Abstract
The current testing of anti-HIV drugs is hampered by the lack of a small animal that is readily available and easy to handle; can be infected systemically with HIV type 1 (HIV-1); harbors the major HIV-1 target cells in a physiological frequency, organ distribution, and activation state; and is established as a pharmacological model. Here, we explored the potential of outbred Sprague–Dawley rats that transgenically express the HIV-1 receptor complex on CD4 T cells and macrophages as a model for the preclinical evaluation of inhibitors targeting virus entry or reverse transcription. The concentrations of the peptidic fusion inhibitor enfuvirtide or the nonnucleoside reverse transcriptase inhibitor efavirenz required to inhibit HIV-1 infection of cultured primary CD4 T cells and macrophages from human CD4 and CCR5-transgenic rats differed by no more than 3-fold from those required for human reference cultures. Prophylactic treatment of double-transgenic rats with a weight-adapted pediatric dosing regimen for either enfuvirtide (s.c., twice-daily) or efavirenz (oral, once-daily) achieved a 92.5% or 98.8% reduction, respectively, of the HIV-1 cDNA load in the spleen 4 days after i.v. HIV-1 challenge. Notably, a once-daily dosing regimen for enfuvirtide resulted in a ≈5-fold weaker inhibition of infection, unmasking the unfavorable pharmacokinetic characteristics of the synthetic peptide in the context of an efficacy trial. This work provides proof of principle that HIV-susceptible transgenic rats can allow a rapid and predictive preclinical evaluation of the inhibitory potency and of the pharmacokinetic properties of antiviral compounds targeting early steps in the HIV replication cycle.
Keywords: animal model, drugs, efficacy trial
Despite major achievements in HIV pharmacotherapy over the past decade, there remains an urgent need for more potent, less toxic, and conceptually novel antiretroviral drugs. The process of anti-HIV drug development is facilitated by efficient and predictive models capable of selecting the best compounds at each decision point. The preclinical validation in animal models is a critical complementation of in vitro efficacy and toxicity assays to facilitate the prioritization of antiviral compounds for phase I clinical trials. This notion is emphasized by the fact that a number of compounds with relatively good efficacy against HIV-1 in vitro, including dextran sulfate and the nonnucleoside reverse transcriptase inhibitor (NNRTI) atevirdine, have failed in clinical trials (1, 2).
Native mice and rats are nonpermissive for HIV-1 (3). The preclinical testing of anti-HIV drugs has thus concentrated on various xenotransplant models, in which human hematopoietic cells or tissues are transplanted into immunodeficient strains of mice. These models include severe combined immunodeficiency (SCID) mice, in which human fetal thymus and liver are simultaneously implanted under the kidney capsule [SCID-hu (Thy/Liv) model] (3, 4). Three to five months posttransplantation, successfully vascularized grafts can be directly challenged with HIV-1 during a second surgical procedure. Three to six weeks postinfection, end point analyses of grafts include the quantification of the proviral load, CD4 T cell depletion, and overall graft viability, permitting an assessment of drug efficacy and toxicity (5). As alternative yet related mouse models for HIV-1 infection, human peripheral blood lymphocytes (PBL) can be adoptively transferred i.p. into either SCID mice (hu-PBL-SCID model) or into sublethally irradiated mice reconstituted with SCID bone marrow (Trimera mouse model) (4, 6). However, all of these xenotransplant models do not recapitulate the full spectrum, frequency, and physiological organ distribution of HIV-1 target cells found in humans and are technically very challenging and time-consuming. The simian immunodeficiency virus (SIV)/HIV (SHIV) rhesus macaque model has been of some value for preclinical testing (7–11), but non-human primate studies are severely limited by ethical concerns and high cost, resulting in small animal group sizes and restricted accessibility.
In a conceptually different approach to the development of an HIV animal model (12), we have recently generated immunocompetent transgenic rats on an outbred Sprague–Dawley background that express the HIV-1 receptor complex selectively on CD4 T cells, macrophages, and microglia (13). In humans, these cell types are the primary targets for productive HIV infection. Ex vivo cell cultures from human CD4 and CCR5 (hCD4/hCCR5)-transgenic rats were susceptible to infection by HIV-1, leading to expression of early viral gene products. Furthermore, primary macrophages and microglia supported a productive infection by various recombinant and primary strains of HIV-1, including YU-2, JR-CSF, Ada-M, Ba-L, as well as the primary, patient-derived HIV-1 isolates C1 and O3. In contrast, T cell cultures from double-transgenic rats did not allow a spreading infection (13). After systemic HIV-1 challenge, lymphatic organs from hCD4/hCCR5-transgenic rats contained different HIV-1 cDNA species and early viral gene products, demonstrating HIV-1 susceptibility in vivo. Specifically, CD4 T lymphocytes and macrophages residing in spleen, thymus, as well as in peripheral blood from hCD4/hCCR5-transgenic rats expressed EGFP from the nef locus after infection with a replication-competent HIV-1 reporter virus. Furthermore, double-transgenic rats infected with the CCR5-using strain HIV-1YU-2 displayed a low-level plasma viremia up to 7 weeks postchallenge as well as two jointed LTR (2-LTR) circles in spleen and thymus 6 months postinfection, consistent with several rounds of low-level replication (13). These rats have been valuable for experimental studies on aspects of HIV-1- and protease inhibitor-induced peripheral neuropathy (14, 15).
In this work, we explored the suitability of hCD4/hCCR5-transgenic rats to serve as a model for a rapid, quantitative, and predictive evaluation of anti-HIV compounds. Ex vivo and in vivo proof-of-principle efficacy studies were conducted for two clinically approved antiretroviral drugs, the NNRTI efavirenz (Sustiva) and the peptidic fusion inhibitor enfuvirtide (T-20, Fuzeon).
Results
Anti-HIV Efficacies of an NNRTI or of a Fusion Inhibitor in Primary T Cells and Macrophages from hCD4/hCCR5-Transgenic Rats and Humans Are Comparable.
We first investigated potential species differences for anti-HIV drug susceptibility studies by comparing the efficacy of the prototypic NNRTI efavirenz and of the fusion inhibitor enfuvirtide for blocking HIV-1 infection in cultured primary target cells that were derived from either HIV-susceptible hCD4/hCCR5-transgenic rats or healthy human donors.
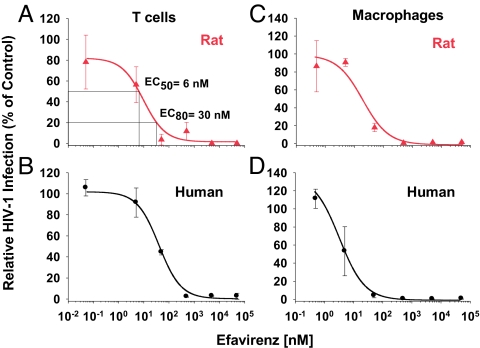
Activated primary T cells were pretreated with different concentrations of efavirenz, challenged with an HIV-1NL4–3 EGFP reporter virus, and analyzed for early viral gene expression on day 3 postinfection. In T cell cultures from both species, the relative percentage of HIV-1-infected cells decreased in a concentration-dependent manner (Fig. 1 A and B), and the effective concentration, 50% (EC50), and EC80 values for efavirenz were in the nanomolar range and statistically indistinguishable for rats and humans (Table 1). Similarly, the EC50 and EC80 values for efavirenz on HIV-1-infected spleen-derived macrophages from double-transgenic rats and human monocyte-derived macrophages were quite comparable (Fig. 1 C and D and Table 1). The infection of rat macrophages by a vesicular stomatitis virus glycoprotein (VSV-G) pseudotyped HIV-2ROD-A EGFP reporter virus or an HIV-1NL4–3-based reporter virus carrying multiple NNRTI resistance mutations in the reverse transcriptase gene could not or only very inefficiently be inhibited by efavirenz, respectively (Table 1), consistent with analogous studies in human cells (16).
Fig. 1.
Dose–response curves for the antiviral potency of the NNRTI efavirenz in ex vivo cultures of primary T cells and macrophages from hCD4/hCCR5-transgenic rats and humans. (A and B) Activated T cells from double-transgenic rats and human donors were pretreated with efavirenz for 1 h and subsequently challenged with VSV-G HIV-1NL4–3 EGFP viruses. (C and D) Macrophages from double-transgenic rats and human monocyte-derived macrophages were pretreated with efavirenz overnight and then challenged with VSV-G HIV-1NL4–3 EGFP viruses. On day 3 postinfection, the percentage of EGFP-positive cells was scored by flow cytometry. The percentage of infected cells in untreated controls was set at 100%, and relative levels of HIV-1 infection are shown. Each experiment was performed in triplicate, and one to four experiments were conducted. Given are arithmetic means ± SD of one experiment. EC50 and EC80 values were determined by using Prism software (GraphPad, San Diego, CA) and are shown in A and in Table 1.
Table 1.
Comparable anti-HIV efficacy of the NNRTI efavirenz and of the fusion inhibitor enfuvirtide in cultured primary cells from hCD4/hCCR5-transgenic rats and humans
| Drug and species | Virus | Inhibitory concentration, nM |
|||
|---|---|---|---|---|---|
| Macrophages |
T cells |
||||
| EC50* | EC80* | EC50* | EC80* | ||
| Efavirenz | |||||
| Human | HIV-1NL4-3 | 6 (1) | 20 (1) | 24 ± 14 (3) | 93 ± 57 (3) |
| Rat | HIV-1NL4-3 | 13 ± 6 (4) | 50 ± 17 (4) | 21 ± 13 (2) | 112 ± 83 (2) |
| Rat | HIV-1NL4-3RTmut | 3,100 ± 600 (2) | 15,000 ± 5,000 (2) | ||
| Rat | HIV-2ROD-A | >50,000 (2) | >50,000 (2) | ||
| Enfuvirtide | |||||
| Human | HIV-1YU-2 | 76 ± 65 (2) | 352 ± 308 (2) | 144 ± 32 (2) | 434 ± 99 (2) |
| Rat | HIV-1YU-2 | 32 ± 7 (2) | 227 ± 135 (2) | 116 ± 3 (2) | 460 ± 105 (2) |
*EC50 and EC80 values were derived from the dose–response experiments described in Figs. 1 and 2 and from experiments conducted in an analogous manner. Shown is the arithmetic mean ± SEM from independent experiments performed employing cells from one to four donors (shown in parentheses); each infection was done in triplicate.
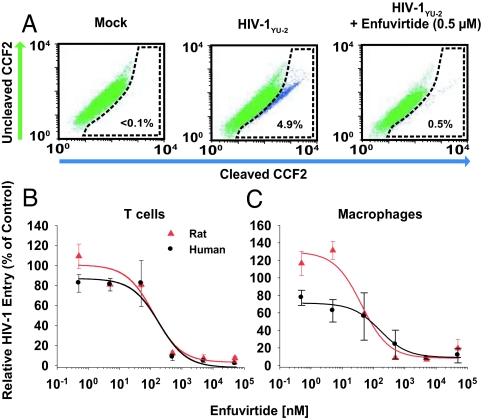
To test the antiviral ex vivo efficacy of the entry inhibitor enfuvirtide, a synthetic peptide corresponding to a region in the transmembrane subunit of the HIV-1 envelope glycoprotein (17), we performed a virion-fusion assay (18, 19). Cells were exposed to enfuvirtide and then challenged with HIV-1YU-2 virions carrying β-lactamase-Vpr chimeric fusion proteins (BlaM-Vpr). The change in fluorescence emission of the cell-permeable CCF2 substrate after cleavage by BlaM-Vpr, which enters the target cell cytoplasm by virion fusion, is a sensitive marker for virus entry and can be quantified by flow cytometry. The dose–response curves for enfuvirtide-mediated inhibition of HIV-1 entry in T cells from hCD4/hCCR5-transgenic rats and human donors almost superimposed (Fig. 2B), and as a result, the EC50 and EC80 values were very similar (Table 1). This virion-fusion assay could also be adapted to primary macrophage cultures (Fig. 2A), and the results obtained demonstrated a similar antiviral potency of enfuvirtide in these HIV-susceptible phagocytic scavengers from both species (Fig. 2C and Table 1). In summary, the inhibitory efficacies of a prototypic NNRTI and of a fusion inhibitor in primary HIV target cells from hCD4/hCCR5-transgenic rats and humans are comparable.
Fig. 2.
HIV-1 virion fusion is efficiently blocked in T cells and macrophages from hCD4/hCCR5-transgenic rats by enfuvirtide. (A) HIV-1YU-2 virion fusion in rat macrophages was analyzed by multiparameter flow cytometry as described in Materials and Methods. Shown are representative FACS dot plots for the detection of CCF2 substrate cleavage in rat macrophages which were either mock-infected (Left) or HIV-1YU-2-infected either in the absence (Middle) or presence (Right) of enfuvirtide. (B and C) Dose–response curves on T cells (B) and macrophages (C) from both species. The percentage of infected cells in untreated control cells was set at 100%. Two experiments were performed, each in triplicate. EC50 and EC80 values were determined by using Prism software and are shown in Table 1.
Efavirenz Inhibits HIV-1 Infection in hCD4/hCCR5-Transgenic Rats in Vivo.
Next, we investigated the antiviral in vivo potency of the NNRTI efavirenz in HIV-susceptible transgenic rats. Efavirenz was administered to six hCD4/hCCR5-transgenic rats at a weight-adjusted, pediatric dose of 25 mg/kg per day by once-daily oral gavage. Six control group animals received an equivalent volume of PBS. Rats were dosed 3 days before and 4 days after tail vein challenge with HIV-1YU-2. On day 7, animals were killed, and the HIV-1 cDNA load in DNA extracts from splenocytes was determined by quantitative duplex PCR analysis.
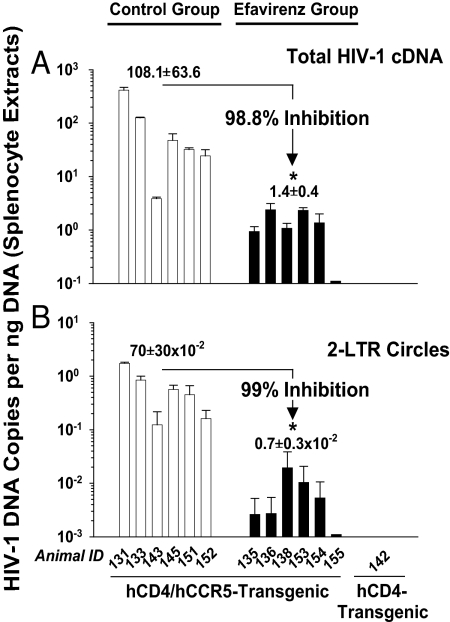
HIV-1 cDNA was readily detected in splenocyte extracts from PBS-treated, HIV-1YU-2-challenged hCD4/hCCR5-transgenic rats, ranging from 3 to 414 HIV-1 cDNA copies per ng of DNA (Fig. 3A). As a specificity control, no HIV-1 cDNA could be amplified from samples of a hCD4-single transgenic animal (animal 142) challenged with the identical infectious dose, demonstrating that the amplified HIV cDNA in samples from double-transgenic rats had been generated de novo after a receptor complex-mediated infection in vivo. Importantly, oral efavirenz treatment had a drastic antiviral activity: the HIV-1 cDNA load in splenocytes from HIVYU-2-infected animals from the efavirenz group was reduced by 98.8% (1.92 log10) relative to the control group (Fig. 3A). This antiviral activity was highly significant according to the Mann–Whitney U test (P < 0.0025). To confirm this result independently, DNA extracts from splenocytes were reanalyzed in a second duplex PCR, this time amplifying circularized HIV-1 cDNA genomes containing two joined LTRs. The presence of these 2-LTR circles in a cell is an established surrogate for successful HIV entry, reverse transcription, and nuclear import (20). In good agreement with the results for total proviral DNA, splenocyte samples from the efavirenz group showed a concordant reduction in 2-LTR circles by 99.0% (2.0 log10, P < 0.0025) (Fig. 3B). Thus, HIV-1-susceptible hCD4/hCCR5-transgenic rats allowed a rapid and quantitative drug testing, and they demonstrated a high antiviral activity of an orally bioavailable reverse transcriptase inhibitor at a pediatric dosage.
Fig. 3.
Efavirenz inhibits HIV-1 infection in vivo. hCD4/hCCR5-transgenic rats were treated with either efavirenz at 25 mg/kg per day (efavirenz group) or PBS (control group) by once-daily oral gavage for 3 days. On day 3, animals from both experimental groups as well as a hCD4-single transgenic rat (rat 142) were challenged i.v. with HIV-1YU-2. Dosing was continued in both treatment groups for 4 more days, and then all animals were killed, and their spleens were removed. The antiviral efficacy was assessed by determining the load of either total HIV-1 proviral DNA (A) or HIV-1 2-LTR circles (B), relative to a rat GAPDH standard in DNA extracts from splenocytes by duplex PCR. Results given for animal groups are the arithmetic mean ± SEM. Nonparametric statistical analyses were performed by using the Mann–Whitney U test; the asterisks indicate P < 0.0025.
Prophylactic Treatment with Enfuvirtide Markedly Diminishes the Proviral Load, but Its in Vivo Potency Depends on the Dosing Regimen.
The antiviral efficacy of the entry inhibitor enfuvirtide in hCD4/hCCR5-transgenic rats was tested in an experimental setup analogous to the efavirenz study. In two treatment groups, enfuvirtide was administered by s.c. injection at 4 mg/kg per day, corresponding to the dosage generally applied in HIV-infected children (21). The first group of rats received this daily enfuvirtide dosage divided into two injections at ≈12-h intervals, as recommended in humans, whereas the second group was injected only once daily. All rats were challenged i.v. on day 3 with HIV-1YU-2 at a 10-fold lower infectious dose than was used in the efavirenz study.
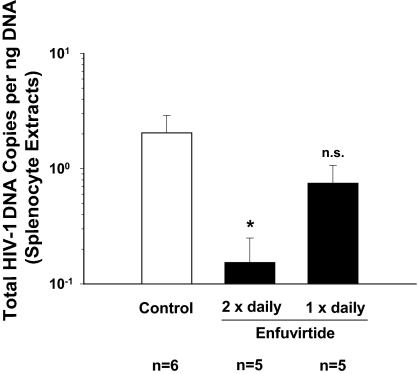
The twice-daily enfuvirtide treatment group showed a >10-fold reduction (92.8%; 1.14 log10) of the HIV-1 cDNA load in splenocyte extracts relative to the water-injected control group (Mann–Whitney U test; P < 0.018) (Fig. 4), demonstrating a high antiviral potency of a prophylactic application of the fusion inhibitor in this small animal model. In contrast, the reduction of the HIV-1 cDNA load in the once-daily treatment group was only modest (≈2-fold) and did not reach statistical significance (P = 0.33). Again, no HIV-1 cDNA could be amplified from samples of the hCD4-single transgenic animal (animal 53) (data not shown). In summary, enfuvirtide treatment markedly diminished the proviral load in a secondary lymphoid organ in HIV-1-infected hCD4/hCCR5-transgenic rats, but the antiviral in vivo potency of the synthetic peptide strongly depended on the dosing regimen.
Fig. 4.
Enfuvirtide blocks HIV-1 entry in vivo. hCD4/hCCR5-transgenic rats were treated s.c. with either enfuvirtide at 4 mg/kg per day either twice daily or once daily. The control group was treated twice daily with distilled water. On day 3, animals from all three groups and one hCD4-single transgenic rat (rat 53; data not shown) were challenged i.v. with HIV-1YU-2. Dosing was continued, and the experiment was terminated on day 4 postinfection. The antiviral efficacy of enfuvirtide was assessed by duplex PCR as described for Fig. 3A. Results given for animal groups are the arithmetic mean ± SEM. Nonparametric statistical analyses were performed by the Mann–Whitney U test; the asterisk indicates P < 0.018; n.s., not significant (P = 0.33).
Discussion
Information on the antiviral in vitro and in vivo potency, toxicity, and pharmacological properties, including bioavailability and pharmacokinetics, guides the development and advancement of anti-HIV compounds. A rapid preclinical validation in a readily available HIV-susceptible small animal would facilitate an evidence-based prioritization of compounds for phase I clinical trials. In this work, we provide proof of principle that rats, which transgenically express the HIV-1 receptor complex on biologically relevant target cells, recapitulate the antiviral potency and basic pharmacological properties of two different anti-HIV drugs that are widely used in HIV-infected patients. Both the orally administered NNRTI efavirenz and the s.c.-injected peptidic fusion inhibitor enfuvirtide had a high antiviral efficacy in this small animal model.
First, the potencies of these prototypic anti-HIV drugs on cultured primary T cells and macrophages from transgenic rats and humans were comparable, with EC50 and EC80 concentrations differing by no more than 3-fold. Similarly, rat cells revealed a lack of antiviral efficacy of the NNRTI when a drug-resistant HIV-1 or HIV-2 strain was tested. On the level of HIV-infected primary cells ex vivo, this observation indicates conserved modes of drug metabolism and action and supports the validity of a rat–human cross-species extrapolation of drug efficacy also in an in vivo context. Second, the overall reduction in the spleen-associated HIV cDNA load achieved by enfuvirtide or efavirenz treatment in a 7-day trial in the transgenic rat model, 1.14 log10 and 1.92 log10, respectively, was within the range seen for the reduction in plasma viral load in monotherapy studies in HIV-infected adults. Kilby and colleagues (17) reported that patients receiving 200 mg of enfuvirtide per day by twice-daily injection experienced a ≈1.6 log10 median decline in plasma HIV RNA in a 7-day dosing period (17). Similarly, oral efavirenz monotherapy in an HIV-infected patient at 200 mg per day for 7 days resulted in a ≈1.5 log10 decline in HIV RNA in plasma (22). Although the cross-species comparison for the antiviral in vivo potency of these two drugs is promising, the testing of additional compounds in the transgenic rat model will be required to define more carefully the accuracy of its predictive value for HIV-infected humans.
Outbred Sprague–Dawley rats are the most widely used and best validated small animal model for basic studies on bioavailability, pharmacokinetics, and pharmacodynamics of virtually all drug candidates, and pharmaceutical companies use these rodents for large proportions of their mandatory toxicity testing (23). This fact greatly enhances the utility of the HIV-susceptible transgenic rats for preclinical drug efficacy studies. Furthermore, the antiviral in vivo efficacy of the synthetic enfuvirtide peptide in hCD4/hCCR5-transgenic Sprague–Dawley rats was strongly dependent on the dosing regimen; a once-daily administration was markedly inferior (≈5-fold lower inhibition) in its antiviral potency compared with a twice-daily regimen. This result is consistent with the known unfavorable pharmacokinetic properties of enfuvirtide, with a short median half-life of the peptide in rats as well as in humans (median t1/2 = ≈2 h) (17). The results from the two treatment groups in our enfuvirtide study suggest that unfavorable pharmacokinetic properties affecting the trough concentration of a compound can be reflected in a loss of antiviral efficacy, which further validates the model. In principle, established pharmacokinetic analyses could be conveniently conducted in the course of an efficacy evaluation because repeated blood draws of up to 1.5 ml are possible in HIV-infected rats. Thus, a comprehensive preclinical evaluation of anti-HIV compounds, addressing antiviral in vivo potency with a dynamic range of up to 3 orders of magnitude as well as toxicity and pharmacokinetics, can be performed in hCD4/hCCR5-transgenic rats in <2 weeks. As an additional advantage over current xenotransplant models, HIV-susceptible hCD4/hCCR5-transgenic rats harbor both CD4 T cells and macrophages in a physiological frequency, organ distribution, and activation state. Notably, in HIV-infected patients these major HIV target cell populations provide distinct milieus with respect to the effectiveness of antiretroviral therapy (24–27). Furthermore, because of the ease of breeding and handling large numbers of these immunocompetent transgenic rats, even medium-throughput in vivo drug screening approaches appear feasible. Despite only transient HIV-1 replication in vivo (13), the current study suggests that the hCD4/hCCR5-transgenic rat model is well suited to evaluate new antiviral lead compounds targeting viral entry or reverse transcription, and they may thus help guide the selection of effective antiviral compounds to treat HIV-1 disease in humans. In addition, the testing of alternative anti-HIV strategies, such as integrase inhibitors as well as gene therapy approaches, can be envisioned. Building on such exciting applications for the current form of the model, we are now pursuing different strategies, including virus modifications, adjuvant pharmacostimulation, and additional genetic manipulations of the host, to enhance HIV replication in this rodent model further.
Materials and Methods
Transgenic Rat Model.
The generation and initial characterization of hCD4/hCCR5-transgenic rats has been reported in ref. 13. Animal experiments were conducted according to the German Animal Welfare Act and with authorization of the Regierungspräsidium Karlsruhe (35-9185.81/G-100/02); experiments were supervised by animal welfare officers of Heidelberg University.
Primary Cells.
Cultures of primary T lymphocytes and macrophages from transgenic rats and random human donors were generated as reported in refs. 12, 28, and 29.
Virus Stocks.
The HIV-1YU-2 proviral DNA was cloned directly from brain tissue of a patient who died of AIDS dementia complex (30). The generation of replication-competent HIV-1YU-2 stocks for in vivo infection studies has been reported in ref. 13. Virus stocks were characterized for p24 concentration and for infectious titer (TZM-BL IU) as described in ref. 29. The molecular clones HIV-1NL4–3 E− EGFP, which carries an egfp gene within the nef locus (31), HIV-2ROD-A E− EGFP (32), or HIV-1NL4–3 RTmut(K20R, K32R, V35L, K65R, L100I, K103N, L214F, P272A, I293V) E− EGFP (kind gift from Hans-Georg Kräusslich, University of Heidelberg, Germany) were pseudotyped with VSV-G as reported in ref. 12. HIV-1YU-2 virions containing BlaM-Vpr were produced by triple-transfection of 293T cells by calcium phosphate DNA precipitation (18). Two days posttransfection, the supernatant was concentrated by using Centricon Plus-70 spin columns (Millipore, Billerica, MA), and then virus particles were purified through a 20% sucrose cushion (27,000 × g, 4°C, 60 min). The virion-enriched pellet was resuspended in medium and stored at −80°C.
Antiviral Drugs.
Enfuvirtide (Roche, Indianapolis, IN) was freshly dissolved in distilled H2O at 9 mg/ml and further diluted in H2O. Efavirenz (Bristol–Myers Squibb, Jacksonville, FL) was purchased as a drinking solution at 30 mg/ml and diluted in PBS.
Ex Vivo HIV-1 Virion-Fusion Assay.
The flow cytometry-based HIV-1 virion-fusion assay was conducted as described in refs. 18 and 19. Briefly, primary T cells and macrophages were pretreated with the indicated concentrations of enfuvirtide for 30 min. Subsequently, cells were challenged with HIV-1YU-2 BlaM-Vpr virions (50 ng per 2 × 106 T cells or per ≈2 × 105 macrophages) for 3–4 h, washed, and then loaded with CCF2/AM dye overnight. Fusion was monitored with a three-laser BD FACSAria Cell Sorting System (BD Biosciences, San Jose, CA).
Ex Vivo Efficacy Testing of Efavirenz.
Macrophages were pretreated overnight and T cells for 1 h with efavirenz at the indicated concentrations, and then they were subsequently challenged with single-round VSV-G HIV-1NL4–3 EGFP reporter virions (200–500 ng of p24 per 3 × 106 T cells or per ≈2 × 105 macrophages). On day 3 postinfection, the percentage of EGFP-positive T cells and macrophages was scored by flow cytometry on a FACSCalibur by using BD CellQuest Pro 4.0.2 software (BD PharMingen, San Jose, CA).
In Vivo Efficacy Testing.
Two independent efficacy studies with a similar design were performed (animal age, 8 weeks; weight range, 140–306 g). For the NNRTI study, groups consisting of six hCD4/hCCR5-transgenic rats were treated with either efavirenz at 25 mg/kg per day or PBS by once-daily oral gavage for 3 days. On day 3, rats from both experimental groups were anesthesized and challenged by tail vein injection through a plastic catheter with HIV-1YU-2 (6.7 × 106 TZM-BL IU; 5,000 ng of p24 per rat) as described in ref. 13. For the fusion inhibitor study, groups consisting of five or six hCD4/hCCR5-transgenic rats were treated with enfuvirtide at 4 mg/kg per day, applied by either twice-daily or once-daily s.c. injection. The control group was injected twice daily with an equivalent volume of distilled water. On day 3, all rats were challenged i.v. with HIV-1YU-2 (6.7 × 105 TZM-BL IU; 500 ng of p24 per rat). In both efficacy studies, one HIV-nonsusceptible, hCD4-single transgenic rat was challenged with the identical virus inoculum. Dosing was continued for 4 more days postchallenge, and then all animals were killed. Total DNA was prepared from single-cell suspensions of splenocytes by using DNeasy tissue kits (Qiagen, Valencia, CA), and it was analyzed by quantitative duplex PCR.
Quantification of HIV-1 DNA Species.
The amount of total HIV-1 cDNA and HIV-1 2-LTR circles in splenocyte extracts was analyzed by quantitative duplex PCR by using the ABI 7500 sequence detection system (Applied Biosystems, Foster City, CA). Total HIV-1 cDNA was amplified by using a primer pair specific for LTR U5 and gag and the probe 5′-[fluorescein (FAM)]-CAGTGGCGCCCGAACAGGGA-[rhodamine (TAMRA)]-3′ as reported in refs. 33 and 34. For quantification of 2-LTR circle junctions a forward primer annealing at U5, a reverse primer annealing at U3 and the probe 5′-(FAM)-TCCACACTGACTAAAAGGGTCTGGGGATCTCT-(TAMRA)-3′ were used (33, 34). For standard curves, dilutions of pHIV-1NL4–3 E− EGFP and pU3U5 covering 5 logs were used, supplemented with DNA from uninfected cells. Results obtained for HIV-1 cDNA species were normalized to the amount of cellular DNA, which was quantified in the same reaction by amplification of the rat GAPDH gene DNA (reagents from Applied Biosystems). For the latter, dilutions of genomic DNA extracted from primary rat T cells were used for standard curves. The cycling program was as follows: 2 min at 50°C; 10 min at 95°C; 40 cycles of 15 s at 95°C then 1 min at 60°C. All samples were run in duplicate. Data analysis was performed by using the 7500 system software. The lowest detection limit ranged from 0.02 to 0.03 copies per ng of DNA and from 0.05 copies per ng of DNA for total HIV-1 cDNA and for 2-LTR circles, respectively. Some values for extracts of the treatment groups had to be extrapolated.
Acknowledgments
We thank Drs. Hans-Georg Kräusslich, Warner Greene, and Mark Goldsmith for continuous support and Drs. Jochen Bodem, Matthias Dittmar, Beatrice Hahn, Martin Hartmann, Hans-Georg Kräusslich, Nathaniel Landau, and Mrs. Lisa Black for providing reagents. We thank Drs. Zeger Debyser, Myriam Witvrouw, and Bénédicte Van Maele (Katholieke Universiteit Leuven, Leuven, Belgium) for assistance in setting up the quantitative PCR analyses. We thank Mrs. Julia Lenz and Dr. Blanche Schwappach (ZMBH, Heidelberg, Germany) for BD FACSAria analysis. We thank Dr. Cheryl Stoddart (Gladstone Institute of Virology and Immunology, San Francisco, CA) and Dr. Peter Larson (Trimeris, Morrisville, NC) for helpful discussions. We thank Drs. Valerie Bosch, Oliver Fackler, Jason Kreisberg, Nico Michel, and Mrs. Hanna-Mari Tervo for critical reading of the manuscript. This work was supported by Deutsche Forschungsgemeinschaft Grant Ke 742/2 (to O.T.K.), European TRIoH Consortium EU Project LSGH-2003-503480 (to O.T.K.), and J. David Gladstone Institutes Subcontract R0051-B (to O.T.K.) of National Institutes of Health Grant R01-MH64396.
Abbreviations
- BlaM-Vpr
β-lactamase-Vpr chimeric fusion protein
- hCD4/hCCR5
human CD4 and CCR5
- HIV-1
HIV type 1
- 2-LTR
two joined LTRs
- NNRTI
nonnucleoside reverse transcriptase inhibitor
- SCID
severe combined immunodeficiency
- VSV-G
vesicular stomatitis virus type G.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Demeter LM, Meehan PM, Morse G, Fischl MA, Para M, Powderly W, Leedom J, Holden-Wiltse J, Greisberger C, Wood K, et al. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;19:135–144. doi: 10.1097/00042560-199810010-00006. [DOI] [PubMed] [Google Scholar]
- 2.Flexner C, Barditch-Crovo PA, Kornhauser DM, Farzadegan H, Nerhood LJ, Chaisson RE, Bell KM, Lorentsen KJ, Hendrix CW, Petty BG, et al. Antimicrob Agents Chemother. 1991;35:2544–2550. doi: 10.1128/aac.35.12.2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Maanen M, Sutton RE. Curr HIV Res. 2003;1:121–130. doi: 10.2174/1570162033352075. [DOI] [PubMed] [Google Scholar]
- 4.Borkow G. IUBMB Life. 2005;57:819–823. doi: 10.1080/15216540500459642. [DOI] [PubMed] [Google Scholar]
- 5.Rabin L, Hincenbergs M, Moreno MB, Warren S, Linquist V, Datema R, Charpiot B, Seifert J, Kaneshima H, McCune JM. Antimicrob Agents Chemother. 1996;40:755–762. doi: 10.1128/aac.40.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mosier DE. Virology. 2000;271:215–219. doi: 10.1006/viro.2000.0336. [DOI] [PubMed] [Google Scholar]
- 7.Veazey RS, Klasse PJ, Schader SM, Hu Q, Ketas TJ, Lu M, Marx PA, Dufour J, Colonno RJ, Shattock RJ, et al. Nature. 2005;438:99–102. doi: 10.1038/nature04055. [DOI] [PubMed] [Google Scholar]
- 8.Hazuda DJ, Young SD, Guare JP, Anthony NJ, Gomez RP, Wai JS, Vacca JP, Handt L, Motzel SL, Klein HJ, et al. Science. 2004;305:528–532. doi: 10.1126/science.1098632. [DOI] [PubMed] [Google Scholar]
- 9.Hu SL. Curr Drug Targets Infect Disord. 2005;5:193–201. doi: 10.2174/1568005054201508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.North TW, Van Rompay KK, Higgins J, Matthews TB, Wadford DA, Pedersen NC, Schinazi RF. J Virol. 2005;79:7349–7354. doi: 10.1128/JVI.79.12.7349-7354.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giuffre AC, Higgins J, Buckheit RW, Jr, North TW. Antimicrob Agents Chemother. 2003;47:1756–1759. doi: 10.1128/AAC.47.5.1756-1759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keppler OT, Yonemoto W, Welte FJ, Patton KS, Iacovides D, Atchison RE, Ngo T, Hirschberg DL, Speck RF, Goldsmith MA. J Virol. 2001;75:8063–8073. doi: 10.1128/JVI.75.17.8063-8073.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keppler OT, Welte FJ, Ngo TA, Chin PS, Patton KS, Tsou CL, Abbey NW, Sharkey ME, Grant RM, You Y, et al. J Exp Med. 2002;195:719–736. doi: 10.1084/jem.20011549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones G, Zhu Y, Silva C, Tsutsui S, Pardo CA, Keppler OT, McArthur JC, Power C. Virology. 2005;334:178–193. doi: 10.1016/j.virol.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 15.Pettersen JA, Jones G, Worthington C, Krentz HB, Keppler OT, Hoke A, Gill MJ, Power C. Ann Neurol. 2006;59:816–824. doi: 10.1002/ana.20816. [DOI] [PubMed] [Google Scholar]
- 16.Witvrouw M, Pannecouque C, Switzer WM, Folks TM, De Clercq E, Heneine W. Antivir Ther. 2004;9:57–65. [PubMed] [Google Scholar]
- 17.Kilby JM, Hopkins S, Venetta TM, DiMassimo B, Cloud GA, Lee JY, Alldredge L, Hunter E, Lambert D, Bolognesi D, et al. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 18.Venzke S, Michel N, Allespach I, Fackler OT, Keppler OT. J Virol. 2006;80:11141–11152. doi: 10.1128/JVI.01556-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cavrois M, De Noronha C, Greene WC. Nat Biotechnol. 2002;20:1151–1154. doi: 10.1038/nbt745. [DOI] [PubMed] [Google Scholar]
- 20.Sharkey ME, Teo I, Greenough T, Sharova N, Luzuriaga K, Sullivan JL, Bucy RP, Kostrikis LG, Haase A, Veryard C, et al. Nat Med. 2000;6:76–81. doi: 10.1038/71569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Church JA, Hughes M, Chen J, Palumbo P, Mofenson LM, Delora P, Smith E, Wiznia A, Hawkins E, Sista P, Cunningham CK. Pediatr Infect Dis J. 2004;23:713–718. doi: 10.1097/01.inf.0000133045.45316.6a. [DOI] [PubMed] [Google Scholar]
- 22.Bacheler LT, Anton ED, Kudish P, Baker D, Bunville J, Krakowski K, Bolling L, Aujay M, Wang XV, Ellis D, et al. Antimicrob Agents Chemother. 2000;44:2475–2484. doi: 10.1128/aac.44.9.2475-2484.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abbott A. Nature. 2004;428:464–466. doi: 10.1038/428464a. [DOI] [PubMed] [Google Scholar]
- 24.Venzke S, Keppler OT. Expert Rev Clin Immunol. 2006;2:613–626. doi: 10.1586/1744666X.2.4.613. [DOI] [PubMed] [Google Scholar]
- 25.Jong A, Huang SH. Curr Drug Targets Infect Disord. 2005;5:65–72. doi: 10.2174/1568005053174672. [DOI] [PubMed] [Google Scholar]
- 26.Ghosn J, Chaix ML, Peytavin G, Rey E, Bresson JL, Goujard C, Katlama C, Viard JP, Treluyer JM, Rouzioux C. AIDS. 2004;18:1958–1961. doi: 10.1097/00002030-200409240-00014. [DOI] [PubMed] [Google Scholar]
- 27.Garbuglia AR, Zaccarelli M, Calcaterra S, Cappiello G, Marini R, Benedetto A. J Chemother. 2001;13:188–194. doi: 10.1179/joc.2001.13.2.188. [DOI] [PubMed] [Google Scholar]
- 28.Goffinet C, Keppler OT. FASEB J. 2006;20:500–502. doi: 10.1096/fj.05-4651fje. [DOI] [PubMed] [Google Scholar]
- 29.Keppler OT, Allespach I, Schuller L, Fenard D, Greene WC, Fackler OT. J Virol. 2005;79:1655–1665. doi: 10.1128/JVI.79.3.1655-1665.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Kappes JC, Conway JA, Price RW, Shaw GM, Hahn BH. J Virol. 1991;65:3973–3985. doi: 10.1128/jvi.65.8.3973-3985.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He J, Chen Y, Farzan M, Choe H, Ohagen A, Gartner S, Busciglio J, Yang X, Hofmann W, Newman W, et al. Nature. 1997;385:645–649. doi: 10.1038/385645a0. [DOI] [PubMed] [Google Scholar]
- 32.Reuter S, Kaumanns P, Buschhorn SB, Dittmar MT. Virology. 2005;332:347–358. doi: 10.1016/j.virol.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 33.Van Maele B, De Rijck J, De Clercq E, Debyser Z. J Virol. 2003;77:4685–4694. doi: 10.1128/JVI.77.8.4685-4694.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brussel A, Sonigo P. J Virol. 2003;77:10119–10124. doi: 10.1128/JVI.77.18.10119-10124.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]