteins. Immunol Rev. 1995;147:5–29. doi: 10.1111/j.1600-065x.1995.tb00085.x. [DOI] [PubMed] [Google Scholar]
- 37.Porcelli S, Gerdes D, Fertig AM, Balk SP. Human T cells expressing an invariant V alpha24-JalphaQ TCR alpha are CD4- and heterogeneous with respect to TCR beta expression. Hum Immunol. 1996;48:63–7. doi: 10.1016/0198-8859(96)00090-0. 10.1016/0198-8859(96)00090-0. [DOI] [PubMed] [Google Scholar]
- 38.Koseki H, Imai K, Nakayama F, Sado T, Moriwaki K, Taniguchi M. Homogenous junctional sequence of the V14+ T-cell antigen receptor alpha chain expanded in unprimed mice. Proc Natl Acad Sci USA. 1990;87:5248–52. doi: 10.1073/pnas.87.14.5248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smiley ST, Kaplan MH, Grusby MJ. Immunoglobulin E production in the absence of interleukin-4-secreting CD1-dependent cells. Science. 1997;275:977–9. doi: 10.1126/science.275.5302.977. [DOI] [PubMed] [Google Scholar]
- 40.Mendiratta SK, Martin WD, Hong S, Boesteanu A, Joyce S, Van Kaer L. CD1d mutant mice are deficient in natural T cells that promptly produce IL-4. Immunity. 1997;6:469–77. doi: 10.1016/s1074-7613(00)80290-3. [DOI] [PubMed] [Google Scholar]
- 41.Chen YH, Chiu NM, Mandal M, Wang N, Wang CR. Impaired NK1+ T cell development and early IL-4 production in CD1-deficient mice. Immunity. 1997;6:459–67. doi: 10.1016/s1074-7613(00)80289-7. [DOI] [PubMed] [Google Scholar]
- 42.Park SH, Chiu YH, Jayawardena J, Roark J, Kavita U, Bendelac A. Innate and adaptive functions of the CD1 pathway of antigen presentation. Seminars Immunol. 1998;10:391–8. doi: 10.1006/smim.1998.0139. [DOI] [PubMed] [Google Scholar]
- 43.Cardell S, Tangri S, Chan S, Kronenberg M, Benoist C, Mathis D. CD1-restricted CD4+ T cells in major histocompatibility complex class II-deficient mice. J Exp Med. 1995;182:993–1004. doi: 10.1084/jem.182.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park SH, Roark JH, Bendelac A. Tissue-specific recognition of mouse CD1 molecules. J Immunol. 1998;160:3128–34. [PubMed] [Google Scholar]
- 45.Chiu YH, Jayawardena J, Weiss A, Lee D, Park SH, Dautry-Varsat A, Bendelac A. Distinct subsets of CD1d-restricted T cells recognize self-antigens loaded in different cellular compartments. J Exp Med. 1999;189:103–10. doi: 10.1084/jem.189.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gumperz J, Roy C, Makowska A, Lum D, et al. Murine CD1d-restricted T cell recognition of cellular lipids. Immunity. 2000;12:211–21. doi: 10.1016/s1074-7613(00)80174-0. [DOI] [PubMed] [Google Scholar]
- 47.Shamshiev A, Donda A, Carena I, Mori L, De Kappos L, Libro G. Self glycolipids as T-cell autoantigens. Eur J Immunol. 1999;29:1667–75. doi: 10.1002/(SICI)1521-4141(199905)29:05<1667::AID-IMMU1667>3.0.CO;2-U. 10.1002/(sici)1521-4141(199905)29:05<1667::aid-immu1667>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 48.Carroll KK, Guthrie N, Ravi K. Dolichol: function, metabolism, and accumulation in human tissues. Biochem Cell Biol. 1992;70:382–4. doi: 10.1139/o92-059. [DOI] [PubMed] [Google Scholar]
- 49.Barry CE, Lee RE, Mdluli K, Sampson AE, Schroeder BG, Slayden RA, Yuan Y. Mycolic acids: structure, biosynthesis and physiological functions. Prog Lipid Research. 1998;37:143–79. doi: 10.1016/s0163-7827(98)00008-3. [DOI] [PubMed] [Google Scholar]
- 50.Brennan PJ, Besra GS. Structure, function and biogenesis of the mycobacterial cell wall. Biochem Soc Transactions. 1997;25:188–94. doi: 10.1042/bst0250188. [DOI] [PubMed] [Google Scholar]
- 51.Moody DB. Polyisoprenyl glycolipids as targets of CD1-mediated immune responses. Cellular Molec Life Sci. 2001;58 doi: 10.1007/PL00000789. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shinkai K, Locksley RM. CD1, tuberculosis, and the evolution of major histocompatibility complex molecules [comment] J Exp Med. 2000;191:907–14. doi: 10.1084/jem.191.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Porcelli S, Morita CT, Brenner MB. CD1b restricts the response of human CD4–8– T lymphoyctes to a microbial antigen. Nature. 1992;360:593–7. doi: 10.1038/360593a0. [DOI] [PubMed] [Google Scholar]
- 54.Rosat JP, Grant EP, Beckman EM, et al. CD1-restricted microbial lipid antigen-specific recognition found in the CD8+ alpha-beta cell pool. J Immunol. 1999;162:366–71. [PubMed] [Google Scholar]
- 55.Schofield L, McConville MJ, Hansen D, Campbell AS, Fraser-Reid B, Grusby MJ, Tachado SD. CD1d-restricted immunoglobulin G formation to GPI-anchored antigens mediated by NKT cells. Science. 1999;283:225–9. doi: 10.1126/science.283.5399.225. 10.1126/science.283.5399.225. [DOI] [PubMed] [Google Scholar]
- 56.Molano A, Park SH, Chiu YH, Nosseir S, Bendelac A, Tsuji M. Cutting edge: the IgG response to the circumsporozoite protein is MHC class II-dependent and CD1d-independent: exploring the role of GPIs in NK T cell activation and antimalarial responses. J Immunol. 2000;164(10):5005–9. doi: 10.4049/jimmunol.164.10.5005. [DOI] [PubMed] [Google Scholar]
- 57.Romero JF, Eberl G, MacDonald HR, Corradin G. CD1d-restricted NK T cells are dispensable for specific antibody responses and protective immunity against liver stage malaria infection in mice. Parasite Immunol. 2001;23(5):267–9. doi: 10.1046/j.1365-3024.2001.00381.x. [DOI] [PubMed] [Google Scholar]
- 58.Exley MA, Bigley NJ, Cheng O, et al. CD1d-reactive T-cell activation leads to amelioration of disease caused by diabetogenic encephalomyocarditis virus. J Leukoc Biol. 2001;69(5):713–18. [PubMed] [Google Scholar]
- 59.Behar SM, Dascher CC, Grusby MJ, Wang CR, Brenner MB. Susceptibility of mice deficient in CD1D or TAP1 to infection with Mycobacterium tuberculosis. J Exp Med. 1999;189:1973–80. doi: 10.1084/jem.189.12.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sieling PA, Porcelli SA, Duong BT, Spada F, Bloom BR, Diamond B, Hahn BH. Human double-negative T cells in systemic lupus erythematosus provide help for IgG and are restricted by CD1c. J Immunol. 2000;165(9):5338–44. doi: 10.4049/jimmunol.165.9.5338. [DOI] [PubMed] [Google Scholar]
- 61.Jackman RM, Stenger S, Lee A, et al. The tyrosine-containing cytoplasmic tail of CD1b is essential for its efficient presentation of bacterial lipid antigens. Immunity. 1998;8:341–51. doi: 10.1016/s1074-7613(00)80539-7. [DOI] [PubMed] [Google Scholar]
- 62.Porcelli S, Brenner MB, Greenstein JL, Balk SP, Terhorst C, Bleicher PA. Recognition of cluster of differentiation 1 antigens by human CD4–CD8– cytolytic T lymphocytes. Nature. 1989;341:447–50. doi: 10.1038/341447a0. [DOI] [PubMed] [Google Scholar]
- 63.Spada FM, Koezuka Y, Porcelli SA. CD1d-restricted recognition of synthetic glycolipid antigens by human natural killer T cells. J Exp Med. 1998;188:1529–34. doi: 10.1084/jem.188.8.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Prigozy TI, Naidenko O, Qasba P, et al. Glycolipid antigen processing for presentation by CD1d molecules. Science. 2001;291:664–7. doi: 10.1126/science.291.5504.664. 10.1126/science.291.5504.664. [DOI] [PubMed] [Google Scholar]
- 65.Baba T, Natsuhara Y, Kaneda K, Yano I. Granuloma formation activity and mycolic acid composition of mycobacterial cord factor. Cellular Mol Life Sci. 1997;53:227–32. doi: 10.1007/PL00000595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Moody DB, Reinhold BB, Reinhold VN, Besra GS, Porcelli SA. Uptake and processing of glycosylated mycolates for presentation to CD1b-restricted T cells. Immunol Lett. 1999;65:85–91. doi: 10.1016/s0165-2478(98)00129-1. [DOI] [PubMed] [Google Scholar]
- 67.Cresswell P. Assembly, transport, and function of MHC class II molecules. Annu Rev Immunol. 1994;12:259–93. doi: 10.1146/annurev.iy.12.040194.001355. [DOI] [PubMed] [Google Scholar]
- 68.Stenger S, Mazzaccaro RJ, Uyemura K, et al. Differential effects of cytolytic T cell subsets on intracellular infection. Science. 1997;276:1684–7. doi: 10.1126/science.276.5319.1684. 10.1126/science.276.5319.1684. [DOI] [PubMed] [Google Scholar]
- 69.Stenger S, Hanson DA, Teitelbaum R, et al. An antimicrobial activity of cytolytic T cells mediated by granulysin. Science. 1998;282:121–5. doi: 10.1126/science.282.5386.121. [DOI] [PubMed] [Google Scholar]
- 70.Babbitt BP, Allen PM, Matsueda G, Haber E, Unanue ER. Binding of immunogenic peptides to Ia histocompatibility molecules. Nature. 1985;317:359. doi: 10.1038/317359a0. [DOI] [PubMed] [Google Scholar]
- 71.Parham P. Antigen presentation. Chewing the fat. Nature. 1994;372:615–16. doi: 10.1038/372615a0. [DOI] [PubMed] [Google Scholar]