Abstract
The Providencia stuartii AarA protein is a member of the rhomboid family of intramembrane serine proteases and is required for the production of an unknown quorum-sensing molecule. In a screen to identify rhomboid-encoding genes from Proteus mirabilis, tatA was identified as a multicopy suppressor and restored extracellular signal production as well as complementing all other phenotypes of a Prov. stuartii aarA mutant. TatA is a component of the twin-arginine translocase (Tat) protein secretion pathway and likely forms a secretion pore. By contrast, the native tatA gene of Prov. stuartii in multicopy did not suppress an aarA mutation. We find that TatA in Prov. stuartii has a short N-terminal extension that was atypical of TatA proteins from most other bacteria. This extension was proteolytically removed by AarA both in vivo and in vitro. A Prov. stuartii TatA protein missing the first 7 aa restored the ability to rescue the aarA-dependent phenotypes. To verify that loss of the Tat system was responsible for the various phenotypes exhibited by an aarA mutant, a tatC-null allele was constructed. The tatC mutant exhibited the same phenotypes as an aarA mutant and was epistatic to aarA. These data provide a molecular explanation for the requirement of AarA in quorum-sensing and uncover a function for the Tat protein export system in the production of secreted signaling molecules. Finally, TatA represents a validated natural substrate for a prokaryotic rhomboid protease.
In a process known as quorum-sensing, bacterial cells communicate information to each other by using diffusible chemical signals (1–3). This signaling controls biological responses as diverse as antibiotic synthesis, virulence, and biofilm development (1–3). In the Gram-negative human pathogen Providencia stuartii, quorum-sensing represses the expression of an acetyltransferase that modifies peptidoglycan (4, 5). Additional lacZ fusions to various genes were found to be up-regulated by an extracellular signaling molecule in culture supernatants (6). For the above genes, production of this extracellular signal requires AarA, a member of the rhomboid family of intramembrane serine proteases (6–10). In Drosophila, rhomboid proteins cleave the transmembrane domain of the epidermal growth factor receptor ligands, thereby releasing them to activate signaling (10). Drosophila Rhomboid-1 and AarA are able to functionally substitute for each other, indicating that the specificity of these rhomboids has been conserved across evolution (11). However, despite these similarities, the physiological substrate of AarA and the mechanism mediating AarA-dependent quorum-sensing in Prov. stuartii has remained unknown.
In a search for Proteus mirabilis genes that can complement the loss of aarA in Prov. stuartii, the tatA gene in multicopy was found to rescue aarA-dependent phenotypes. The twin-arginine translocase (Tat) system for protein export transports prefolded, cofactor-containing proteins with a twin-arginine motif within the signal sequence (12–14). In Escherichia coli, four proteins, TatA–TatC and TatE, comprise the Tat system, although only three subunits, TatA–TatC, are required for protein export (13, 15). The Tat system also appears to be required for the insertion of certain proteins into the cytoplasmic membrane (16), and it has not previously been implicated in quorum-sensing. Comprehensive reviews of the Tat system have been compiled (17–19).
Unexpectedly, despite complementation by the Prot. mirabilis tatA gene, the native tatA gene from Prov. stuartii did not rescue the phenotypes of an aarA mutant. In this study, we show that Prov. stuartii is one of a small number of bacteria with an extended TatA protein and that AarA is required for the proteolytic activation of TatA by removal of this N-terminal extension. Thus, quorum-sensing in Prov. stuartii requires the Tat protein export system and is activated by rhomboid cleavage. The Prov. stuartii TatA protein also represents a validated natural substrate for a prokaryotic rhomboid protease.
Results
Identification of a High-Copy Suppressor That Restores Extracellular Signal Production to an aarA Mutant of Prov. stuartii.
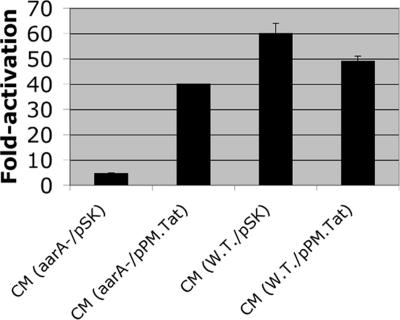
Prov. stuartii aarA mutations are pleiotropic, and the resulting phenotypes include loss of an extracellular signaling molecule, abnormal cell division (cell chaining), inability to grow on MacConkey agar, and loss of a diffusible yellow pigment (6, 7, 20). In a search for rhomboid-like proteins in the related organism Prot. mirabilis, a pACYC184-based genomic library was electroporated into Prov. stuartii XD37.A ΔaarA cma37-lacZ and colonies with restored production of yellow pigment were identified. Plasmids conferring this phenotype contained related inserts, and sequence analysis indicated that the only complete genes present were tatA alone or tatA and tatB, both of which encode products involved in the Tat export system required for secretion of cofactor-containing proteins with twin-arginine residues in the signal peptide (12–14). To determine whether tatA alone also could restore extracellular signal production, a fragment containing only tatA was generated by PCR and cloned to yield pPM.TatPm. The control vector and pPM.TatPm were electroporated into Prov. stuartii XD37.A (ΔaarA, cma37::lacZ), and conditioned medium (CM) was prepared from each strain. To assay CM for signal production, the wild-type strain XD37 containing the cma37::lacZ fusion was used. This lacZ fusion is activated by an AarA-dependent extracellular signal and serves as a biosensor (6). As seen in Fig. 1, CM from XD37.A (ΔaarA) cells containing pPM.TatPm resulted in a 40-fold activation of cma37::lacZ expression, relative to a 4-fold activation in CM from XD37.A (ΔaarA) cells that contained the control vector pSK. The expression of pPM.TatPm in the wild-type background did not increase signal production above normal levels (Fig. 1).
Fig. 1.
Overexpression of tatA from Prot. mirabilis restores signal production to Prov. stuartii aarA mutants. Cultures of Prov. stuartii XD37 wild-type or XD37.A ΔaarA (designated aarA-) containing pBluescript SK(−) (vector) or pPM.tatA containing the tatA gene were grown in LB with 200 μg/ml ampicillin and CM was harvested at an optical density of A600 = 0.9. Signal activity was tested by using Prov. stuartii XD37 cma37-lacZ as a signal biosensor. The values represent fold-activation relative to the same cells grown in control LB and were determined from quadruplicate samples from two independent experiments.
The identification of tatA from Prot. mirabilis as a multicopy suppressor of an aarA mutation was not unique. In a search for E. coli genes that restored aarA mutant phenotypes, we previously reported identification of the rhomboid GlpG (20). However, additional E. coli genes that restored pigment production and extracellular signal were tatA and tatE (data not shown). The TatA and TatE proteins of E. coli are highly similar and functionally redundant (13).
AarA Mutants Are Defective in Tat Function and Rescued by tatA in Multicopy.
The identification of tatA as a high-copy suppressor of the aarA-dependent production of an extracellular signal suggested a close relationship between the AarA rhomboid and the Tat system. Tat-dependent phenotypes that have been observed in E. coli include cell chaining, detergent sensitivity, and the inability to use trimethylamine-N-oxide (TMAO) as an electron acceptor during anaerobic growth with glycerol as a carbon source (21, 22). Prov. stuartii aarA mutants exhibit a prominent chaining phenotype (7, 20) and are similar in this respect to E. coli tat mutants (22). A Prov. stuartii aarA mutant (XD37.A) was therefore tested for the additional Tat-dependent phenotypes and was found to be unable to grow anaerobically on glycerol TMAO plates or aerobically on MacConkey agar plates containing the detergent sodium deoxycholate (Table 1). The presence of pSK.aarA rescued all of these phenotypes indicating that loss of aarA was responsible for these phenotypes (Table 1). In addition, the presence of tatA from Prot. mirabilis (pPM.TatPm) rescued the ability of a Prov. stuartii aarA mutant to grow on MacConkey agar, restored anaerobic growth on TMAO glycerol agar plates, and restored normal cell morphology (Table 1).
Table 1.
Tat phenotypes of various strains
| Strain | MacConkey agar | TMAO agar | Morphology |
|---|---|---|---|
| XD37 wild-type | + | + | Rods |
| XD37.A ΔaarA | − | − | Chains |
| XD37.A ΔaarA/pSK | − | − | Chains |
| XD37.A ΔaarA/pSK.aarA | + | + | Rods |
| XD37.A ΔaarA/pPM.TatAPm | + | + | Rods |
| XD37.A ΔaarA/pBC.SK | − | − | Chains |
| XD37.A ΔaarA/pBC.TatAPs | − | ± | Chains |
| XD37.A ΔaarA/pBC.TatAΔ2-8Ps | + | + | Rods |
| PR50 wild type | + | + | Rods |
| PR51 ΔaarA | − | − | Chains |
| PR50.C tatC::SmR | − | − | Chains |
| PR51.C ΔaarA, tatC::SmR | − | − | Chains |
The data for MacConkey agar indicate normal growth (+) and no growth (−). TMAO agar data were determined after 4 days of incubation at 37°C (± indicates pinpoint colonies). Morphology data were determined by phase contrast microscopy.
Isolation and Expression of the Prov. stuartii tat Operon.
To understand how overexpression of heterologous TatA proteins could suppress an aarA mutation, we isolated the native tat locus from Prov. stuartii. Despite repeated attempts, we were unable to isolate plasmids with the Prov. stuartii tat locus by restored pigment production, a strategy used to identify tatA from Prot. mirabilis and E. coli. Complementation based on restoring growth of an aarA mutant on MacConkey plates also failed. Therefore, an alternative strategy was used based on the location of the tat operon adjacent to the ubiB (formerly yigR) gene in both E. coli and Prov. stuartii. Plasmids that complement the ubiB mutation were isolated from a Prov. stuartii genomic library (see Materials and Methods), and sequence analysis of several clones indicate that genes corresponding to tatABC were present.
Unexpectedly, a clone containing the entire tat operon from Prov. stuartii was unable to rescue the aarA-dependent phenotypes. To determine whether the tatA gene alone from Prov. stuartii could rescue the aarA-dependent phenotypes in a manner seen with tatA from Prot. mirabilis or E. coli, we subcloned only the tatA gene resulting in pBC.TatAPs. However, when pBC.TatAPs was introduced into the Prov. stuartii aarA mutant XD37.A, there was no rescue of pigment production, and cells were still unable to grow on MacConkey agar. In addition, these cells still exhibited the chaining morphology (Table 1). However, the ability of XD37.A/pBC.TatAPs to grow anaerobically on glycerol TMAO plates was partially rescued, and cells formed pinpoint colonies. To test whether the Prov. stuartii TatA protein was functional in a heterologous organism, we introduced this plasmid into a tatA/tatE double mutant of E. coli. The Prov. stuartii TatA protein corrected both the cell-chaining phenotype and the ability to grow on glycerol TMAO plates (data not shown).
Expression of the tat Operon Is Not Dependent on aarA.
To determine whether the basis of suppression of an aarA mutation by increased TatA was due to lowered levels of tat operon expression in an aarA mutant, we examined the accumulation of the tat operon by Northern blot analysis of RNA prepared from wild-type PR50 or PR51 ΔaarA. However, the accumulation of tat operon mRNA was similar in both backgrounds (data not shown).
The Prov. stuartii TatA Protein Is Processed by AarA.
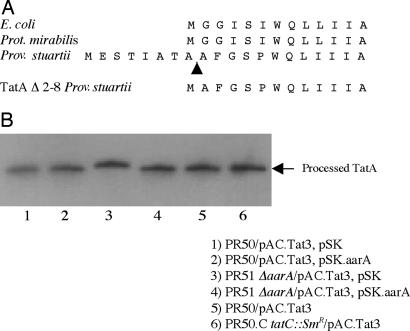
Examination of the TatAPs protein revealed an unusual feature when compared with the Prot. mirabilis TatA (TatAPm) and E. coli (TatAEc) proteins. The TatAPs was 7 aa longer at the N terminus, immediately adjacent to the transmembrane domain (Fig. 2A). In fact, examination of TatA proteins from other organisms in the GenBank database revealed that Prov. stuartii was one of a few organisms containing a TatA protein with a similar extension. This unusually extended transmembrane domain region of TatAPs combined with the inability of the native TatAPs to rescue the aarA mutation prompted us to test the possibility that the TatAPs required cleavage by the AarA protease to remove the extension to function properly. We hypothesized that the TatAPm and TatAEc proteins function in an AarA-independent manner because they are naturally missing this N-terminal extension. To test this possibility, a His6 epitope tag was placed at the C terminus of TatAPs and a pACYC184 derivative encoding the TatAPs-His6 protein (pAC.Tat3) was introduced into wild-type Prov. stuartii PR50 and the isogenic aarA mutant PR51. Cell extracts from the PR50 expressing TatAPs-His6 and PR51 expressing TatAPs-His6 were analyzed by Western blot analysis with an antibody to the His6 tag (Fig. 2B). The predicted molecular mass of the mature TatAPs-His6 protein is 11,250 Da. In PR51 ΔaarA, the size of the TatAPs-His6 hybrid protein was ≈0.5 kDa larger than in wild-type PR50 (Fig. 2B, lanes 1 and 3). Introduction of the aarA gene on a compatible pBluescript SK(−) plasmid into PR51 ΔaarA resulted in a TatAPs-His6 protein that was ≈0.5 kDa smaller and identical in size compared with that seen in wild-type cells (Fig. 2B, lane 4). To determine the basis of the reduced size in wild-type cells, the processed and unprocessed forms of the TatAPs-His6 protein were purified by nickel affinity chromatography. Analysis of the N terminus by Edman degradation revealed that the N-terminal sequence of the smaller processed form of TatA was AFGSPWQLI, an exact match to amino acids 9–17 of TatA (Fig. 2A). The N-terminal sequence of the unprocessed TatA protein yielded the expected sequence of MESTIATA. Therefore, the TatA protein is processed in an AarA-dependent manner by removal of the N-terminal 8 aa.
Fig. 2.
TatA processing in wild-type and an aarA mutant. (A) The TatA proteins from E. coli, Prot. mirabilis, and Prov. stuartii are aligned. The arrowhead designates the site of TatA processing in wild-type Prov. stuartii. Also shown is the sequence of the truncated form of TatA (TatAΔ2-8) that was constructed by site-directed mutagenesis. (B) Total cell protein from the indicated strains was separated on by 16.5% tris/tricine SDS/PAGE and transferred to nitrocellulose. For the Western blot analysis of TatA, filters were probed with an alkaline phosphatase-conjugated His5 antibody.
Purified AarA Cleaves TatA in Vitro.
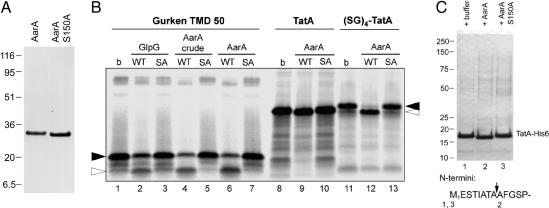
To determine whether the AarA-dependent cleavage of TatA was direct, the ability of purified AarA to process TatA was investigated in vitro. We expressed in E. coli C-terminal His-tagged forms of wild-type AarA and a predicted inactive mutant in which the active site serine (Ser-150) was mutated to alanine (Fig. 3A) (9, 23). The isolated, detergent-solubilized cellular membranes containing AarA showed robust proteolytic activity against a model substrate derived from Drosophila Gurken, whereas, as predicted, no activity was detected for its S150A active-site mutant (Fig. 3B). Next, the detergent-solubilized AarA was purified to homogeneity (Fig. 3A) by nickel affinity chromatography. This purified enzyme maintained its proteolytic activity against Gurken-derived peptide (Fig. 3B). Purified AarA, but not its active-site mutant, caused a minor increase in electrophoretic mobility of the in vitro translated TatA, consistent with the proteolytic removal of the small N-terminal extension (Fig. 3B). To enhance the change in mobility of the processed TatA, we added an artificial N-terminal (SerGly)4 extension to the in vitro translated TatA. As expected, the cleaved product of this extended TatA was more easily distinguished from the uncleaved starting material. Notably, in Fig. 3C, incubation of substoichiometric amounts of purified AarA with purified full-length TatAPs-His6 led to its complete conversion to the faster-migrating species. Chemical protein sequencing confirmed that the cleavage was identical to that in vivo: The N terminus of the TatA cleavage product started with amino acids AFGSPWQLI, whereas incubation of TatAPs-His6 with buffer only or AarA S150A did not lead to any change of the original N terminus (MESTIATA). Therefore the AarA rhomboid protease is necessary and sufficient for TatA proteolytic maturation, and TatA is a direct substrate of AarA.
Fig. 3.
In vitro cleavage of TatA by purified AarA protease. (A) Coomassie-stained SDS/PAGE of NiNTA agarose-purified wild-type AarA-His6 (1 μg) and its active-site mutant S150A (1.5 μg). (B) The model substrate derived from Drosophila Gurken (23), full-length Prov. stuartii TatA and its variant, which were N-terminally extended by four SerGly repeats [(SG)4-TatA], were in vitro translated and labeled with [35S]Met. Substrate peptides were incubated with the indicated purified enzymes or crude detergent-solubilized membranes from E. coli expressing AarA (lanes 4 and 5) for 60 min at 30°C, separated by SDS/PAGE, and autoradiographed (buffer only; SA, GlpG-S201A or AarA-S150A). The sizes of the uncleaved substrate and its C-terminal cleavage product are denoted by filled and open arrowheads, respectively. In all cases, the substrate cleavage by AarA depended on the presence of the predicted catalytic serine residue. Cleavage of the Gurken peptide by purified E. coli rhomboid GlpG is shown for comparison. Reactions shown in lanes 2, 3, 6, 7, 9, 10, 12, and 13 contained 3.8 μg of enzyme per 40 μl of reaction, and those in lanes 4 and 5 contained 5.4 μg of enzyme per 40 μl of reaction. (C) Nickel-affinity chromatography-purified TatA-His6 (190 μg/ml) was incubated in the presence of buffer only (b), purified AarA, or its active site mutant S150A (both at 15 μg/ml) at 30°C for 2.5 h. Mixtures were then separated by SDS/PAGE and stained by Coomassie brilliant blue. Sequences of the N termini of the resulting TatA bands were determined by automated Edman degradation. The AarA cleavage site in Prov. stuartii TatA is indicated by an arrow.
A Truncated Form of TatA Missing Seven N-Terminal Amino Acids Is Able to Restore Tat Function in an AarA-Independent Manner.
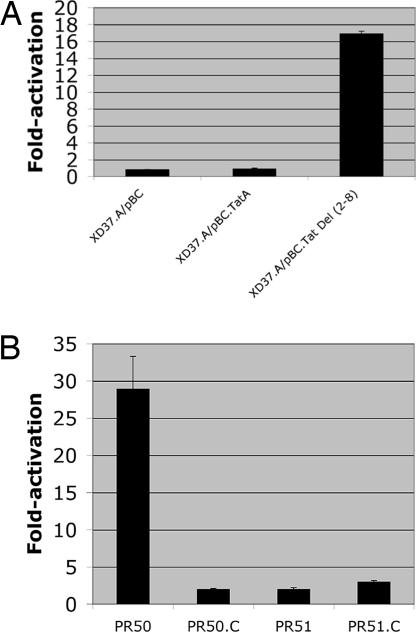
The requirement for AarA in processing the TatAPs protein suggested that the dependence for AarA in Tat function could be bypassed by expressing a TatAPs that was artificially truncated from amino acids 2–8 (Fig. 2A). A plasmid containing this truncated TatAΔ2–8Ps was introduced into the aarA mutant XD.37.A along with the wild-type TatAPs and the vector control. The presence of TatAΔ2–8Ps restored all of the aarA-dependent phenotypes back to wild-type, including pigment production, cell morphology, growth on MacConkey plates, and anaerobic growth on glycerol/TMAO plates (Table 1). In addition, production of the extracellular activating signal was restored by the presence of TatAΔ2–8Ps (Fig. 4A).
Fig. 4.
Role of the Tat transport system in extracellular signal production. The ability of CM from the indicated strains to activate the cma37::lacZ fusion in XD37 is shown. (A) The ability of a TatAPs protein missing amino acids 2–8 to restore signal production is shown. (B) The production of signal from tatC and aarA mutants is shown. For these experiments, CM was harvested at an OD of 0.9. The values represent fold-activation relative to the same cells grown in control LB and were determined from quadruplicate samples from two independent experiments.
Role of the Prov. stuartii Tat System in Extracellular Signal Production.
The original observation that AarA was required for signal production, coupled with the data presented in this study that AarA is required for Tat function, suggested that AarA works through the Tat export system for production of this extracellular signal. To test this possibility, a tatC::SmR mutation was created as described in Materials and Methods; TatC is an essential component of the Tat export system. Strain PR50.C (tatC::SmR) exhibited the same phenotypes as the aarA mutant PR51 (Table 1), although TatA was processed normally in the tatC::SmR background (Fig. 2B, lane 6). CM prepared from the tatC::SmR mutant exhibited a markedly reduced ability to activate the cma37::lacZ fusion (2-fold) relative to CM from wild-type PR50 (29-fold) (Fig. 4B). The level of activation by CM from the tatC::SmR mutant was the same as that observed with CM from the aarA mutant PR51 (2-fold) and from the tatC::SmR, aarA double mutant PR51.C (3-fold). We conclude that the Tat export system has an essential role in Prov. stuartii quorum-sensing.
Discussion
Previously, we identified a role for AarA, a rhomboid-type protease, in the production of an extracellular quorum-sensing molecule in Prov. stuartii (6). However, aarA mutants also possess many of the phenotypes associated with loss of Tat function in E. coli (21, 22, 24), including cell chaining, detergent sensitivity, and the inability to grow anaerobically with TMAO as an electron acceptor (Table 1). This observation, together with our finding that overexpression of TatA from Prot. mirabilis can suppress the loss-of-signal production in aarA mutants, suggested a close relationship between AarA and the Tat system in Prov. stuartii. The work reported here demonstrates that this relationship is based on the requirement for a functional AarA rhomboid to process the Prov. stuartii TatA between the eighth and ninth amino acids from the N terminus. This processing is required for TatA function, because an artificially truncated form of TatA missing amino acids 2–8 (TatAΔ2–8) restored Tat function in an AarA-independent manner (Table 1 and Fig. 4), which explains why TatA from Prot. mirabilis or E. coli, both naturally missing an N-terminal extension, restored Tat function in an AarA-independent manner in Prov. stuartii (Table 1 and Fig. 1). Interestingly, TatA proteins with similar N-terminal extensions have been observed in a number of species, for example, the halophilic archaeon Natronomonas pharaonis and the bacteria Bacteroides fragilis and Leptospira interrogans. This information suggests the intriguing possibility that rhomboid-dependent activation of Tat-dependent protein export may be conserved in other organisms.
The ability of purified AarA to direct TatA cleavage in vitro, combined with the genetic data, demonstrates that AarA is directly responsible for TatA processing in vivo. Therefore, we have identified a rhomboid protease/substrate pair in a prokaryotic system. Why must the TatA N-terminal extension be removed for activity? Studies in E. coli have demonstrated that TatA can exist in complexes with TatB and TatBC and as a separate TatA homooligomer (25–28). In addition, the TatA protein itself may form the actual pore for protein secretion (29). Based on this information, the simplest explanation is that the unprocessed form of TatA is unable to interact with itself or one or more of the Tat proteins, possibly because of an unfavorable conformation. Preliminary studies indicate that TatA is membrane-associated in an aarA mutant, indicating that processing is not required for insertion into the cytoplasmic membrane. These data are consistent with mechanistic and structural data about rhomboids, which have their active site within the lipid bilayer (30).
This work provides a functional explanation for the requirement of AarA in Prov. stuartii quorum-sensing. Additionally, we show that the Tat system is required for production/activity of a quorum-sensing molecule. One obvious possibility that we have ruled out in this study is that the N-terminal peptide released by TatA cleavage is the signaling molecule itself. This statement is based on the observation that there is no signal activity in a tatC mutant (Fig. 4), although TatA is still processed normally (Fig. 2). In addition, the artificially truncated TatA protein of Prov. stuartii and the native TatA of Prot. mirabilis are able to restore signal production to an aarA mutant yet are missing this N-terminal extension (Figs. 1 and 2). Therefore, the production or activity of signal may require the Tat-dependent transport of an enzyme that is involved in signal production. Alternatively, the signal itself may be transported by the Tat system. Identification of the Prov. stuartii signaling molecule is vital to understanding this mechanism further.
Materials and Methods
Bacterial Growth Conditions.
Growth media was Luria broth (LB) containing 10 g of tryptone, 5 g of yeast extract, and 5 g of NaCl per liter. Antibiotics were used at the following concentrations: for Prov. stuartii, 100 μg/ml chloramphenicol; for Prot. mirabilis, 100 μg/ml chloramphenicol and 15 μg/ml tetracycline.
Preparation of CM.
CM were prepared by inoculation of 30 ml of LB with antibiotics as needed with 3 μl of an overnight culture or a single colony followed by shaking (250 rpm) at 37°C. Cultures were harvested at an OD600 of 0.9. The cells were pelleted at 4,300 × g for 10 min, and supernatants were harvested. To control for pH changes and nutrient deprivation of the CM, the pH was adjusted to 7.5 and tryptone-yeast extract was added to a final concentration of 0.5× relative to the concentration present in LB. CM was filter-sterilized with a syringe filter (pore size, 0.22 μm). The first 3 ml of CM that passed through the filter was discarded, and the rest was stored at −80°C until use. To assay CM, a dilute suspension of the cells to be tested was inoculated at a dilution of 1:1,000 and grown to an OD600 of 0.35. β-Galactosidase assays were done on SDS/chloroform-treated cells grown to mid-logarithm phase according to the method of Miller (31).
Isolation of tatA from Prot. mirabilis.
A genomic library consisting of partial Sau3A fragments of Prot. mirabilis cloned into the BamHI site of pACYC184 (32) was introduced into Prov. stuartii XD37.A by electroporation. Plasmids that complemented aarA were first identified as colonies with restored production of a diffusible yellow pigment absent in aarA mutants. Plasmids with apparent complementation of aarA were sequenced by subcloning a 1.8-kb EcoRV fragment into pBluescript SK(−) and by using the T3 and T7 primers. The tatA coding region was amplified from Prot. mirabilis 7002 by PCR with the primers 5′-ACATGTCTAGAAAAGTATAACTCCTAAAATAG-3′ and 5′-TAGGCTACTCGAGCTAAAACCAATGTCAAACAC-3′ and cloned into pBluescript SK(−) by using the XbaI and XhoI sites.
Isolation of the Prov. stuartii tatA Region.
The tat region of Prov. stuartii was cloned based on linkage to the ubiB gene (formerly yigR). A genomic Prov. stuartii library of partially digested Sau3A fragments in pACYC184 was introduced into E. coli DM123 ubiB::KmR by standard electroporation. Plasmids that complemented the ubiB mutation were identified as normal-sized colonies in the background of microcolonies formed by the ubiB mutant. Plasmids from large colonies were retransformed to verify that the plasmid restored normal growth. Restriction mapping and DNA sequencing with sets of primers that were used to “walk” along the sequence were used to identify the tatABC operon immediately adjacent to ubiB. The sequence of the tat region from Prov. stuartii has been deposited in the GenBank database (accession no. DQ989793).
Northern Blot Analysis.
The accumulation of tat operon mRNA from Prov. stuartii was assayed by Northern blot analysis. Cell samples were collected at an OD of 0.6, and RNA was extracted by using the MasterPure RNA Purification kit (Epicentre, Madison, WI). RNA was separated on a formaldehyde agarose (1.2%) gel and blotted onto a nitrocellulose membrane. A digoxigenin-labeled tatABC probe was synthesized by PCR with the primers 5′-GTTGAGAGCAAAAATAAAGAG-3′ and 5′-ATGCGGATCCCCATCCCTAAATAGAAAAGG-3′.
Construction of pET.TatA-His.
PCR was used to amplify the tatA gene from Prov. stuartii and engineer a His6 tag at the C terminus by using the primers 5′-GGTACAGGATCCGTTAAACCTTGTTTTGCGAC-3′ and 5′-CGTACGGGATCCCACGGTTTAATGATGATGATGATGATGACCCTGCTCTTTATTTTTGC-3′. These primers engineered BamH1 restriction sites at both ends of the PCR product. In addition, the TatA-His6 is missing its native tat promoter but contains the native ribosome-binding site. This fragment was cloned into BamHI-digested pACYC184 (32), resulting in plasmid pAC.Tat3. To construct pET.TatA-His, the BamHI fragment from pAC.Tat3 was cloned into pET21a such that TatA-His6 was transcribed from the T7 promoter. The pET.TatA-His6 plasmid was electroporated into PR50 or PR51, which contained p184.T7 and encoded an IPTG-inducible T7 RNA polymerase gene (33). Transformants were selected on LB agar plates containing 400 μg/ml ampicillin and 100 μg/ml chloramphenicol.
Western Blot Analysis.
Bacterial cultures were grown with shaking at 37°C in LB and the appropriate antibiotics. Cells were pelleted at an OD600 of 0.6 and resuspended in tricine sample buffer (Bio-Rad, Hercules, CA). Bacterial pellets were lysed in tricine sample buffer by heating at 95°C for 10 min. Protein samples were heated at 95°C for 10 min and subjected to SDS/PAGE by using a Bio-Rad ReadyGel system and 16.5% tris/tricine peptide gels. Gels were electrotransferred to nitrocellulose and probed at 1:1,700 with a Tetra-His HRP conjugate antibody (Qiagen, Valencia, CA). Detection was carried out by using enhanced chemiluminescence (ECL Western blotting detection reagents; GE Healthcare, Piscataway, NJ) in accordance with standard procedures (Amersham Biosciences, Uppsala, Sweden).
Purification of TatA-His6 and N-Terminal Sequencing.
An overnight culture of PR50 pET.TatA-His6/p184.T7 was diluted 1:100 in 200 ml of LB with 400 μg/ml ampicillin and 100 μg/ml chloramphenicol and grown with shaking at 37°C. IPTG was added at 1 mM and at an OD600 of 0.6, and cells were grown for an additional 90 min before pelleting. The pellet was resuspended in 10 ml of lysis buffer (200 mM Tris, pH 8.0/1% Triton X-100) and incubated at room temperature for 15 min before being filtered through a 0.22-μm filter. A Talon resin column (BD Biosciences, Franklin Lakes, NJ) was prewashed three times in wash buffer (50 mM NaH2PO4/300 mM NaCl, pH 7.0) before addition of the lysate and washed three times before elution in 3 ml of elution buffer (50 mM NaH2PO4/300 mM NaCl, pH 7.0/150 mM imidazole, pH 7.0). Samples were concentrated by using a Microcon YM-3 centrifugal filter device (Millipore, Billerica, MA) before being run on a 16.5% tris/tricine peptide gel (Bio-Rad Ready Gel). A prominent band was seen at ≈20 kDa. For sequencing, an unstained gel was blotted onto a poly(vinylidene difluoride) membrane and stained with Amido Black staining solution (Sigma–Aldrich, St. Louis, MO). N-terminal sequencing was done with a 491A Pulsed-Liquid Sequencer (PE Biosystems, Foster City, CA) with a PE Biosystems 140S PTH Analyzer (Procise-HT).
Expression and Purification of AarA.
Wild-type His6-tagged AarA (23) was expressed from the pET25+-based construct (pET.AarA.His6). The predicted catalytically inactive AarA mutant S150A was constructed by QuikChange mutagenesis (Stratagene, La Jolla, CA) by using pET.AarA.His6 as a template and primers 5′-ACTATCGGTGTTGGGGCTGCAGGCGCGATTATGGG-3′ and 5′-CCCATAATCGCGCCTGCAGCCCCAACACCGAT-AGT-3′ to generate pET25.AarAS150A.His6. Mutation was verified by DNA sequencing. Both AarA-His6 and AarAS150A-His6 were overexpressed in E. coli BL21(DE3)Gold, and total membrane fractions were isolated as described (23). The whole purification procedure was carried out at 4°C. Membrane pellets originating from 1 liter of bacterial culture were resuspended in 4 ml of buffer B [20 mM Hepes-NaOH (pH 7.4)/10% (vol/vol) glycerol/0.3 M NaCl/10 mM imidazole), and total protein concentration was determined with a DC protein assay (Bio-Rad). If necessary, protein concentration was adjusted to 5 mg/ml and 20% (wt/vol) n-dodecyl-β-d-maltoside (DDM) was added to a final concentration of 1.5% to solubilize membrane proteins. The resulting solutions were rocked at 4°C for 1 h, and then they were centrifuged at 100,000 × g for 30 min at 4°C. The supernatants were loaded onto 0.5-ml, Ni-NTA agarose gravity-flow columns (Qiagen) preequilibrated in buffer B containing 0.05% DDM. Columns were washed with 4 ml of buffer B plus 0.05% DDM and then 4 ml of buffer B with 0.05% DDM and 50 mM imidazole. Purified proteins were eluted by 1.5 ml of buffer B with 0.05% DDM and 250 mM imidazole, dialyzed into buffer B containing 0.05% DDM, flash-frozen in liquid nitrogen, and stored at −80°C. Protein concentration was determined with BCA protein assay (Pierce, Rockford, IL).
In Vitro Rhomboid Cleavage Assays.
Radiolabeled TatA variants were generated by cell-free in vitro translation with wheat-germ extract (Promega, Fitchburg, WI) and [35S]Met (GE Healthcare) as described (34). Templates for in vitro transcription were generated from pET.TatA-His6 by using forward primers 5′- CGATTAGGTGACACTATAGAATACCATGGAATCAAC-TATTGCAACGG-3′ (to produce full-length TatA) or 5′-CGATTAGGTGACACTATAGAATACCATGTCAG-GTTCAGGTTCAGGTTCAGGTGAATCAACTATTGCAA-CGGCC-3′ [to add (SG)4 peptide to the N terminus of TatA] and reverse primer 5′-AAGAAGCTACATCATCATACCCTGCTCTTTATTTTTGCTCTC-3′ (adding -MMM to the C terminus of TatA). Rhomboid cleavage assays were conducted in 50 mM Hepes-NaOH (pH 7.4)/10% glycerol /50 mM EDTA at 30°C. Typically, 1–4 μl of translation mixtures and the indicated amount of enzyme (Fig. 3B) were used in 40 μl of reaction volume. Cleavage reactions were stopped by protein precipitation with 10% (wt/vol) trichloroacetic acid, and samples were prepared for SDS/PAGE, electrophoresed, and autoradiographed as described (34).
Site-Directed Mutagenesis of tatAPs.
Amino acids 2–8 were deleted from pBC.TatAPs by oligonucleotide-directed mutagenesis with the QuikChange kit (Stratagene). The primers used for this deletion were 5′-GAGGTATAAACAATGGCTTTTGGTAGCCCTTGG-3′ and 5′-CCAAGGGCTACCAAAAGCCATTGTTTATACCTC-3′.
Construction of the tatC-Null Allele.
An internal sequence of the tatA gene from Prov. stuartii was amplified by PCR with the primers 5′-ATGCTCTAGAATTGAACTCAGGAAGCGGCTG-3′ and 5′-ATGCGGATCCCCATCCCTAAATAGAAAAGG-3′. The PCR product was cloned into the suicide vector pKNG101 (35) by using XbaI and BamHI sites engineered in the primers, resulting in plasmid pPTA1. The pPTA plasmid was introduced into the chromosome of Prov. stuartii PR50 or PR51 ΔaarA by a plate-mating with E. coli SM10/pPTA. Prov. stuartii exconjugants were selected on LB media containing 15 μg/ml tetracycline and 35 μg/ml streptomycin. All streptomycin-resistant colonies were sucrose-sensitive, and Southern blot analysis indicated that they contained pPTA inserted into the tatC gene.
Acknowledgments
This work was supported by National Science Foundation Award 0406047 (to P.N.R.) and a Merit Review Award from the Department of Veterans Affairs. P.N.R. was supported by a Research Career Scientist award from the Department of Veterans Affairs. K.S. was a recipient of a Marie Curie Intraeuropean Fellowship.
Abbreviations
- CM
conditioned media
- DDM
n-dodecyl-β-d-maltoside
- Tat
twin-arginine translocase
- TMAO
trimethylamine-N-oxide.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. DQ989793).
References
- 1.Fuqua C, Winans SC, Greenberg EP. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 2.Dunny GM, Leonard BA. Annu Rev Microbiol. 1997;51:527–564. doi: 10.1146/annurev.micro.51.1.527. [DOI] [PubMed] [Google Scholar]
- 3.Miller MB, Bassler BL. Annu Rev Microbiol. 2001;55:165–199. doi: 10.1146/annurev.micro.55.1.165. [DOI] [PubMed] [Google Scholar]
- 4.Payie KG, Rather PN, Clarke AJ. J Bacteriol. 1995;177:4303–4310. doi: 10.1128/jb.177.15.4303-4310.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rather PN, Parojcic MM, Paradise MR. Antimicrob Agents Chemother. 1997;41:1749–1754. doi: 10.1128/aac.41.8.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rather PN, Ding X, Baca-DeLancey RR, Siddiqui S. J Bacteriol. 1999;181:7185–7191. doi: 10.1128/jb.181.23.7185-7191.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rather PN, Orosz E. J Bacteriol. 1994;176:5140–5144. doi: 10.1128/jb.176.16.5140-5144.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallio M, Kylsten P. Curr Biol. 2000;10:693–694. doi: 10.1016/s0960-9822(00)00722-3. [DOI] [PubMed] [Google Scholar]
- 9.Urban S, Lee JR, Freeman M. Cell. 2001;107:173–182. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 10.Urban S, Lee JR, Freeman M. EMBO J. 2002;21:4277–4286. doi: 10.1093/emboj/cdf434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallio M, Sturgill G, Rather P, Kylsten P. Proc Natl Acad Sci USA. 2002;99:12208–12213. doi: 10.1073/pnas.192138799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santini CL, Ize B, Chanal A, Muller M, Giordano G, Wu LF. EMBO J. 1998;17:101–112. doi: 10.1093/emboj/17.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sargent F, Bogsch EG, Stanley NR, Wexler M, Robinson C, Berks BC, Palmer T. EMBO J. 1998;17:3640–3650. doi: 10.1093/emboj/17.13.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weiner JH, Bilous PT, Shaw GM, Lubitz SP, Frost L, Thomas GH, Cole JA, Turner RJ. Cell. 1998;93:93–101. doi: 10.1016/s0092-8674(00)81149-6. [DOI] [PubMed] [Google Scholar]
- 15.Wexler M, Sargent F, Jack RL, Stanley NR, Bogsch EG, Robinson C, Berks BC, Palmer T. J Biol Chem. 2000;275:16717–16722. doi: 10.1074/jbc.M000800200. [DOI] [PubMed] [Google Scholar]
- 16.Hatzixanthis K, Palmer T, Sargent F. Mol Microbiol. 2003;49:1377–1390. doi: 10.1046/j.1365-2958.2003.03642.x. [DOI] [PubMed] [Google Scholar]
- 17.Wu LF, Ize B, Chanal A, Quentin Y, Fichant G. J Mol Microbiol Biotechnol. 2000;2:179–189. [PubMed] [Google Scholar]
- 18.Berks BC, Sargent F, Palmer T. Mol Microbiol. 2000;35:260–274. doi: 10.1046/j.1365-2958.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 19.Lee PA, Tullman-Ercek D, Georgiou G. Annu Rev Microbiol. 2006;60:373–395. doi: 10.1146/annurev.micro.60.080805.142212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clemmer KM, Sturgill GM, Veenstra A, Rather PN. J Bacteriol. 2006;188:3415–3419. doi: 10.1128/JB.188.9.3415-3419.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bogsch EG, Sargent F, Stanley NR, Berks BC, Robinson C, Palmer T. J Biol Chem. 1998;273:18003–18006. doi: 10.1074/jbc.273.29.18003. [DOI] [PubMed] [Google Scholar]
- 22.Stanley NR, Findlay K, Berks BC, Palmer T. J Bacteriol. 2001;183:139–144. doi: 10.1128/JB.183.1.139-144.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemberg MK, Menendez J, Misik A, Garcia M, Koth CM, Freeman M. EMBO J. 2005;24:464–472. doi: 10.1038/sj.emboj.7600537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ize B, Porcelli I, Lucchini S, Hinton JC, Berks BC, Palmer T. J Biol Chem. 2004;279:47543–47554. doi: 10.1074/jbc.M406910200. [DOI] [PubMed] [Google Scholar]
- 25.Bolhuis A, Bogsch EG, Robinson C. FEBS Lett. 2000;472:88–92. doi: 10.1016/s0014-5793(00)01428-9. [DOI] [PubMed] [Google Scholar]
- 26.Sargent F, Gohlke U, de Leeuw E, Stanley NR, Palmer T, Saibil HR, Berks BC. Eur J Biochem. 2001;268:3361–3367. doi: 10.1046/j.1432-1327.2001.02263.x. [DOI] [PubMed] [Google Scholar]
- 27.de Leeuw E, Granjon T, Porcelli I, Alami M, Carr SB, Mueller M, Sargent F, Palmer T, Berks BC. J Mol Biol. 2002;322:1135–1146. doi: 10.1016/s0022-2836(02)00820-3. [DOI] [PubMed] [Google Scholar]
- 28.Porcelli I, de Leeuw E, Wallis R, van den Brink-van der Laan E, de Kruijff B, Wallace BA, Palmer T, Berks BC. Biochemistry. 2002;41:13690–13697. doi: 10.1021/bi026142i. [DOI] [PubMed] [Google Scholar]
- 29.Gohlke U, Pullan L, McDevitt CA, Porcelli I, de Leeuw E, Palmer T, Saibil HR, Berks BC. Proc Natl Acad Sci USA. 2005;102:10482–10486. doi: 10.1073/pnas.0503558102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Zhang Y, Ha Y. Nature. 2006;444:179–180. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 31.Miller JH. Experiments in Molecular Genetics. Woodbury, NY: Cold Spring Harbor Lab Press; 1972. [Google Scholar]
- 32.Chang AC, Cohen SN. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macinga DR, Parojcic MM, Rather PN. J Bacteriol. 1995;177:3407–3413. doi: 10.1128/jb.177.12.3407-3413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemberg MK, Martoglio B. Anal Biochem. 2003;319:327–331. doi: 10.1016/s0003-2697(03)00298-7. [DOI] [PubMed] [Google Scholar]
- 35.Kaniga K, Delor I, Cornelis GR. Gene. 1991;109:137–141. doi: 10.1016/0378-1119(91)90599-7. [DOI] [PubMed] [Google Scholar]