Abstract
Pyrrolysine has entered natural genetic codes by the translation of UAG, a canonical stop codon. UAG translation as pyrrolysine requires the pylT gene product, an amber-decoding tRNAPyl that is aminoacylated with pyrrolysine by the pyrrolysyl-tRNA synthetase produced from the pylS gene. The pylTS genes form a gene cluster with pylBCD, whose functions have not been investigated. The pylTSBCD gene order is maintained not only in methanogenic Archaea but also in a distantly related Gram-positive Bacterium, indicating past horizontal gene transfer of all five genes. Here we show that lateral transfer of pylTSBCD introduces biosynthesis and genetic encoding of pyrrolysine into a naïve organism. PylS-based assays demonstrated that pyrrolysine was biosynthesized in Escherichia coli expressing pylBCD from Methanosarcina acetivorans. Production of pyrrolysine did not require tRNAPyl or PylS. However, when pylTSBCD were coexpressed with mtmB1, encoding the methanogen monomethylamine methyltransferase, UAG was translated as pyrrolysine to produce recombinant monomethylamine methyltransferase. Expression of pylTSBCD also suppressed an amber codon introduced into the E. coli uidA gene. Strains lacking one of the pylBCD genes did not produce pyrrolysine or translate UAG as pyrrolysine. These results indicated that pylBCD gene products biosynthesize pyrrolysine using metabolites common to Bacteria and Archaea and, furthermore, that the pyl gene cluster represents a “genetic code expansion cassette,” previously unprecedented in natural organisms, whose transfer allows an existing codon to be translated as a novel endogenously synthesized free amino acid. Analogous cassettes may have served similar functions for other amino acids during the evolutionary expansion of the canonical genetic code.
Keywords: Archaea, methanogenesis, methanosarcina, pyrrolysyl-tRNA, tRNAPyl
Translated in-frame amber (TAG/UAG) codons in the genes encoding methylamine methyltransferases from Methanosarcina barkeri (1–3) were early clues to the existence of pyrrolysine as the 22nd genetically encoded amino acid from nature. Crystallography of MtmB1, the monomethylamine methyltransferase, demonstrated that the UAG codon of the encoding mtmB1 gene corresponds to pyrrolysine, whose structure was proposed as lysine withεN in amide linkage with the d-isomer of 4-methyl-pyrroline-5-carboxylate (4, 5). This structure is consistent with the UAG-encoded residue's mass in three distinct methylamine methyltransferases (6).
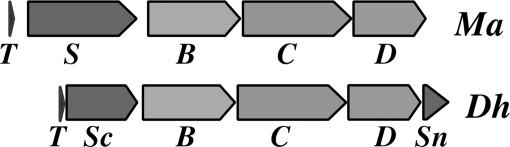
Near the mtmB1 gene in Methanosarcina spp. are the pyl genes (ref. 7; Fig. 1). Two of these genes are known to underlie the genetic encoding of pyrrolysine. The pylT encodes tRNAPyl, whose CUA anticodon complements the UAG codon corresponding to pyrrolysine. Deletion of the pyl gene promoter and pylT leads to loss of pyrrolysine-dependent monomethylamine methyltransferase and methylamine metabolism (8). The pylS gene encodes the pyrrolysyl-tRNA synthetase, PylS, which charges tRNAPyl with chemically synthesized pyrrolysine and further carries out a pyrrolysine-dependent ATP:pyrophosphate exchange reaction (5, 9–11). EF-Tu can bind charged tRNAPyl (12), and recombinant Escherichia coli bearing Methanosarcina acetivorans pylT and pylS is able to decode TAG within an introduced mtmB1 gene as pyrrolysine (9). UAG translation depended on the addition of synthetic pyrrolysine to the medium, because the amino acid is not made in E. coli.
Fig. 1.
The pyl genes from the methanogenic archaeon M. acetivorans (Ma) and the Gram-positive Bacterium D. hafniense (Dh). The gene order of the pyl genetic code expansion cassette is conserved, with the exception that the D. hafniense pylS gene homolog has been split into two genes encoding homologs to the PylS N-terminal domain (pylSn) and the catalytic core domain (pylSc) that now flank pylBCD (7).
Following pylTS are the pylBCD genes. The putative pylBCD gene products have respective similarity to radical SAM proteins, proteins forming amides, and amino acid dehydrogenases and could thereby participate in plausible pathways for biosynthesis of pyrrolysine (7, 13). Cotranscription of pylTSBCD is likely by inspection of the M. acetivorans sequence (8) and has been demonstrated for pylTSBC in M. barkeri (7). The same pylTSBCD arrangement has been maintained in all sequenced Methanosarcina and Methanococcoides genomes (7, 14–16). Surprisingly, this order of genes has also been found in a single genus of Gram-positive bacterium, Desulfitobacterium hafniense (ref. 7; Fig. 1). More recent genomic surveys have indicated no further genomes containing pylTSBCD (17). The restricted distribution of pyl genes to one archaeal family and a distantly related bacterial genus is suggestive of a horizontal gene transfer that involved the entire pyl gene cluster.
The above considerations led us to hypothesize that the driving force underlying the lateral transfer of the intact pyl cluster could be that pylTSBCD are sufficient to encode biosynthesis of pyrrolysine, as well as pyrrolysyl-tRNAPyl, even within very different metabolic backgrounds. If so, the pyl gene cluster might translate UAG as pyrrolysine within organisms by means of amber suppression, which in recent years has been shown to be a powerful avenue to artificially manipulating genetic codes (18–20). However, transfer of a “genetic code expansion cassette” would be unprecedented in natural gene pools. Therefore, we tested this hypothesis by transferring pyl genes from the methanogenic archaeon M. acetivorans to an organism with a distinct metabolism, E. coli.
Results
Vectors Allowing Timed Expression of pyl Genes.
The pylB, pylC, and pylD genes in the genome of M. acetivorans are respectively designated MA0154 (GenBank accession no. NP_615127), MA0153 (GenBank accession no. NP_615126), and MA0152 (GenBank accession no. NP_615125). Our initial attempts to clone pylBCD into E. coli in the presence of pylT and pylS failed to reveal UAG translation. Examination of a pylB and pylC sequence revealed that both annotated frames began with the infrequently used start codon TTG. Alignments of the predicted gene products of the M. acetivorans genes with the archaeal and bacterial homologs, as well as the placement of the ribosomal binding sites, supported TTG as the actual start codon. Because TTG is an infrequent start codon in E. coli (21), the TTG codons were changed to ATG. The M. acetivorans pylB gene also has an annotated UAG stop codon, rendering it possible that the authentic pylB gene product might either end or have pyrrolysine at this position. However, TAG was replaced with a TAA stop in one Archaeon (M. barkeri), and this change was made to M. acetivorans pylB. The modified pylB and pylCD genes were then placed behind separate isopropyl-β-d-thiogalactoside (IPTG)-inducible T7 promoters in pACYCDuet-1, creating plasmid pK13. This same promoter drove pylS, pylT, and mtmB1 expression in pEC03 (9); however, we reasoned that expression of mtmB1 after pylBCD would result in optimal incorporation of biosynthesized pyrrolysine into the UAG-encoded position of recombinant MtmB1. Therefore, pEC03 was modified to create pDLBAD, allowing differential expression of mtmB1 with the inducible E. coli arabinose promoter.
The UAG Translation Product of mtmB1 Is Made in E. coli Expressing pylTSBCD.
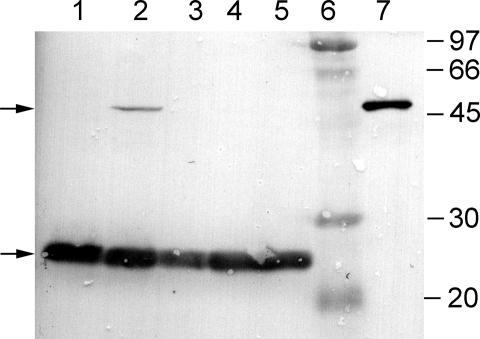
Plasmids pK13 and pDLBAD were cotransformed into E. coli. The pylTSBCD genes were induced at inoculation with IPTG, and the mtmB1 gene was induced with arabinose at midlog phase. Immunoblot analysis revealed that the mtmB1 UAG-termination (23-kDa) and UAG-translation (50-kDa) products were made in the strain containing both plasmids (Fig. 2). In contrast, cells cotransformed with the pACYCDuet-1 vector and pDLBAD produced only the UAG termination product of mtmB1. This result suggested that pylBCD enabled the production of endogenous pyrrolysine that was the used by PylS to charge tRNAPyl.
Fig. 2.
UAG translation in mtmB1 in E. coli bearing the five pyl genes. SDS-solubilized extracts of E. coli transformed with pylT, pylS, and mtmB1 on pDLBAD were analyzed by anti-MtmB1 immunoblot. Additionally, the strains were also transformed with the following: lane 1, the vector pACYCDuet-1; lane 2, pK13 bearing pylB, pylC, and pylD; lane 3, pK14 bearing pylB and pylD; lane 4, pK15 bearing pylC and pylD; and lane 5, pK16 bearing pylB and pylC. Lane 6 contains a set of standard proteins with molecular masses indicated at the right in kilodaltons, whereas lane 7 contains purified MtmB1. The upper arrow indicates the location of the mtmB1 UAG translation product, and the lower arrow indicates the location of the mtmB1 UAG termination product.
UAG Translation in Strains Lacking pylB, pylC, or pylD.
We next tested which of the pylBCD genes were essential for translation of the UAG codon within mtmB1. We modified the pK13 plasmid to create three different plasmids that contained only two of the pylBCD genes. Plasmid pK14 possessed pylB and pylD, and pK15 contained pylC and pylD, whereas pK16 bore pylB and pylC. These plasmids were cotransformed with pDLBAD into E. coli. However, none of these constructs produced significant levels of the UAG translation product of mtmB1 (Fig. 2), indicating that each of the pylBCD genes is essential for UAG translation with mtmB1.
Influence of pylBCD on pylTS-Mediated Amber Suppression During Expression of an E. coli Gene with an In-Frame UAG Codon.
The mtmB1 gene naturally encodes a pyrrolysyl protein within Methanosarcina spp.; this may involve cis-acting elements within the reading frame that promote UAG translation (22, 23), analogous to those required for UGA translation as selenocysteine (24). Because it is possible such elements could even influence UAG translation within a recombinant system (25), we tested whether UAG-dependent translation by pylTSBCD expression would suppress UAG when introduced into an E. coli gene that lacks any such elements. Therefore, we created pDLBADGUS, in which the mtmB1 gene was replaced by the E. coli uidA encoding β-glucuronidase (GUS). The lysine codon (AAA) of uidA at position 286 was changed to UAG. Termination at this codon would truncate the GUS protein by 50%, well before residues essential for catalysis. This plasmid was then transformed into an E. coli strain bearing an internal deletion of the chromosomal uidA gene, which was further lysogenized with λDE3 to provide T7 polymerase. This strain was then further transformed with either pACYCDuet-1 or with pK13. GUS activity was then assessed by direct cell assay. Cells bearing pACYCDuet-1 and pDLBADGUS and induced with IPTG possessed only 0.06 nmol/min per milligram of cellular protein. However, cells bearing pK13 with pDLBADGUS produced 50 times this level of GUS activity upon pylBCD induction, indicating that coexpression of pylTSBCD suppressed the amber codon within the E. coli gene, presumably by UAG translation.
PylS-Based Assays Reveal Pyrrolysine Synthesis Depends on pylBCD Expression.
No chemical test has been developed for pyrrolysine, and therefore we assayed for pyrrolysine in E. coli bearing pylBCD with recombinant PylS, the pyrrolysyl-tRNA synthetase (9). None of the other 20 amino acids can substitute for pyrrolysine in PylS-mediated tRNAPyl aminoacylation or pyrophosphate:ATP exchange reactions (5, 9, 10).
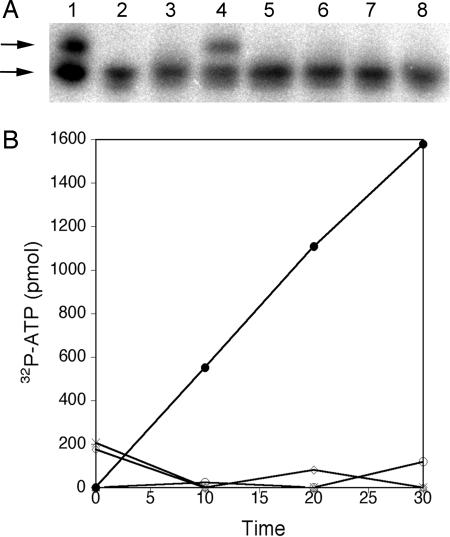
We extracted the methanol-soluble low-molecular-weight fraction from E. coli bearing pACYCDuet-1 and found it did not possess a substrate for tRNAPyl aminoacylation or pyrophosphate:ATP exchange by PylS (Fig. 3). In contrast, Northern blots of acid-urea gels revealed aminoacylation of tRNAPyl that depended upon addition of an extracted metabolite pool from E. coli containing pylBCD on plasmid pK13 (Fig. 3A). Further, a pyrophosphate:ATP exchange reaction was readily carried out by PylS in the presence of this same metabolite extract (Fig. 3B). Using our previously determined kinetic parameters of PylS in the pyrrolysine-dependent pyrophosphate:ATP exchange assay (9), we estimated from the rate of the exchange reaction that E. coli bearing pK13 produced 0.1–0.5 μmol pyrrolysine per gram of dry weight. Production of the PylS substrate for both types of PylS assay was found only with E. coli bearing pylBCD. E. coli transformed with pK14, pK15, or pK16, each of which lacks only one of the pylBCD genes, did not produce a PylS substrate detectable by either assay (Fig. 3). Significantly, the PylS substrate was produced in E. coli lacking pylT and pylS, consistent with the synthesis of pyrrolysine in the absence of tRNAPyl or the pyrrolysyl-tRNA synthetase.
Fig. 3.
Cells bearing pylB, pylC, and pylD produce pyrrolysine detectable by in vitro assays using pyrrolysyl-tRNA synthetase. (A) The direct aminoacylation of tRNAPyl with pyrrolysine present in cell extracts by PylS. Charged and uncharged tRNA species in the isolated cellular pool of tRNAPyl were separated in an acid-urea polyacrylamide gel that was subsequently electroblotted. The blot was then probed with 32P-labeled oligodeoxynucleotide complementary to tRNAPyl and analyzed by phosphorimager. Lane 1 was loaded with cellular tRNA as isolated (9) and has both charged (upper arrow) and uncharged (lower arrow) tRNAPyl, whereas lane 2 is the cellular tRNA pool following deacylation at pH 9 for 30 min and shows only uncharged tRNAPyl. Aminoacylation of tRNAPyl by PylS with pyrrolysine was then tested in 25-μl reactions that contained: 3.2 μM PylS, 50 mM KCl, 1 mM MgCl2, 5 mM ATP, 0.5 mM DTT, 8 μg of M. acetivorans deacylated cellular tRNA, and the metabolite pool from the indicated E. coli strains in 10 mM Hepes buffer, pH 7.2. After incubation for 40 min at 37°C, aminoacylation was tested as above in reactions that also contained the following: lane 3, no metabolite pool; or the pool from, lane 4, pK13 bearing pylB, pylC, and pylD; 5, pK14 bearing pylB and pylD; 6, pK15 bearing pylC and pylD; 7, pK16 bearing pylB and pylC; and 8, pACYCDuet-1 bearing no pyl gene. (B) Cellular amino acid pools were tested for activity in the pyrophosphate:ATP exchange assay mediated by PylS in the presence of pyrrolysine. Exchange of 32P-pyrophosphate into ATP was monitored in 100-μl reactions containing 5.5 μM PylS, 10 mM MgCl2, 25 mM KCl, 1 mM potassium fluoride, 4 mM DTT, 2 mM ATP, and 2 mM 32P-PPi (12 dpm/pmol) in 20 mM Hepes-KOH (pH 7.2) and incubated at 37°C. Aliquots were removed at the time points indicated, and the amount of radiolabel bound to acid-washed activated charcoal was quantified to estimate the amount of 32ATP formed. Shown are illustrated results from averaged duplicate reactions that were supplemented with the extracted amino acid pool from pK13 (●), pK14 (X), pK15 (△), pK16 (◇), or pACYCDuet-1 (○).
Pyrrolysine Is Encoded by UAG in E. coli Expressing pylTSBCD.
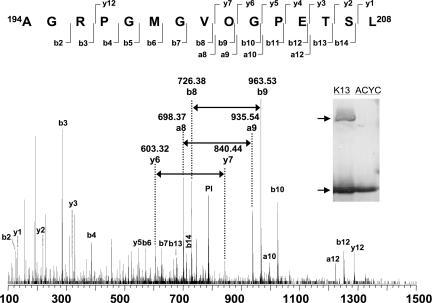
We considered the results of the PylS assays to be presumptive evidence that pyrrolysine was produced by the concerted action of each of the pylBCD gene products in the metabolic background of E. coli. As more definitive proof that pyrrolysine was produced and incorporated into protein in E. coli, we next directly analyzed the UAG-encoded residue in recombinant MtmB1 produced as a result of UAG translation in E. coli bearing pylBCD on pK13 and pDLBAD. The 50-kDa MtmB1 was partially purified (Fig. 4Inset) and subjected to in-gel chymotryptic digestion for subsequent analysis by mass spectrometry. An ion at m/z = 783.432+ was observed in the chymotryptic digest whose mass is consistent with the MtmB1 peptide sequence AGRPGMGVOGPETSL, where O is pyrrolysine. Further fragmentation of this peptide using collision-induced dissociation mass spectrometry [Fig. 4; supporting information (SI) Table 1] confirmed the peptide assignment. Identification of the b-, y-, and a-series ions resulting from fragmentation surrounding the pyrrolysyl residue allowed calculation of the mass of the UAG- encoded residue as 237.15 Da, compared with the theoretical mass of pyrrolysyl residue of 237.16 Da (6).
Fig. 4.
Mass spectrometry demonstrates that the UAG-encoded residue of recombinant mtmB1 is pyrrolysine in cells bearing the five pyl genes. (Inset) An anti-MtmB1 immunoblot of the 7-M urea-solubilized fraction from the cell pellet of French-press-lysed cells transformed with pDLBAD and either pACYCDuet-1 or pK13, as indicated. The upper arrow points to the UAG-translation mtmB1 product, whereas the lower arrow points to the UAG-termination product. Subsequently, the 7-M urea fraction was electrophoresed in an SDS-polyacrylamide gel, and the 50-kDa MtmB1 band was excised and subjected to in-gel digestion with chymotrypsin and analysis by mass spectrometry. The predicted sequence of the peptide ion at m/z = 783.432+ from chymotryptic cleavage of MtmB1 is shown along with the fragmentation sites forming the a-, b-, and y-series ions identified in the ion's collision-induced dissociation spectrum shown below. The peak of the parent ion (PI) is indicated. A horizontal arrow lies between the a-, b-, or y-series ion pairs whose mass differences allow calculation of the mass of the pyrrolysyl residue. The masses of the remaining identified ion peaks are listed in SI Table 1.
Discussion
We have shown here that transfer of the five pyl genes from M. acetivorans to E. coli leads to the translation of UAG as endogenously synthesized pyrrolysine. Recent efforts aimed at producing proteins with specifically modified residues have provided many examples of how the genetic code of Bacteria and Eucarya can be modulated by laboratory mutation of canonical aminoacyl-tRNA synthetase and cognate tRNA pairs to accept unnatural amino acids (18–20). In one case, an additional mutant biosynthetic enzyme enabled autogenous biosynthesis and genetic encoding of aminophenylalanine in E. coli (26). However, a gene cassette has been previously unidentified from nature that is capable of expanding an organism's genetic code by means of amber suppression to include a novel endogenously synthesized amino acid. As such, the pyl cassette gives insight into the biosynthesis and evolutionary history of pyrrolysine and possibly into the evolution of the canonical genetic code.
Previously, we demonstrated that expression of pylS and pylT in E. coli growing in the presence of exogenous pyrrolysine led to UAG translation as pyrrolysine (9). Our current work provides evidence that pylBCD are the key required genes for dedicated pyrrolysine biosynthesis from cellular precursors present in either enteric bacteria or Archaea. This result was unanticipated given the unusual metabolism of methanogenic Archaea and provides genetic proof of pylBCD function in pyrrolysine metabolism that would have otherwise required the inherent difficulties of archaeal genetics. Our results provide strong support that pyrrolysine is synthesized as a free amino acid, because production of pyrrolysine occurred in E. coli in the absence of genes encoding tRNAPyl. Biosynthesis of a PylS substrate without tRNAPyl is a necessary prerequisite if the PylS-catalyzed aminoacylation of tRNAPyl with chemically synthesized pyrrolysine (9–11) is a physiologically significant reaction.
The pylBCD gene products are members of protein families whose activities support several hypothetical pathways of pyrrolysine biosynthesis (ref. 13; see also SI Fig. 5) using intermediates found in either Archaea or Bacteria. PylB has signature residues of the radical SAM family that mediate radical-catalyzed reactions (27) and could be involved in pyrrolysine ring formation or methylation. The pylD gene product possesses the NADH-binding domain of several families of dehydrogenases. PylC has significant similarity to carbamoylphosphate synthetase and d-alanyl-d-alanine ligases, suggesting a potential role in forming the pyrrolysine amide bond. The amounts of pyrrolysine made within E. coli with pylBCD are relatively low compared with other amino acids (for example, tryptophan is ≈50 μmol per g of cells), which may underlie the lower level of UAG translation observed here compared with cells supplemented with high amounts of exogenous pyrrolysine (9). However, the pyrrolysine produced is readily detectable, and the recombinant system we have developed here will be instrumental in the elucidation of the biochemistry of pyrrolysine biosynthesis.
The demonstration that pylBCD are the pyrrolysine biosynthetic genes makes the pyl gene cluster highly unusual in that, to our knowledge, there is no other example of a single cluster of genes that includes the tRNA, aminoacyl-tRNA synthetase, and key biosynthetic genes for synthesis of an amino acid. It is notable that the genes required for the biosynthesis of the selenocystenyl-tRNA are not typically found in a single gene cluster. An exception is the selDABC gene cluster from Eubacterium acidoaminophilum whose products would, with host-supplied seryl-tRNA synthetase, support UGA translation as Sec in the context of genes possessing a SECIS element (28). An mRNA-specific recoding (29) phenomenon also enhances UAG translation as pyrrolysine within mtmB1 within M. acetivorans (30); however, it is as yet unknown whether message context-dependent recoding is operating during translation of UAG as pyrrolysine in mtmB1 in recombinant E. coli (9, 25). However, such recoding is unlikely to underlie the translation of uidA with an in-frame amber codon in pyl cassette-transformed E. coli or in M. acetivorans bearing this same gene (30).
The unique arrangement of the pyl cassette would facilitate the ability to transfer biosynthesis and genetic encoding of pyrrolysine within natural populations. The transmissibility of biosynthesis and genetic encoding of pyrrolysine may underlie its known distribution in nature in a cluster of methanogenic archaeal species and a single Gram-positive bacterial representative. Lateral transfer of the pyl gene cluster provides a ready scenario for movement of the genetically encoded amino acid from one domain to the other. The acquisition of the novel amino acid within one or the other would be stabilized in the genome by the further addition of a metabolic protein whose function depended upon UAG translation as pyrrolysine, such as the trimethylamine methyltransferase homologs common to both D. hafniense and Methanosarcina spp. (1, 7, 8, 31).
Vetsigian et al. (32) have postulated that early stages of the evolution of the genetic code may have been greatly accelerated by horizontal gene transfer events that led to a communal advantage for utilization of a common and increasingly optimized genetic code. Innovations in the genetic code might also move through populations by horizontal gene transfer (32). It has been suggested that, during early cellular evolution, new genetically encoded amino acids joined more ancient ones as novel biosynthetic pathways arose that used older amino acids and their precursors (33–36). Newer amino acids joined the growing genetic code by charging of their own tRNA species that allowed interpretation of a codon as the new recruit to the genetic code, even while the older codon function was maintained. Such a scenario for genetic code expansion requires that the genes for biosynthesis of the new amino acid become present even as a means for joining the genetic code becomes available. In this regard, the pyl genetic code expansion cassette takes on further significance as the first gene cluster identified in nature that simultaneously brings these two requirements for the genetic encoding of a novel free amino acid. Analogous genetic code expansion cassettes may have served useful purposes during the later stages of the development of the modern genetic code. Indeed, the abilities that the pyl cassette can bestow on a recipient organism would even now allow it to be engaged in an evolutionary process, one in which horizontal gene transfer continues to influence the genetic code of modern organisms.
Materials and Methods
Plasmid Constructions.
The pylB, pylC, and pylD genes were amplified by PCR from M. acetivorans C2A genomic DNA (9) using Ex-taq (Takara Mirus Bio, Madison, WI) and were subsequently ligated into pCR2.1-Topo (Invitrogen, Carlsbad, CA). The pylB gene was PCR-amplified by using primers PylBNdeI and PylBBglII (for all primers, see SI Table 2), changing the start codon from TTG to ATG. A silent mutation was made in pylB to remove an internal NcoI site by using primers pylBNcoImut1 and pylBNcoImut2 to facilitate subsequent cloning. The altered pylB gene was ligated as a NdeI and BglII fragment into pACYC-Duet-1 (Novagen, Madison, WI) to create pK11. The pylCD gene sequence was PCR-amplified from genomic DNA by using primers PylCDNcoI and PylCDBamHI that changed the 5′ bases of pylC from TTGA to ATGG. This fragment was then ligated into pK11 at the unique NcoI and BamHI sites to form pK12. The 5′ ATGG sequence of pK12 pylC was then mutated to ATGA by using PylCNcoImut1 and PylCNcoImut2 to create pK13. The pK14 plasmid bearing only pylBD was constructed by amplifying pylD from pK13 by using primers PylDNcoI and PylCDBamHI; this fragment was then ligated into pK13, replacing pylCD. Plasmid pK15 was constructed by excising the pylCD from pK13 and ligating it into pACYC by using the NcoI and BamHI sites. To create pK16 bearing only pylBC, the pylC gene was amplified by using primers PylCMluI and PylCDMluI, ligated into pCR2.1-Topo, and moved into pK11 by using MluI and BamHI.
The pDLBAD plasmid bearing pylT, pylS, and mtmB1 was derived from pEC03 (9). The arabinose promoter from pBAD33 (37) was amplified by using primers pBADF and pBADR and ligated into pCR2.1-Topo. These primers introduced a T7 terminator site at the 5′ end and a ribosomal-binding sequence and NdeI site following the 3′ end of the arabinose promoter. The arabinose promoter was then ligated into pEC03 between NotI and NdeI, thereby replacing the T7 promoter of the pEC03 mtmB1 gene to create pDLBAD. The pDLBAD plasmid was used to create pDLBADGUS, in which mtmB1 was replaced by uidA from E. coli. The uidA gene was obtained from pJK65 (38). Codon 286 (AAA) of uidA was modified with the QuikChange Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA) to TAG with primers UTAGF and UTAGR. The modified gene was then PCR-amplified with primers that introduced NdeI and KpnI sites for insertion into pDLBAD plasmid.
Pyl Gene-Dependent UAG Translation in E. coli.
pDLBAD was cotransformed with the pK13, pK14, pK15, pK16, or pACYCDuet-1 into BL21 Tuner (DE3) (Novagen). Overnight cultures (3 ml) in LB broth supplemented with 100 mg/ml ampicillin and 34 mg/ml chloramphenicol were used as inocula (50 μl) in cultures (1 ml) in the same medium further supplemented with 80 μM IPTG for induction of the pyl genes. Expression of mtmB1 was subsequently induced at OD600 = 1.2 with 0.02% l-arabinose and incubated at 37°C for 2 h. The mtmB1 gene products in equivalent amounts of lysates were then analyzed by immunoblotting a SDS-12.5% polyacrylamide electrophoretic gel by using affinity-purified rabbit anti-MtmB antibody (3). Rainbow Molecular Weight markers (Amersham Biosciences, Piscataway, NJ) were used as molecular size standards.
E. coli BW25141 (39), which bears an internal MluI deletion of the uidA gene (corresponding to codons 200–439), was used to test suppression of the UAG codon at position 286 of the uidA gene carried on pDLBADGUS. The strain was further lysogenized with λDE3 to introduce T7 polymerase. Cells were grown overnight in Terrific Broth containing ampicillin and chloramphenicol, then 50 μl was inoculated into 1 ml of fresh medium containing 80 μM IPTG. At OD = 1.0, 0.2% arabinose was added and, after 3 h, the cells were centrifuged, resuspended in 50 mM KPO4 (pH 7) buffer, centrifuged, and resuspended in 200 μl of the same buffer. GUS activity was measured by using 1.25 mM p-nitrophenolglucuronide at 405 nm by using an extinction coefficient of 18.6 mM−1.
Mass Spectral Identification of Pyrrolysine as the UAG-Encoded Residue.
Recombinant MtmB was isolated for mass spectrometry from a culture (100 ml) of E. coli bearing pDLBAD and pK13. The recombinant MtmB1 was found entirely in inclusion bodies that were centrifuged from a cell lysate. The pellet was then extracted sequentially with 0, 1, 3, 5, and 7 M urea in 50 mM 4-morpholinepropanesulfonic acid, pH 7. The mtmB1 UAG translation product was solubilized in 7 M urea as detected by immunoblotting, and this fraction was further separated by SDS gel electrophoresis. The 50-kDa MtmB1 was subjected to in-gel chymotrypsin digestion and peptide sequencing by tandem mass spectrometry by using previously described methods (6, 9).
Extraction of Metabolite Pools and PylS-Dependent Assays for Pyrrolysine.
Cells carrying pK13–16 were grown on LB with chloramphenicol and induced at OD600 = 0.3 with 1 mM IPTG and incubated for 4 h at 37°C. Harvested cell pellets were then extracted with 2 ml of methanol per gram of cell wet weight for 10 min at room temperature, centrifuged for 5 min at 13,000 × g, and the supernatant was passed through a YM-3 filter (Millipore, Bedford, MA) to completely remove any remaining high-molecular-weight material.
To assay the methanolic extract, aliquots were dried during centrifugation under vacuum, and the extracted metabolites were resuspended in assay buffer appropriate for either aminoacylation or pyrophosphate exchange reactions mediated by PylS. Assays were carried out in the presence of the metabolites extracted from the equivalent of 10 mg of dry weight of E. coli. Aminoacylation of the tRNAPyl in the extracted tRNA pool of M. acetivorans was carried out and detected by Northern blotting of acid-urea gels, as described (9, 40). Pyrophosphate exchange assays also follow previously described protocols (9, 41), except that 5.5 μM PylS in 100 μl of total assay volume was used, and 20 μl of aliquots was taken at each time point.
Supplementary Material
Acknowledgments
We thank John Reeve and Tina Henkin for useful discussions and Marsha Thalhofer for assistance with the pyrophosphate exchange assays. This work was supported by National Institutes of Health Grant GM070663.
Abbreviations
- IPTG
isopropyl-β-d-thiogalactoside
- GUS
β-glucuronidase.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610294104/DC1.
References
- 1.Paul L, Ferguson DJ, Krzycki JA. J Bacteriol. 2000;182:2520–2529. doi: 10.1128/jb.182.9.2520-2529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burke SA, Lo SL, Krzycki JA. J Bacteriol. 1998;180:3432–3440. doi: 10.1128/jb.180.13.3432-3440.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.James CM, Ferguson TK, Leykam JF, Krzycki JA. J Biol Chem. 2001;276:34252–34258. doi: 10.1074/jbc.M102929200. [DOI] [PubMed] [Google Scholar]
- 4.Hao B, Gong W, Ferguson TK, James CM, Krzycki JA, Chan MK. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 5.Hao B, Zhao G, Kang P, Soares J, Ferguson T, Gallucci J, Krzycki J, Chan M. Chem Biol. 2004;11:1317–1324. doi: 10.1016/j.chembiol.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 6.Soares JA, Zhang L, Pitsch RL, Kleinholz NM, Jones RB, Wolff JJ, Amster J, Green-Church KB, Krzycki JA. J Biol Chem. 2005;280:36962–36969. doi: 10.1074/jbc.M506402200. [DOI] [PubMed] [Google Scholar]
- 7.Srinivasan G, James CM, Krzycki JA. Science. 2002;296:1459–1462. doi: 10.1126/science.1069588. [DOI] [PubMed] [Google Scholar]
- 8.Mahapatra A, Patel A, Soares JA, Larue RC, Zhang JK, Metcalf WW, Krzycki JA. Mol Microbiol. 2006;59:56–66. doi: 10.1111/j.1365-2958.2005.04927.x. [DOI] [PubMed] [Google Scholar]
- 9.Blight SK, Larue RC, Mahapatra A, Longstaff DG, Chang E, Zhao G, Kang PT, Green-Church KB, Chan MK, Krzycki JA. Nature. 2004;431:333–335. doi: 10.1038/nature02895. [DOI] [PubMed] [Google Scholar]
- 10.Polycarpo C, Ambrogelly A, Berube A, Winbush SM, McCloskey JA, Crain PF, Wood JL, Söll D. Proc Natl Acad Sci USA. 2004;101:12450–12454. doi: 10.1073/pnas.0405362101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krzycki JA. Curr Opin Microbiol. 2005;8:706–712. doi: 10.1016/j.mib.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Théobald-Dietrich A, Frugier M, Giegé R, Rudinger-Thirion J. Nucleic Acids Res. 2004;32:1091–1096. doi: 10.1093/nar/gkh266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krzycki JA. Curr Opin Chem Biol. 2004;8:484–491. doi: 10.1016/j.cbpa.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 14.Galagan JE, Nusbaum C, Roy A, Endrizzi MG, Macdonald P, FitzHugh W, Calvo S, Engels R, Smirnov S, Atnoor D, et al. Genome Res. 2002;12:532–542. doi: 10.1101/gr.223902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deppenmeier U, Johann A, Hartsch T, Merkl R, Schmitz RA, Martinez-Arias R, Henne A, Wiezer A, Bäumer S, Jacobi C, et al. J Mol Microbiol Biotechnol. 2002;4:453–461. [PubMed] [Google Scholar]
- 16.Goodchild A, Saunders NF, Ertan H, Raftery M, Guilhaus M, Curmi PMG, Cavicchioli R. Mol Microbiol. 2004;53:309–321. doi: 10.1111/j.1365-2958.2004.04130.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Baranov PV, Atkins JF, Gladyshev VN. J Biol Chem. 2005;280:20740–20751. doi: 10.1074/jbc.M501458200. [DOI] [PubMed] [Google Scholar]
- 18.Hendrickson TL, de Crecy-Lagard V, Schimmel P. Annu Rev Biochem. 2004;73:147–176. doi: 10.1146/annurev.biochem.73.012803.092429. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Xie J, Schultz PG. Annu Rev Biophys Biomol Struct. 2006;35:225–249. doi: 10.1146/annurev.biophys.35.101105.121507. [DOI] [PubMed] [Google Scholar]
- 20.Link AJ, Mock ML, Tirrell DA. Curr Opin Biotechnol. 2003;14:603–609. doi: 10.1016/j.copbio.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 21.Blattner FR, Plunkett G, III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 22.Atkins JF, Gesteland R. Science. 2002;296:1409–1411. doi: 10.1126/science.1073339. [DOI] [PubMed] [Google Scholar]
- 23.Namy O, Rousset JP, Napthine S, Brierley I. Mol Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- 24.Böck A, Thanbichler M, Rother M, Resch A. In: The Aminoacyl-tRNA Synthetases. Ibba M, Francklyn C, Cusack S, editors. Austin, TX: Landes Bioscience; 2004. [Google Scholar]
- 25.Schimmel P, Beebe K. Nature. 2004;431:257–258. doi: 10.1038/431257a. [DOI] [PubMed] [Google Scholar]
- 26.Mehl RA, Anderson JC, Santoro SW, Wang L, Martin AB, King DS, Horn DM, Schultz PG. J Am Chem Soc. 2003;125:935–939. doi: 10.1021/ja0284153. [DOI] [PubMed] [Google Scholar]
- 27.Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. Nucleic Acids Res. 2001;29:1097–1106. doi: 10.1093/nar/29.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gursinsky T, Jager J, Andreesen JR, Sohling B. Arch Microbiol. 2000;174:200–212. doi: 10.1007/s002030000196. [DOI] [PubMed] [Google Scholar]
- 29.Gesteland RF, Atkins JF. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 30.Longstaff DG, Blight SK, Zhang L, Green-Church KB, Krzycki JA. Mol Microbiol. 2007;63:229–241. doi: 10.1111/j.1365-2958.2006.05500.x. [DOI] [PubMed] [Google Scholar]
- 31.Ferguson DJ, Jr, Krzycki JA. J Bacteriol. 1997;179:846–852. doi: 10.1128/jb.179.3.846-852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vetsigian K, Woese C, Goldenfeld N. Proc Natl Acad Sci USA. 2006;103:10696–106701. doi: 10.1073/pnas.0603780103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crick FH. J Mol Biol. 1968;38:367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- 34.Wong JT. Proc Natl Acad Sci USA. 1975;72:1909–1912. doi: 10.1073/pnas.72.5.1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong JT. BioEssays. 2005;27:416–425. doi: 10.1002/bies.20208. [DOI] [PubMed] [Google Scholar]
- 36.Yarus M, Caporaso JG, Knight R. Annu Rev Biochem. 2005;74:179–198. doi: 10.1146/annurev.biochem.74.082803.133119. [DOI] [PubMed] [Google Scholar]
- 37.Guzman LM, Belin D, Carson MJ, Beckwith J. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pritchett MA, Zhang JK, Metcalf WW. Appl Environ Microbiol. 2004;70:1425–1433. doi: 10.1128/AEM.70.3.1425-1433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haldimann A, Wanner BL. J Bacteriol. 2001;183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jester BC, Levengood JD, Roy H, Ibba M, Devine KM. Proc Natl Acad Sci USA. 2003;100:14351–14356. doi: 10.1073/pnas.2036253100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cole F, Schimmel PR. Biochemistry. 1970;9:480–489. doi: 10.1021/bi00805a005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.