Abstract
Sleep is one of the most noticeable and widespread phenomena occurring in multicellular animals. Nevertheless, no consensus for a theory of its origins has emerged. In particular, no explicit, quantitative theory exists that elucidates or distinguishes between the myriad hypotheses proposed for sleep. Here, we develop a general, quantitative theory for mammalian sleep that relates many of its fundamental parameters to metabolic rate and body size. Several mechanisms suggested for the function of sleep can be placed in this framework, e.g., cellular repair of damage caused by metabolic processes as well as cortical reorganization to process sensory input. Our theory leads to predictions for sleep time, sleep cycle time, and rapid eye movement time as functions of body and brain mass, and it explains, for example, why mice sleep ≈14 hours per day relative to the 3.5 hours per day that elephants sleep. Data for 96 species of mammals, spanning six orders of magnitude in body size, are consistent with these predictions and provide strong evidence that time scales for sleep are set by the brain's, not the whole-body, metabolic rate.
Keywords: allometric scaling, brain, cellular repair, metabolic rate, sleep times
In contrast to other obvious physiological phenomena such as eating, breathing, and walking, neither the function nor the mechanism by which sleep occurs is well understood, despite its ubiquity (1). Indeed, the quest for a fundamental theory of sleep is considered one of the most important, unsolved problems in science (2–7). Recent neurobiological studies have made great advances in understanding the mechanisms involved in sleep. Hormones, cells, and enzymes whose levels of activity and expression vary during sleep and between sleeping and waking states have been identified (2,8–10). Although these studies have been unable to determine the purpose of sleep, such investigations will play an increasingly important role in determining what processes are specific to sleep.
Among the most studied and best known hypotheses for the purpose of sleep are the following: (i) rest for the body or brain and prevention from overheating (11–13); (ii) cortical reorganization and processing associated with memory and learning (14–17); and (iii) cellular repair in the body or brain (3,18–21,54). Some hypotheses for sleep are easy to eliminate. For example, hypothesis i has been rejected because the energy saved during sleep is minimal, amounting to approximately one frankfurter bun worth of calories per night for humans (11, 12). Moreover, because rates of heating and cooling are set by mass-specific metabolic rate and body size, which should scale identically, equating the heat increase that leads to overheating with the heat lost to return to normal temperatures predicts that the ratio of sleep time to awake time is invariant with respect to body size. This is clearly counter to the observation that sleep times decrease with body size.
Distinguishing among other theories of sleep (e.g., hypotheses i and ii), however, has been much more difficult. Here, we develop a quantitative theory whose general structure can, in principle, be used to test any of these hypotheses for the function of sleep, particularly when combined with interspecific analyses of mammalian sleep data. Recently, Siegel (21) noted that such data could help illuminate and distinguish between different theories of sleep, and he specifically discussed empirical data in relation to sleep as a process of neuronal repair (hypothesis iii).
Our starting point is the observation that all of these suggested underlying processes are related to metabolic rate. By specifying how each of these processes (hypotheses i–iii) is related to metabolic rate, we can derive allometric scaling relationships for several important sleep rates and times, thereby predicting, for example, how total sleep time, sleep cycle time, and the fraction of rapid eye movement (REM) sleep scale with body mass, M. These scaling relationships are usually some combination of simple power laws, ≈aMp, where a is the scaling coefficient and p is the scaling exponent. As we demonstrate, the scaling exponent, p, alone is sufficient to distinguish whether sleep times are set by the whole body's or the brain's metabolic rate and to discern whether hypotheses ii and iii are consistent with empirical data and, thus, are viable theories. Our quantitative theory makes explicit, quantitative predictions regarding scaling exponents, p, which are tested by using empirical data for organisms that span six orders of magnitude in body size. Our analyses show sleep times are set by the brain's, not the whole body's, metabolic rate and that hypotheses ii and iii are both consistent with empirical data.
To distinguish between hypothesis ii (reorganization) and hypothesis iii (repair) requires knowledge regarding the scaling coefficient, a. In the theory, this is related to specific physiological, most likely cellular, processes that are independent of body mass. Unfortunately, little or nothing is known about these, thereby limiting our current ability to make quantitative predictions regarding a. Consequently, experiments that measure the values, or possibly even the magnitudes, of the relevant physiological variables would allow us to further distinguish between reorganization and repair. In Discussion, we address some experiments that would need to be performed to achieve this.
In constructing our theory, we speculate that periods of cortical reorganization and/or repair of neuronal damage are induced primarily by activity during alert wakefulness. Because neurons typically are regenerated slowly or not at all during the lifetime of the organism, faithful repair of cellular damage is critical for maintaining the long-term integrity and functioning of the brain. Moreover, this extremely low turnover of neurons suggests that maintenance of the brain and processes such as learning involve changes within, or connections between, existing neurons, i.e., reorganization.
Cellular repair is especially crucial for the brain because, unlike most other organs and tissues, its cells are not continually being replaced. Dissipative energy is necessarily produced as a by-product of metabolic processes and, inevitably, leads to cellular damage. Dissipation occurs both at the basic biochemical level, as, for example, in the production of free radicals, and at the capillary level in the circulatory system due to hemodynamic analogs of viscous drag forces. Experimental evidence that brain cell damage is caused by sleep deprivation (22, 23) has been reported, although other studies did not corroborate these findings (24, 25). Furthermore, the recent findings of Cirelli et al. (26) demonstrate that Drosophila Shaker mutants with reduced sleep time experience a concurrent and identical decrease in lifespan. Assuming all other factors such as metabolic rate are held constant (consistent with measures of activity; ref. 26), these reduced sleep times result in increased periods of damage and reduced periods of repair. Thus, lifespan for these Drosophila Shaker mutants should be reduced, just as observed, suggesting that sleep indeed is linked to repair (26).
Studies have shown that sleep time is positively correlated to metabolic rate, body weight, brain weight, and other physiological variables (11, 13, 27). Because cellular damage is directly tied to metabolism and its repair is a leading hypothesis for sleep, we develop our theory with this scenario in mind, similar to ideas recently expressed by Siegel (21). However, as we explain, the framework's general structure can accommodate other mechanisms, most notably structural and functional reorganization.
According to these ideas, because smaller animals have proportionately higher metabolic rates per unit mass than larger animals, they require more sleep over a fixed period. Such qualitative predictions that agree with empirical observations are clearly useful, but much more detailed analyses and tests of hypotheses are possible once a quantitative theory has been developed. Below, explicit relationships among sleep, metabolic rate, and body size are derived. These provide a way to distinguish whether cellular repair during sleep occurs solely in the brain or throughout the entire body and whether cellular damage occurs at the same rate during sleep and wakefulness.
Theory
Mass Dependence of the Ratio of Sleep Time to Awake Time.
We begin by assuming that metabolic processes during wakefulness cause most cellular damage and dictate the rate at which sensory input is processed. We also assume that cellular repair and/or neural reorganization occur primarily during sleep. These assumptions will be relaxed below. Dissipative metabolic energy, e.g., viscous-type forces in metabolite transportation networks, including the exchange of oxygen from hemoglobin to tissue, occurs primarily in network terminal units (capillaries and mitochondria), which are approximately constant in size and do not vary appreciably with body mass (28). This invariance implies that both the total rate of metabolic energy dissipation and the metabolic rate itself, B, scale linearly with the number of such terminal units. Consequently, the total rate of cellular damage caused by energy dissipation is a mass-independent fraction, f, of metabolic rate, B. This invariant fraction is calculable at the capillary level (28). At the mitochondrial level, it also holds because mammals share a common biochemistry of metabolism, and the stoichiometry generates a predictable quantity of energy with free radicals as byproducts.
The rate at which metabolic energy is dissipated in an average cell therefore is fBc, where Bc is the average in vivo cellular metabolic rate. The total metabolic rate of the organ, tissue, or whole body being considered is B = NcBc, where Nc is the corresponding total number of cells. During sleep, the total amount of energy causing damage that needs to be repaired therefore is given by fBtA, where tA is total time awake (alert wakefulness) per day, and the number of damaged units is given by Nd = fBtA/ΔEd, where ΔEd is the average energy per damage event. Further, we assume that the process of cellular repair is similar across mammals and occurs at a subcellular level, so that the power density (per unit volume) required for repair, PR, is independent of body size (see Appendix for a more detailed discussion). Thus, the number of repairs is NR = PRvcNctS/ΔER, where vc is the average cell volume, tS is sleep time per day, and ΔER is the average energy required for a single repair. For faithful repair, all damage must be repaired, so Nd ≈ NR. Combining this with our previous relationships gives fBtA ≈ εPRvcNctS, where ε = ΔEd/ΔER. Because ΔEd and ΔER are energies (not rates) that presumably occur at subcellular levels, ε is independent of body size. (Note that our conclusions hold so long as ε ∝ PR−1.) The total volume (of the organ, tissue, or whole body) considered is V = Ncvc, leading to
where M is the corresponding total mass, and ρ ≡ M/V, the tissue density, which is a constant. Eq. 1 can be expressed in terms of purely cellular quantities as tS/tA ≈ (ρf/εPR) Bc/mc, where mc is average cell mass. If neural reorganization is the origin of sleep, Eq. 1 still holds but with PR now interpreted as the power density required for performing such an activity, f the fraction of metabolic energy used for processing information, and ε the measure of efficiency in neural reorganization. In either case, the quantity related to metabolic rate is neither sleep nor awake time but rather, a ratio of the two.
The scaling of whole-body metabolic rate with body size, B = B0M3/4, where B0 is the normalization constant, has been shown to reflect general properties of resource distribution networks. These networks are assumed to be space filling and have invariant terminal units. Minimizing energy expenditure in the circulatory system leads to quarter-power allometric scaling for many physiological rates and times (28). Organs such as the brain, which are supplied by major arteries, behave as nearly autonomous subunits and can effectively be treated as independent systems subject to the constraints of the theory. Their metabolic rate, Bi, therefore is predicted to scale approximately as Mi3/4, where Mi is the organ mass. If organ mass itself scales with total body mass as Mi ∝ Mai, then the metabolic rate of the organ scales with body mass as Bi ∝ M(3/4)ai. From this, it immediately follows that the mass-specific metabolic rate of the organ scales with body mass as Bi/Mi ∝ Mi−1/4 ∝ M−pi, where pi ≡ (1/4)ai. The mass of most organs scales approximately linearly with body mass ai ≈ 1, so mass-specific metabolic rate scales as a typical physiological rate, Bi/Mi ∝ M−1/4. Brains, however, are exceptional. Reported empirical values for ab (b denotes brain) range from 0.65 (≈2/3) to 0.76 (≈3/4) (29–31). Consequently, Bb/Mb ∝ Mb−p with pb ranging from ≈1/6 to 3/16. Unfortunately, no in vivo data are available to test this. However, the limited available data for rates of oxygen consumption in brain tissue in vitro are not inconsistent with these predictions (see Appendix).
Thus, if the primary purpose of sleep is to repair damage done to brain cells, then
with pb in the range 0.16–0.19. This equation explicitly predicts the exponent for how sleep time scales allometrically with body and brain size. Note, however, that the absolute value of tS/tA could be predicted from Eq. 1 if repair and dissipative rates (PR and f) were known. The differences in interpretation between repair and reorganization correspond to different values for the mass-independent constants in Eq. 1. Thus, if the values, or possibly even the magnitudes of PR, f, and ε are known, they can be used to distinguish between reorganization (hypothesis ii) and repair (hypothesis iii) as the function of sleep. It is important to realize that both processes may equally contribute to the function of sleep and that also would be revealed by such a quantitative analysis. Moreover, if the primary purpose were to repair damage to organs other than the brain, then the exponent in Eq. 2 would be −1/4 rather than −1/6 or −3/16. This provides a powerful way to distinguish whether sleep functions to repair molecular and cellular damage or to reorganize cellular connections throughout the body or primarily in the brain.
Extension to Include Damage During Sleep.
Eqs. 1 and 2 can be further generalized to include damage occurring during sleep itself:
where α ≡ (fB)S/(fB)A is the damage rate during sleep relative to that during alert wakefulness.
If power devoted to repair far exceeds that leading to damage during sleep itself, i.e., εPRM/ρ ≫ (fB)S = α(fB)A, the additional contribution, −α, can be ignored. Because tA/tS > 0, it follows that εPRMb1/4/ρ > α(fB0)A, so the contribution of α decreases with increasing Mb (or M), only being important for small mammals. For example, if α ≈ 1/3, as suggested by data (see Appendix), then damage during sleep is significant only for very small animals that sleep >≈18 h/day.
On the other hand, if damage rates were unchanged between sleeping and waking states (α ≈ 1), then, because tS + tA = 1 day, Eq. 3 reduces to a pure power law for tS: tS ∝ Mpb. This is inconsistent with fits to data (see below) and with the previously mentioned value of α ≈ 1/3. However, it is known that whole-body metabolic rate decreases by only ≈10–15% in sleep versus resting states, and the brain's metabolic rate decreases by much less (4). Thus, for damage rates to decrease significantly during sleep, f must decrease, driven presumably by mechanisms that, for example, suppress radical production or increase antioxidants. Regardless of mechanism, Eq. 3 provides a framework for probing this important question by determining α.
This analysis suggests that the primary quantity to consider is the ratio tS/tA rather than either tS or tA separately, as has been typically done in the literature. This is to be contrasted with typical physiological rates (such as heart and respiratory rates), which scale as mass-specific metabolic rate, B/M ∝ M−1/4, and to most physiological times (such as blood circulation time and time to maturity), which scale as its inverse, M1/4. These scaling behaviors originate from generic constraints on distribution networks (28, 32) and pertain to quantities that are linked to the primary beneficiary function of metabolism, namely to supply energy and nutrients. This is in distinct contrast to sleep time, tS, whose primary purpose is hypothesized either to counteract detrimental, secondary effects of metabolic processes or to reorganize neural circuitry to incorporate sensory input obtained during alert wakefulness, another secondary effect. This critical difference is reflected in the scaling of tS, which not only does not follow a simple power law, but, unlike almost all physiological times, decreases, rather than increases, with body size.
Mass Dependence of Sleep Cycle Time and Fraction of Sleep Time Spent in REM.
Two of the most intriguing aspects of sleep are the division between REM and non-REM states and the oscillations between them known as the sleep cycle. Understanding these phenomena could help reveal the role that each plays in brain repair and functioning. If, unlike sleep itself, they do not counteract the detrimental effects of metabolism or help process sensory input during alert wakefulness but are driven by the primary beneficiary function of metabolism, then all times associated with them are expected to scale like typical physiological times. As such, sleep cycle time, tc, the time between the endings of periods of REM sleep, is predicted to scale as tc ∝ Mb1/4 ∝ Mpb. Consequently, if nC is the number of sleep cycles per day, so total sleep time tS = nCtc, then nc(∝tS/tc) decreases with M but does not follow a simple power law. The average length of both REM sleep per cycle, tR, and of non-REM sleep per cycle, tNR, are likewise predicted to scale as tR ∝ tNR ∝ tc ∝ Mb1/4 ∝ Mpb. So, during a time interval that spans several sleep cycles, the fraction of sleep time spent in REM, R = nCtR/tS = tR/tC, is predicted to be independent of mass. A possible mechanism for the division between REM and non-REM states is that they represent distinct periods devoted primarily to reorganizing or repairing different functional components or cell types or tissue within the brain. Local regions that become activated during REM cycles have been identified, and knowledge of their size would allow an estimate for R.
Results
Ratio of Sleep Time to Awake Time.
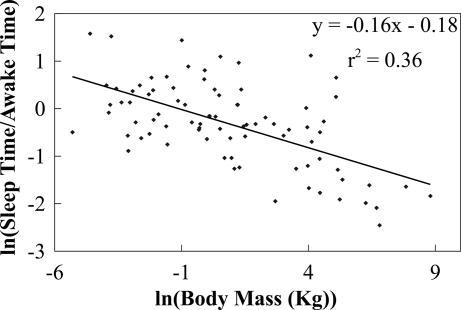
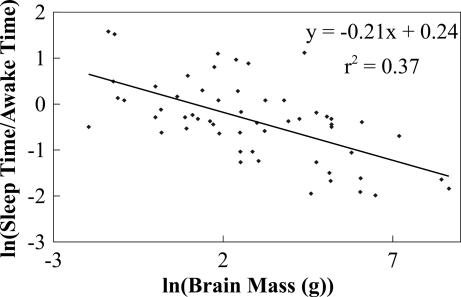
We now compare our predictions with data [see supporting information (SI) Table 1]. Fig. 1 shows daily values of ln(tS/tA) versus ln(M) for 83 taxa, representing 96 species and 79 genera of mammals and spanning six orders of magnitude in body mass. The slope, obtained by ordinary least squares (OLS) regression, is –0.16 [P < 0.0001; n = 83, 95% confidence interval (CI): –0.21, −0.11], in agreement with Eq. 2. Note that the 95% CI includes both −1/6 and −3/16 (values linked to repair and reorganization in the brain) but exclude −1/4 (the value predicted if repair and reorganization occur throughout the body), consistent with the hypothesis that the major function of sleep is related to the brain. This exponent is in clear disagreement with +1/4, the naive expectation if tS behaves like a typical physiological time. Not only does it have the wrong magnitude but, more significantly, the wrong sign. In Fig. 2, using data for brain mass, we plot ln(tS/tA) versus ln(Mb), which has a slope of −0.21 (P < 0.0001; n = 56, 95% CI: −0.28, −0.14), in agreement with Eq. 2 and the −1/4 predicted if the brain's metabolic rate drives sleep. Recall that tS satisfies a pure power law when damage is uniform throughout sleeping and waking periods (α ≈ 1). Data give an exponent for tS of −0.1 with a 95% CI that does not include the values −1/6, −3/16, or −1/4 predicted if α ≈ 1. Moreover, by fixing pb ≈ 1/6 or 3/16, α is determined to be small (see Appendix). These results are consistent with damage during sleep being negligible. Although gathering more data would improve estimates for α, the scatter in Figs. 1 and 2 is not just measurement error but is due to inherent biological effects such as alternate life histories or predator–prey relationships (27, 33).
Fig. 1.
Plot of the logarithm of the ratio of total sleep time, tS, to total awake time per day, tA, versus the logarithm of body mass, ln(M), in kilograms. The slope computed by using OLS regression is –0.16 (P < 0.0001; n = 83, 95% CI: –0.21, −0.11). Note that the 95% CI includes –1/6 and –3/16 predicted by the theory but excludes –1/4, supporting the hypothesis that cellular repair or neural reorganization during sleep occurs primarily in the brain.
Fig. 2.
Plot of the logarithm of the ratio of total sleep time, tS, to total awake time per day, tA, versus the logarithm of brain mass, ln(Mb), in grams. The slope computed by using OLS regression is –0.21 (P < 0.0001; n = 56, 95% CI: −0.28, −0.14). Note that the 95% CI includes the predicted value of –1/4, providing more evidence that sleep is driven primarily by the brain's metabolic rate.
Sleep Cycle Time and Fraction of Sleep Time Spent in REM.
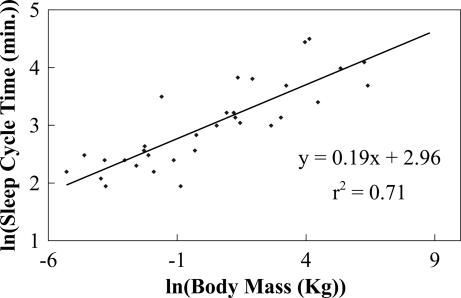
In Fig. 3, we show ln(tc) versus ln(M) for 32 species of mammals, spanning five orders of magnitude. Using OLS regression, we find a slope of 0.19 (P < 0.0001; n = 32, 95% CI: 0.14, 0.23), which is close to the predicted value of 3/16. The 95% CI includes the predicted 1/6 and 3/16 but excludes 1/4, thereby providing support that sleep cycle time is set directly by the brain's metabolic rate. Finally, in Fig. 4, we show ln(R) versus ln(M), for 61 species of mammals, spanning five orders of magnitude. The mean value of R is 0.17, with a slope of −6 × 10−3 (P = 0.74; n = 61, 95% CI: −0.04, 0.03), consistent with zero and REM sleep being a constant fraction of total sleep time, as predicted. See Elgar et al. (13) for additional evidence.
Fig. 3.
Plot of the logarithm of sleep cycle time in minutes, the period between REM and non-REM sleep, versus the logarithm of body size, ln(M), in kilograms. The slope computed by using OLS regression is 0.19 (P < 0.0001; n = 32, 95% CI: 0.14, 0.23). Note that the 95% CI includes the predicted values of 3/16 and 1/6 but excludes 1/4 [Reproduced with permission from ref. 53 (Copyright 2006, Springer).].
Fig. 4.
Plot of the logarithm of the ratio of REM sleep time to total sleep time per day, ln(R), versus the logarithm of body size, ln(M), in kilograms. The slope computed by using OLS regression is −6e−3 (P = 0.74; n = 61, 95% CI: −0.04, 0.03). The average value for R is 0.17. Note that the 95% CI for the slope is narrow and includes zero. The large outlier on the right side of the plot corresponds to the pilot whale (G. scammoni), a marine mammal.
Discussion
Based on these and others results, we speculate that the process identified as sleep is a special state of the brain that is devoted primarily to the critical activities of repair and reorganization. This leads to the conclusion that other organs or tissues do not require an analogous special state because they can be repaired or reorganized during waking or resting periods without significantly interfering with “normal” functionality (5).
Because lifespan scales like a typical physiological time (≈M1/4 or slightly shallower) (30), whereas sleep times scale according to Eqs. 1–3 [≈M−1/6/(1 + aM−1/6)], smaller mammals spend less time asleep on average during a lifetime than larger ones. Conversely, larger animals spend a much smaller fraction of their lives asleep than smaller ones. These observations raise interesting questions regarding the possible ecological and evolutionary consequences of sleep (e.g., predator–prey effects) and the corresponding advantages and/or disadvantages it confers on different size mammals (27, 34, 35). Indeed, the fraction of time spent asleep must eventually become limiting for small animals because some fraction of time is necessary just to forage and to reproduce (13, 34, 35).
For example, separating carnivores, herbivores, and omnivores suggests differences. Performing regressions of the sleep time to awake time ratio on body size, as in Fig. 1, for each of these groups gives the allometric exponents: −0.12 for carnivores (P = 0.02; n = 27, 95% CI: –0.22, −0.03), −0.20 for herbivores (P < 0.005; n = 26, 95% CI: –0.20, −0.13), and −0.10 for omnivores (P = 0.03; n = 30, 95% CI: –0.18, −0.01). [Note that our results for carnivores differ from Siegel (21) because he excludes marine mammals.] Importantly, for all of these cases, the 95% CIs include our prediction. The variability, however, is quite large because the separation of these groups decreases both the number of data points and the mass range covered. Notably, the variability for herbivores is significantly less than that for carnivores or omnivores. This difference may have arisen because carnivores and omnivores experience less selective pressure, because of lower risks of predation and a higher calorie diet, to maximize awake time and, thus, to optimize the sleep time (21). Including such effects could enhance this theory.
Another natural extension of our analyses is to mammals during ontogenetic growth and to other taxa. This is particularly interesting for birds because their brain allometry and partial use of unihemispheric sleep distinguish them from most mammals (1, 2, 30), and for ectotherms because the temperature dependence of metabolic rate as well as the variation in cell and genome size adds other parameters to the theory (32). Unfortunately, in both cases there are, at present, insufficient data for definitive analyses.
We have presented a theory to address sleep-related questions in a well defined, general framework whose quantitative predictions can be compared directly with existing data. Our results demonstrate that there is substantial evidence that the function of sleep is related to metabolic processes in the brain and, in particular, to the repair of neuronal damage and/or reorganization (3, 18–21). The theory is able to explain sleep times that differ by a factor of seven across organisms that differ in mass by six orders of magnitude, revealing why a mouse sleeps four times longer per day than an elephant. Further work needs to be done to understand the detailed mechanisms underlying damage and repair and their relationship to metabolic rate and sleep. Our work suggests several avenues of related experimental investigation. These include (i) tests of whether the power density of cellular repair and/or neural organization is body mass independent, (ii) measurements of the fraction of metabolic energy given to damage or neural reorganization, and (iii) determination of the scaling of brain metabolic rate with body size. Our results strongly suggest that metabolic processes in the brain control the underlying mechanisms for the function of sleep and are a major determinant of why sleep cycle time increases with body size and why mammals need the amounts of sleep they do.
Methods
All of the sleep data are from Zepelin (33), and methods are detailed therein. Data for body masses were not given by Zepelin (33), so most values for body mass were taken from Meddis (27), which is an earlier compilation that Zepelin (33) draws from heavily. When sleep measurements listed in Zepelin (33) were not given or did not match sleep data in Meddis (27), we used the masses given in Smith et al. (36), and when masses could not be found there, we used the average of the range of values given in Nowak (37). All of the data and the sources for the body mass values are listed in SI Table 1. Some of the original sources given in Zepelin (33) were consulted to determine which species were used. In a few cases, the logarithmic averages of body masses were calculated for groups of species (e.g., four species of Microtus and five species of Peromyscus). This was done to be consistent with the original sleep data in Zepelin (33). Data for brain mass were taken from Meddis (27), but two values were excluded because the sleep values given did not match those given in Zepelin (33).
Certain marine mammals sleep with one hemisphere of the brain at a time. Because in Eq. 1 the variable tS is the amount of sleep time per cell or per tissue, total sleep time for these marine mammals must be divided by two to obtain the appropriate sleep time. [There is often an asymmetry between amounts of sleep for the left and right hemisphere in marine mammals, so dividing by two should be regarded as the average sleep per neuron (38).] The original sleep data from Zepelin (33) were adjusted accordingly for the three species: Tursiops truncates, Globiocephalus scammoni, and Phocoena phocoena. Moreover, marine mammals are known to have very small or perhaps nonexistent amounts of REM sleep, and in accord with several other studies, the data for REM sleep for T. truncates and P. phocoena were excluded (33). Data for the REM sleep of G. scammoni was included and corresponds to the large outlier in Fig. 3. Finally, REM sleep time and sleep cycle time for Elephas maximus were excluded because Zepelin (33) denotes those values as doubtful, and the REM sleep time of 0.0 for Tachyglossus aculeatus was excluded because recent studies have shown that differentiating between REM and non-REM sleep for this primitive species is especially difficult (39).
Allometric exponents were determined by using OLS regression on ln-ln plots of the data. CI (95%) and P values were computed by using Mathematica (Wolfram Research, Inc., Champaign, IL). It is most appropriate to perform regressions and calculate statistics in ln-ln space because that is the space in which the error for each data point is approximately the same, thus satisfying one of the central assumptions of linear regression. For example, within a species, the variance in body mass increases approximately linearly with the mean body mass, so that the relative error is approximately constant (≈0.23) (40). Consequently, in linear space, the error for each data point increases systematically with body mass (the x axis), but in ln-ln space, the error for each data point, which is the relative error of the linear values, is approximately constant and independent of the logarithm of body mass (the x axis). Similarly, the relative error of physiological rates and times, such as metabolic rates and sleep times, should be approximately constant, and, therefore, plots and regressions should be done in ln-ln space.
To test for the normality of the residuals in our plots, we used Matlab (The MathWorks, Natick, MA) to compute the Lilliefors (41) and Jarque–Bera (42) tests for our data. Both tests choose a normal distribution with unknown variance and mean as the null hypothesis. The Lilliefors test determines how much the cumulative distribution function for the residuals differs from that for a normal distribution, and the Jarque–Bera test determines how much the skew and kurtosis of the residuals differ from that of a normal distribution (with a skew and kurtosis of zero). Because these tests depend on different characteristics of the data, we applied both tests to the residuals in our plots. Data often are considered normal if the null hypothesis of normality is not rejected for a significance level <0.05. For the Lilliefors test, normality was not rejected for a significance level of αS < 0.16 for the residuals in Fig. 1 and αS < 0.13 for the residuals in Fig. 2. Using Matlab, the maximum computable significance level for the Lilliefors test is 0.2, and within this bound, normality was not rejected for the residuals in Figs. 3 or 4. For the Jarque–Bera test, normality was not rejected for a significance level of αS < 0.21 for the residuals in Fig. 1, αS < 0.38 for the residuals in Fig. 2, αS < 0.60 for the residuals in Fig. 3, and αS < 0.47 for the residuals in Fig. 4. Thus, the residuals passed normality tests for all cases considered.
Supplementary Material
Acknowledgments
We thank Morgan Ernest for help with taxonomy, James Brown and Jon Wilkins for constructive comments and clarifying discussions, Jason Kaufman for references on the scaling of metabolic rate with brain size, Dan Reuman for help with statistics, and the “Scaling Group” at the University of New Mexico and the Santa Fe Institute for helpful discussions, especially Alex Herman. V.M.S. and G.B.W. were supported by the Thaw Charitable Trust, a Packard Interdisciplinary Science Grant, the National Science Foundation, and Los Alamos National Laboratory (LANL) for LANL/Laboratory Directed Research and Development (LDRD) Grant 20030050DR. V.M.S. also acknowledges support from the National Institutes of Health Grant 1 P50 GM68763-02 through the Bauer Center for Genomics Research and the Fontana laboratory at Harvard Medical School.
Abbreviations
- CI
confidence interval
- OLS
ordinary least squares
- REM
rapid eye movement.
Appendix
Dependence of Power Density Given to Repair, PR, on Body Size, M.
We assume that the power density (per unit volume) given to repair or reorganization, PR, is constant. This is because we are assuming that the repair or reorganization processes are occurring locally, at a level significantly smaller than that of the cell, and because we assume that the entire cell, including cell walls, proteins, and the cytoskeleton, requires repair.
Depending on the exact mechanisms of cellular damage and repair, however, it may be most appropriate to focus on other quantities. For example, if DNA is the dominant site of damage and repair, it is most appropriate to consider the power given to repair per DNA content. If power given to repair per nucleotide or per codon is constant because of the limited spatial access to these domains, then it follows that power given to repair per DNA content is constant. For mammals, DNA content and glial cell size are roughly invariant, but brain neuronal cell size appears to vary as Mb1/4 (43–47). Consequently, within this scenario, power given to repair per unit volume, PR, would still be a constant for glial cells but would decrease as Mb−1/4 for neuronal cells. Thus, if glial cells require the most repair, the theory is exactly the same as presented in Theory. However, if brain neuronal cells require the most repair during sleep, most repair at the cellular level is to DNA, and if power given to repair per DNA content is constant, then the mass dependence of PR decreases as Mb−1/4, so the ratio of sleep time to awake time would be predicted to be mass-independent. This prediction is in clear disagreement with data and suggests that this scenario is not correct. However, it does provide a potential means for either using our theory to support reorganization (hypothesis ii) as the dominant function of sleep over repair (hypothesis iii) or for falsifying our theory altogether. That is, if this mass dependence for PR was supported by empirical findings at the cellular level, it would suggest that our theory for repair is incorrect and, depending on the analogous findings for neural reorganization, that the influence of metabolic rate on sleep time requires reevaluation. Alternatively, if Bb/Mb was constant, which contradicts our assumption and limited empirical data (see below), and PR decreased as Mb−1/4, the predictions of our theory would be exactly the same as presented in Theory, but the source of the mass dependence would come from the power density given to repair and not from the mass-specific metabolic rate of the brain. Measurements of PR and pb therefore are crucial for determining both the source of mass dependencies in our theory and for assessing the validity of our theory.
Within ectotherms, cell size and genome size vary over several orders of magnitude, and this variation may be related to mass-specific metabolic rate (48, 49). Therefore, to extend this framework to ectotherms, it will be crucial to discern between the different mechanisms given above. This problem is somewhat simplified by the fact that cell size and genome size appear to vary together linearly.
Because we do not know the exact nature of the repair occurring at the cellular level (either what is being repaired or the mechanism by which it is repaired), we focus on the power per unit volume in Theory because its generality does not depend on a specific repair mechanism. As explained above, however, the framework can be modified to accommodate different and/or more complicated repair mechanisms.
In Vitro Data for Scaling Exponent of Brain Metabolic Rate with Body Mass, pb.
Qualitative support for a negative value for pb is found in Elliott and Henderson (50), Davies (51), and Tower and Young (52). Elliott and Henderson (50) measured rates of oxygen consumption in cortical tissue for three species of mammals (rat, cat, and cattle). Their measurements were taken in vitro within 1 h of killing of the animals. They concluded that pb ≈ 0.1. Subsequent studies by Davies (51) concluded that in vitro measurements are usually an underestimate of in vivo values, but using a limited amount of in vitro data for rates of oxygen consumption in brain tissue, he obtained a value of pb ≈ 0.07. Later, Tower and Young (52) performed the same measurements as Elliott and Henderson (50) with inclusion of data for fin and sperm whale and obtained similar results. Tower and Young also used data for seven species to show that the activity of acetylcholinesterase in the dorsal cerebral cortex decreases with brain mass as Mb−0.2. Although these findings for the scaling of oxygen consumption rates in the brain are based on very limited amounts of in vitro data (as opposed to the in vivo data needed to test directly this theory), the scaling exponents are not inconsistent with the theoretical values of –1/6 and –3/16.
Estimate of Fraction of Metabolic Rate Given to Damage During Sleep, α.
We performed a linear plot for tA/tS versus M−3/16 and versus M−1/6. From Eq. 3 the intercept of these plots corresponds to α. Using OLS regression, we obtained the value α = 0.35 (P = 0.074; n = 83, 95% CI: −0.04, 0.75) for pb = −3/16 and α = 0.27 (P = 0.20; n = 83, 95% CI: −0.15, 0.70) for pb = −1/6. The 95% CI is large but suggests α < 1. Note that OLS regression assumes the same error for each datum, which is not strictly satisfied in these plots (see Methods). The mass dependence for the standard deviation of each datum in this plot behaves approximately like σ/M19/16 and σ/M7/6, respectively, where σ is the standard deviation in mass for each species. The relative standard deviation, σ/M, is roughly constant for mammals (40), so the standard deviation for each datum in this plot slightly decreases with M. Consequently, the errors for α are even larger than the ones given here, but the data likely still suggest α < 1.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610080104/DC1.
References
- 1.Campbell SS, Tobler I. Neurosci Biobehav Rev. 1984;8:269–300. doi: 10.1016/0149-7634(84)90054-x. [DOI] [PubMed] [Google Scholar]
- 2.Kryger MH, Roth T, Dement WC, editors. Principles and Practice of Sleep Medicine. 3rd Ed. Philadelphia: Saunders; 2000. [Google Scholar]
- 3.Siegel JM. Sci Amer. 2003;289:92–97. doi: 10.1038/scientificamerican1103-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rechtschaffen A. Perspect Biol Med. 1998;41:359–390. doi: 10.1353/pbm.1998.0051. [DOI] [PubMed] [Google Scholar]
- 5.Horne JA. Why We Sleep, The Function of Sleep in Humans and Other Mammals. Oxford: Oxford Univ Press; 1988. [Google Scholar]
- 6.Lawton G. New Sci. 2004;183:28–29. [Google Scholar]
- 7.Hobson JA. Nature. 2005;437:1254–1256. doi: 10.1038/nature04283. [DOI] [PubMed] [Google Scholar]
- 8.Pace-Schott EF, Hobson JA. Nat Rev Neurosci. 2002;3:591–605. doi: 10.1038/nrn895. [DOI] [PubMed] [Google Scholar]
- 9.Tononi G, Cirelli C. Neuropsychopharmacology. 2001;25:S28–S35. doi: 10.1016/S0893-133X(01)00322-0. [DOI] [PubMed] [Google Scholar]
- 10.Gallopin T, Fort P, Eggermann E, Cauli B, Luppi PH, Rossier J, Audinat E, Muhlethaler M, Serafin M. Nature. 2000;404:992–995. doi: 10.1038/35010109. [DOI] [PubMed] [Google Scholar]
- 11.Zepelin H, Rechtschaffen A. Brain Behav Evol. 1974;10:425–470. doi: 10.1159/000124330. [DOI] [PubMed] [Google Scholar]
- 12.Mamelak M. J Geriatr Psych Neurol. 1997;10:29–32. doi: 10.1177/089198879701000106. [DOI] [PubMed] [Google Scholar]
- 13.Elgar MA, Pagel MD, Harvey PH. Anim Behav. 1988;36:1407–1419. [Google Scholar]
- 14.Stickgold R, Hobson JA, Fosse R, Fosse M. Science. 2001;294:1052–1057. doi: 10.1126/science.1063530. [DOI] [PubMed] [Google Scholar]
- 15.Maquet P. Science. 2001;294:1048–1052. doi: 10.1126/science.1062856. [DOI] [PubMed] [Google Scholar]
- 16.Tononi G, Cirelli C. Brain Res Bull. 2003;62:143–150. doi: 10.1016/j.brainresbull.2003.09.004. [DOI] [PubMed] [Google Scholar]
- 17.Benington JH, Frank MG. Prog Neurobiol. 2003;69:71–101. doi: 10.1016/s0301-0082(03)00018-2. [DOI] [PubMed] [Google Scholar]
- 18.Reimund E. Med Hypotheses. 1994;43:231–233. doi: 10.1016/0306-9877(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 19.Adam K, Oswald I. J R Coll Physicians Lond. 1977;11:376–388. [PMC free article] [PubMed] [Google Scholar]
- 20.Inoue S, Honda K, Komoda Y. Behav Brain Res. 1995;69:91–96. doi: 10.1016/0166-4328(95)00014-k. [DOI] [PubMed] [Google Scholar]
- 21.Siegel JM. Nature. 2005;437:1264–1271. doi: 10.1038/nature04285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramanathan L, Gulyani S, Nienhuis R, Siegel JM. NeuroReport. 2002;13:1387–1390. doi: 10.1097/00001756-200208070-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eiland MM, Ramanathan L, Gulyani S, Gilliland M, Bergmann BM, Rechtschaffen A, Siegel JM. Brain Res. 2002;945:1–8. doi: 10.1016/s0006-8993(02)02448-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gopalakrishnan A, Ji L, Cirelli C. Sleep. 2003;26:A10. [Google Scholar]
- 25.Cirelli C, Shaw PJ, Rechtschaffen A, Tononi G. Brain Res. 1999;840:184–193. doi: 10.1016/s0006-8993(99)01768-0. [DOI] [PubMed] [Google Scholar]
- 26.Cirelli C, Bushey D, Hill S, Huber R, Kreber R, Ganetzky B, Tononi G. Nature. 2005;434:1087–1092. doi: 10.1038/nature03486. [DOI] [PubMed] [Google Scholar]
- 27.Meddis R. In: Sleep Mechanisms and Functions in Humans and Animals. Mayes A, editor. Cambridge, UK: Van Nostrand Reinhold; 1983. [Google Scholar]
- 28.West GB, Brown JH, Enquist BJ. Science. 1997;276:122–126. doi: 10.1126/science.276.5309.122. [DOI] [PubMed] [Google Scholar]
- 29.Pagel MD, Harvey PH. Am Nat. 1988;132:344–359. [Google Scholar]
- 30.Peters RH. The Ecological Implications of Body Size. Cambridge, UK: Cambridge Univ Press; 1983. [Google Scholar]
- 31.Armstrong E. Neurosci Lett. 1982;34:101–104. doi: 10.1016/0304-3940(82)90159-8. [DOI] [PubMed] [Google Scholar]
- 32.Gillooly JF, Charnov EL, West GB, Savage VM, Brown JH. Nature. 2002;417:70–73. doi: 10.1038/417070a. [DOI] [PubMed] [Google Scholar]
- 33.Zepelin H. In: Principles and Practice of Sleep Medicine. 2nd Ed. Kryger MH, Roth T, Dement WC, editors. Philadelphia: Saunders; 1989. pp. 30–49. [Google Scholar]
- 34.Allison T, Cicchetti D. Science. 1976;194:732–734. doi: 10.1126/science.982039. [DOI] [PubMed] [Google Scholar]
- 35.Lima SL, Rattenborg NC, Lesku JA, Amlaner CJ. Anim Behav. 2005;70:723–736. [Google Scholar]
- 36.Smith FA, Lyons SK, Ernest SKM, Jones KE, Kaufman DM, Dayan T, Marquet PA, Brown JH, Haskell JP. Ecology. 2003;84:3403. [Google Scholar]
- 37.Nowak RN. Walker's Mammals of the World. Baltimore: Johns Hopkins Univ Press; 1991. [Google Scholar]
- 38.Mukhametov LM. Exp Brain Res. 1984;8:227–238. [Google Scholar]
- 39.Siegel JM, Manger PR, Nienhuis R, Fahringer HM, Pettigrew JD. J Neurosci. 1996;16:3500–3506. doi: 10.1523/JNEUROSCI.16-10-03500.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Savage VM. J Theor Biol. 2004;227:525–534. doi: 10.1016/j.jtbi.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 41.Lilliefors HW. J Am Stat Assoc. 1967;62:399–402. [Google Scholar]
- 42.Jarque CM, Bera AK. Int Stat Rev. 1987;55:163–172. [Google Scholar]
- 43.Friede RL. Proc Natl Acad Sci USA. 1963;49:187–193. doi: 10.1073/pnas.49.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maldonado R, San Jose H, Martinoya C, Gunther B. Acta Physiol Lat Am. 1974;24:328–335. [PubMed] [Google Scholar]
- 45.Morgado E, Ocqueteau C, Cury M, Becker L, Gonzalez U, Muxica L, Gunther B. Arch Biol Med Exp (Santiago) 1990;23:21–27. [PubMed] [Google Scholar]
- 46.Savage VM, Allen AP, Brown JH, Gillooly JF, Herman AB, Woodruff WH, West GB. Proc Natl Acad Sci USA. 2007 doi: 10.1073/pnas.0611235104. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tower DB, Young OM. J Neurochem. 1973;20:269–278. doi: 10.1111/j.1471-4159.1973.tb12126.x. [DOI] [PubMed] [Google Scholar]
- 48.Cavalier-Smith T, editor. The Evolution of Genome Size. Chichester, UK: Wiley; 1985. [Google Scholar]
- 49.Gregory TR. Biol Rev. 2001;76:65–101. doi: 10.1017/s1464793100005595. [DOI] [PubMed] [Google Scholar]
- 50.Elliott KAC, Henderson N. J Neurophysiol. 1948;11:473–484. doi: 10.1152/jn.1948.11.6.473. [DOI] [PubMed] [Google Scholar]
- 51.Davies M. J Cell Comp Physiol. 1961;57:135–147. doi: 10.1002/jcp.1030570302. [DOI] [PubMed] [Google Scholar]
- 52.Tower DB, Young OM. J Neurochem. 1973;20:253–267. doi: 10.1111/j.1471-4159.1973.tb12125.x. [DOI] [PubMed] [Google Scholar]
- 53.Savage VM, West GB. In: Complex System Science in Biomedicine. Deisboeck TS, Kresh JY, editors. New York: Springer; 2006. [Google Scholar]
- 54.Gally JA, Edelman GM. C R Biol. 2004;327:721–727. doi: 10.1016/j.crvi.2004.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.