Abstract
Studies of flower development in core eudicot species have established a central role for B class MADS-box genes in specifying petal and stamen identities. Similarly in maize and rice, B class genes are essential for lodicule and stamen specification, suggesting homology of petals and lodicules and conservation of B class gene activity across angiosperms. However, lodicules are grass-specific organs with a morphology distinct from petals, thus their true homology to eudicot and nongrass monocot floral organs has been a topic of debate. To understand the relationship of lodicules to the sterile floral organs of nongrass monocots we have isolated and observed the expression of B class genes from a basal grass Streptochaeta that diverged before the evolution of lodicules, as well as the outgroups Joinvillea and Elegia, which have a typical monocot floral plan. Our results support a conserved role for B function genes across the angiosperms and provide additional evidence linking the evolution of lodicules and second whorl tepal/petals of monocots. The expression data and morphological analysis suggest that the function of B class genes should be broadly interpreted as required for differentiation of a distinct second floral whorl as opposed to specifying petal identity per se.
Keywords: flower development, MADS-box, petal evolution, B-function, Streptochaeta
The ABC model of floral patterning, developed from studies of the model species Arabidopsis and Antirrhinum, proposes that three classes of genes act alone or in combination to establish the identities of the four concentric whorls of floral organs (1, 2). A class genes alone establish sepal identity, A class genes combine with B class genes to establish petal identity, B and C class genes combine to establish stamen identity, and C class genes act alone to confer carpel identity and floral meristem determinacy. As these genes were cloned, they were found to belong, with the exception of APETALA2, to the conserved MADS-box family of transcription factors. Since establishment of this simple model, there has been interest in determining the degree to which it applies to other more distantly related angiosperms (3). However, there remains little functional evidence that ABC MADS-box genes are playing conserved roles in flower development outside of the core eudicots. Recent work in grasses has shown that mutations in B and C class genes result in phenotypes similar to those observed in B and C class mutants of higher eudicot species like Arabidopsis and Antirrhinum (4–7). Thus, these genetic analyses suggest that the B and C class functions of the ABC model may have been established early in the history of the angiosperms, but the lack of genetic knockout or knockdown data in many nonmodel species of angiosperms makes testing this model difficult.
The origin of the petal and the number of times it has evolved have long been of interest to botanists (8–10), as has the origin of novel structures in the grasses such as the palea, lemma, and lodicules (11–13). Current research has explored the use of B class gene expression as a marker for petal identity and on the role of B class genes specifying petal identity outside the core eudicots. The term petal refers to organs with a combination of morphological characteristics: (i) position, in a whorl just outside the stamens but internal to sepals; (ii) appearance, compared with the sepals, generally larger, colored or otherwise nongreen and more delicate; and (iii) epidermal cell morphology, characteristic conical cells (P. Endress, personal communication). However, these three characteristics do not always co-occur in angiosperms, making a general definition of petals difficult.
The original B class mutants described in Arabidopsis and Antirrhinum undergo homeotic transformations in second whorl (petal) and third whorl (stamen) organs into sepals and carpels, respectively (14, 15). Antirrhinum and Arabidopsis both have two B class genes that ultimately were shown to encode a pair of closely related MADS-box genes: APETALA3 (AP3) (14) and PISTILLATA (PI) (15) in Arabidopsis and their Antirrhinum orthologs DEFECIENS (DEF) (16) and GLOBOSA (GLO) (17), respectively. The paralogous AP3/DEF and PI/GLO lineages are the result of a duplication that occurred near the base of the angiosperms (18). Furthermore, a duplication in the AP3/DEF lineage at the base of the core eudicots gave rise to the euAP3 and paleoAP3 lineages, which have distinct motifs within their C terminus (19). Rescue of the Arabidopsis ap3 mutant with a chimeric AP3 protein containing a paleoAP3 C terminus from a basal eudicot resulted in stamen rescue but no rescue of petal identity (20). These results suggest that the paleoAP3 functions primarily in stamen identity, and, perhaps, the euAP3 evolved a distinct role in petal identity in the core eudicots.
Further evidence that the euAP3 lineage evolved to specify core eudicot petal identity comes from a study in which expression of B class genes in noncore eudicots was shown to be weak or patchy in petals (21). This contrasts with core eudicots, where expression is strong throughout petal development. In light of morphological and anatomical evidence that petals evolved multiple times independently during the evolution of angiosperms (8–10), it was proposed that B class genes were recruited to a central role in petal identity in a common ancestor of the core eudicots, but that in other angiosperm lineages, B class genes are not necessarily specifying petaloidy (21, 22).
A critical test of this hypothesis would be to disrupt B class gene function in a noncore eudicot species. To date, such a disruption of B class gene function has been described only for the paleoAP3 genes of the grass species maize and rice, which both have highly derived floral organs (5, 6). These mutants show transformation of stamens to carpels, as seen in higher eudicots, in addition to transformation of a grass-specific organ, the lodicule, into a lemma/palea-like organ. This phenotype is consistent with an interpretation of lodicules as modified grass petals and palea as grass sepals. This, if a correct interpretation, would suggest that B class gene function is conserved in the common ancestor of monocots and eudicots and is consistent with a subsequent analysis indicating conservation of the biochemical function of the maize and Arabidopsis B class proteins (23). Other than position, however, little in the mature morphology of lodicules indicates homology with petals, raising the possibility that B class genes were recruited independently to specify lodicule fate in the grasses (24, 25). Remane's well known criteria for homology (26) include homology of position, homology of what he called “special characteristics” (color, texture, and cell structure), and homology via intermediate forms. The latter criterion suggests that we may view two structures as homologous if they are connected by a series of transitional forms. To help clarify the relationship between petals and lodicules, we have isolated and observed the expression pattern of B class genes from Streptochaeta angustifolia, a basal grass species that diverged before the evolution of lodicules, as well as from nongrass outgroups Joinvillea ascendens and Elegia elephas that have a typical monocot floral plan. Our results are consistent with Remane's criteria for homology and indicate that lodicules indeed are modified second whorl organs and that B class genes of a basal grass species and nongrass outgroups mark the fate of the second and third floral whorls. Furthermore, these expression patterns suggest that B class gene activity specifies a second whorl identity independent of the showy characteristics commonly interpreted as petaloid. These results provide further evidence that B class control of second and third whorl organ identities is conserved between monocots and eudicots.
Results
Isolation of B Class Genes from Basal Grass Species and Outgroups.
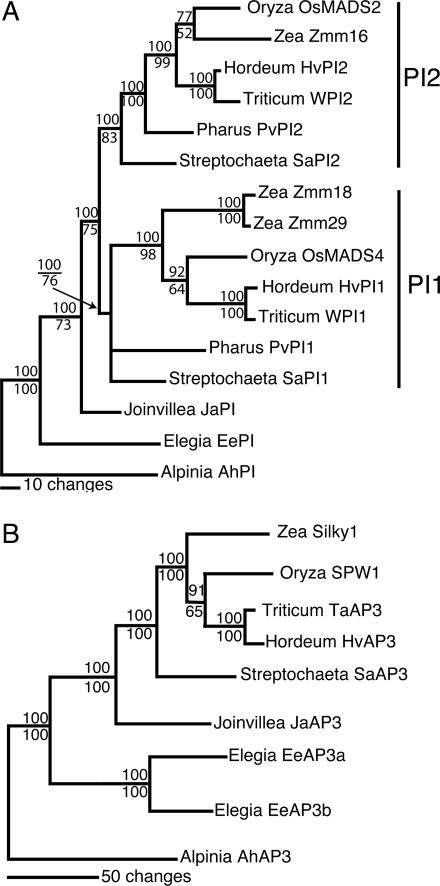
Grass B class genes were isolated originally from maize (Zea mays) and rice (Oryza sativa) (5, 27–29). In both species, there appears to be a single AP3 ortholog, Silky1 (Si1) in maize and SUPERWOMAN1 (SPW1) in rice, mutations of which result in a strong B class homeotic phenotype (5, 6, 29). However, a previous analysis of the grass PI-like genes indicated that a duplication event in a common ancestor of maize and rice lead to two paralogous lineages, one containing the rice OsMADS2 and maize Zmm16 and the other containing rice OsMADS4 and the maize genes Zmm18 and Zmm29 (29). To examine expression of AP3 and PI orthologs, as well as to more confidently place the duplication event in the grass PI genes, we isolated AP3 and PI orthologs from the basal grass species Pharus and Streptocheata, as well as from two closely related outgroup species, Joinvillea and Elegia. A Bayesian phylogenetic analysis of the grass AP3 genes closely matches the consensus topology published by the Grass Phylogeny Working Group (30), and is in agreement with a single lineage for AP3-like genes in the grasses (Fig. 1B). This is consistent with a single AP3 ortholog in the complete rice genome sequence and a nonredundant (i.e., strong) B class phenotype when this gene is disrupted in rice spw1 and maize si1 mutants. However, two AP3-like genes were isolated from Elegia, which appear to be the result of a duplication event sometime in the evolution of the Restionaceae.
Fig. 1.
Phylogenetic analysis of B class sequences from the Poaceae and close outgroups. Trees are 50% majority rule consensus, and phylogenetic analysis was performed as described in Materials and Methods. Bayesian posterior probabilities are indicated above the branches, with maximum likelihood bootstrap values below. Taxa from which the genes were isolated are as follows: HvPI1, HvPI2, and HvAP3 Hordeum vulgare (barley), WPI1, WPI2, and TaAP3, Triticum aestivum (wheat); OsMADS2, OsMADS4, and SPW1, Oryza sativa (rice); Zmm16, Zmm18, Zmm29, and Si1, Zea mays (corn); PvPI1, PvPI2, and PvAP3, Pharus virescens; SaPI1, SaPI2, and SaAP3, Streptochaeta angustifolia; JaPI and JaAP3, Joinvillea ascendens; EePI, EeAP3a, and EeAP3b, Elegia elephas (cape rush); AhPI and AhAP3, Alpinia hainanensis (ginger). (A) PI orthologs from the grass family. Two well supported clades of grass PI orthologs exist and are named PI1 and PI2 as indicated. (B) AP3 orthologs from the grass family.
In the PI phylogeny, two well supported clades of grass PIs, named here PI1 and PI2, are apparent with Joinvillea ascendens PI (JaPI) as sister to both clades (Fig. 1A). Each clade has a PI ortholog from each grass species, and the topology matches the Grass Phylogeny Working Group topology with only slight, unsupported variations. These results are consistent with a PI duplication event occurring at or near the base of the grass family, coinciding with a putative genomewide duplication event early in the evolution of the grass family (31). A maximum likelihood analysis gave AP3 and PI trees with the same topology as the Bayesian analysis, but with weaker support for some of the clades, particularly in the PI tree (Fig. 1A).
Morphology of Second Whorl Organs in Streptochaeta, Joinvillea, and Elegia Is Distinct.
The grass flower, relative to other monocots, is a derived structure, in which the sterile organs are of uncertain homology and have a grass specific nomenclature (13). The lodicule is such an organ unique to the grasses that occurs in a whorl just outside the stamens. Lodicules can be fleshy or scale-like and generally swell at anthesis to allow the stamens to extend and the lemma and palea to separate. The two grasses Anomochloa and Streptochaeta are sisters to all other grasses and do not have lodicules (32, 33). In Streptochaeta, three leaf or bract-like organs surround the stamens, whereas in Anomochloa, a hairy “perigonate anulus” surrounds the stamens, indicating that lodicules evolved after the lineages for Streptochaeta and Anomochloa diverged (34). Outgroups to the grasses including Joinvillea and Restionaceae have a typical monocot floral plan in which the sterile organs occur in two separate whorls, the inner and outer tepals. Considering the hypothesis that lodicules are modified petals (inner tepals), we observed the floral ontogeny of Streptochaeta, Joinvillea, and Elegia (Restionaceae) to understand the morphology of their second whorl organs.
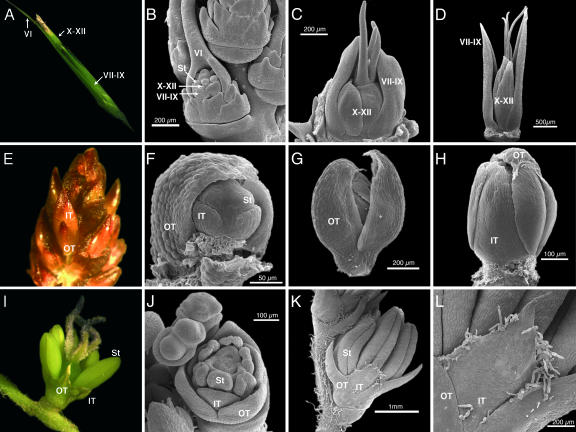
The Streptocheata spikelet equivalent has been described as a complex arrangement of twelve bracts (I–XII) that initiate before the reproductive organs (32). Bracts I–V initiate in a spiral, are small, and occasionally can develop axillary spikelet equivalents of their own. Bract VI is large with a long curled awn that can entangle passing animals for seed dispersal. After bract VI, an apparent whorl of smaller bracts develops, with the two bracts (VII and VIII) opposite VI developing into shorter pointed structures, and the third member of this whorl (IX), adjacent to VI, is either reduced or absent. Bracts X–XII are similar and develop into an overlapping whorl that elongates and hardens at maturity to enclose the ovary and developing seed. Early developmental stages show that bracts VII and VIII initiate in an apparent whorl and quickly grow to cover the inner organs (Fig. 2 A–C). Bracts X–XII initiate as a whorl outside the stamens and inside the VII-VIII-(IX) whorl, and because of their position often are interpreted as lodicules (12), although they have none of the other morphological characteristics of lodicules. Dissecting the large bract VI from the flower and imaging from behind shows that the X-XI-XII whorl overlaps and surrounds the developing stamens and is distinct in shape from the VII-VIII-(IX) whorl (Fig. 2 C and D). If the VII-VIII-(IX) bracts are interpreted as the first or outer whorl and X-XI-XII as the second or inner whorl of a Streptochaeta flower that is subtended by the large bract VI, then there is a clear differentiation in morphology between the first and second floral whorls in this basal grass.
Fig. 2.
Early floral development in Streptochaeta, Elegia, and Joinvillea. (A–D) Streptochaeta spikelet equivalent development. (A) Mature spikelet equivalent with anthers emerging from the overlapping whorl of bracts X–XII, which are distinct in shape and size from the pointed bracts VII–IX. (B) Early floral development showing the long-awned bract VI, initiation of the “outer tepal” bracts VII–IX, one of the “inner tepal” bracts X–XII, as well as stamen (St) and carpel primordia. (C) The entire spikelet equivalent was detached from the inflorescence, the large enclosing bract VI was removed, and the flower was viewed from behind. Three stigmas are emerging from the overlapping whorl of bracts X–XII. Outside of this whorl, two of the bracts from the VII–IX whorl are developing their pointed tips, whereas the third apparently has aborted. (D) Later stage flower dissected from the inflorescence as in C showing the distinct development of the pointed outer tepal whorl VII–IX and the overlapping inner tepal whorl X–XII. (E–H) Elegia floral development. (E) A mature inflorescence containing flowers subtended by bracts just before anthesis. Labeled flower has the bract removed, showing that the inner tepals (IT) are longer than the outer tepals (OT) and each is morphologically distinct. (F) Young floral meristem with one outer tepal removed showing the initiation of inner tepal and St primordia. (G) Maturing flower showing hooded outer tepals. (H) Same as in G but with two outer tepals removed, showing the inner tepals as more laminar in shape than the young outer tepals in F and G. (I–L) Joinvillea floral development. (I) Mature flower showing the apparently similar morphology of the inner and outer tepals. (J) Early floral development clearly showing characteristic monocot floral morphology. (K) Later developmental stage showing distinct shape of the inner and outer tepal whorls. (L) Close view of the inner tepal in K showing flat broad triangular shape with a papillate margin as opposed to the narrowly pointed, curved outer tepal with a smoother margin.
The typical monocot floral plan has whorls of organs occurring in multiples of three, with the first two whorls generally composed of three members each (35). These first two whorls can be either distinct or similar in adult morphology. If they are distinct, and the second whorl is clearly modified to attract pollinators, they are called sepals and petals, respectively, similar to flowers of eudicots. When they are similar, they are referred to as tepals, which can be either large and showy, “petaloid,” or nonshowy, “sepaloid.” The closest extant relatives to the grasses include Joinvillea and the Restionaceae, which have two similar whorls of nonshowy tepals. However, the development of these flowers shows that, although similar, the first and second whorls do have distinct morphologies (Fig. 2 E–L). In Elegia, the outer tepals initiate sequentially, rather than in a whorl, and are hooded (Fig. 2 F and G), whereas the inner tepals are more laminar and develop as an overlapping whorl that surrounds the stamens (Fig. 2H). As the flower matures, the inner tepals elongate and continue to enclose the reproductive organs (Fig. 2E). In Joinvillea, the outer tepal whorl is hooded and elongated with a smooth margin, whereas the inner tepal whorl is laminar and triangular with a papillate margin (Fig. 2 K and L).
Thus, in both Joinvillea and Elegia, the inner and outer tepals are distinct in morphology, although the second whorl does not have the characteristic showy features of petals. A distinct morphology for the first and second whorl also has been described for other taxa in the same family as Elegia (36). Additionally, the Streptochaeta inner whorl (interpreting bracts VII-VIII-IX as the outer whorl) has a distinct morphology, as do lodicules in the grasses. Although it is not clear what organs should be interpreted as the grass first whorl (13), some evidence suggests the palea and/or lemma could be (5, 6). The position and distinct morphology of the second whorl in Streptochaeta, Joinvillea, and Elegia, along with the grass lodicules, naturally suggests the hypothesis that lodicules are modified second whorl organs or inner tepals and that the apparent second whorl of Streptochaeta may be a transitional form (sensu Remane, ref. 26) in the evolution of lodicules.
B Class MADS-Box Genes Mark the Second and Third Whorls of Streptochaeta, Elegia, and Joinvillea.
Our observations of the distinct morphology of the second whorl organs of Streptochaeta and nongrass outgroups, in addition to genetic data from maize and rice showing that B class genes control the identity of the second whorl lodicules, suggest that B class genes control second whorl organ identity in a broader sense than just petal identity, as seen in eudicots or lodicule identity in the grasses. If true, one would expect to see B class gene expression in the developing second whorl organ primordia of Streptochaeta and grass outgroups. Such expression would further support interpretation of lodicules as modified second whorl organs. Consequently, we performed RNA in situ hybridization on developing flowers of these species by using probes derived from the B class genes we had isolated.
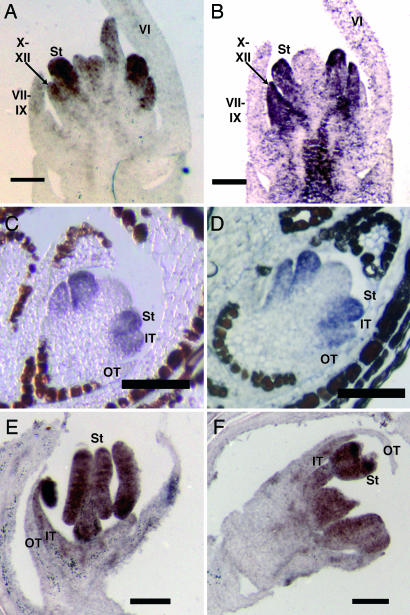
In Streptochaeta, we examined expression of the AP3 ortholog SaAP3 and the PI ortholog SaPI2. SaAP3 was expressed strongly in the stamens and second whorl primordia. Additionally, weaker expression was observed in the developing carpel and ovules (Fig. 3A). Such expression in the fourth whorl is not uncommon and has been reported for PI and DEF at early stages of floral development (15, 37) as well as for the maize PI orthologs at later stages (29). For SaPI2, expression was observed only in the stamen whorl and the second whorl (Fig. 3B). In Elegia, we performed in situ hybridizations with both of the AP3 orthologs, EeAP3a and EeAP3b. For EeAP3a, we observed strong expression in the developing stamen and second whorl primordia (Fig. 3C). A similar result was seen with EeAP3b (Fig. 3D), although expression possibly was weaker overall compared with EeAP3a (data not shown). We were able to obtain tissue of Joinvillea, although the floral stages were not as young as for Streptochaeta and Elegia, and fresh tissue was not available for fixing. For these reasons, in situ hybridization of the Joinvillea ascendes AP3 (JaAP3) and PI (JaPI) was not as robust. Nevertheless, for both JaAP3 and JaPI, expression was observed in the stamen whorl and the second whorl, but apparently absent from the first whorl (Fig. 3 E and F).
Fig. 3.
In situ RNA hybridization of B class genes in Streptochaeta, Elegia, and Joinvillea. (A) SaAP3 is strongly expressed in the developing stamens (St) and the whorl containing bracts X–XII. Weaker expression is evident in the carpel and possibly the VII–VIII whorl. (B) SaPI2 is expressed strongly in the stamen whorl and the X–XII whorl, as is SaAP3. (C) EeAP3a is strongly expressed in the emerging stamens and inner tepals (IT) but absent from the outer tepals (OT). (D) EeAP3b expression is very similar to EeAP3a: strong in stamens and inner tepals but absent from outer tepals. (E) JaAP3 expression can be seen in the stamens and inner tepal, but the adjacent outer tepal is relatively lacking in JoinAP3 expression. (F) JaPI expression, like JaAP3, is seen in the stamens and inner tepals but relatively absent from the outer tepals. (Scale bars: 100 μm.)
Discussion
B Class Gene Expression Supports a Second Whorl Origin for the Grass Lodicule.
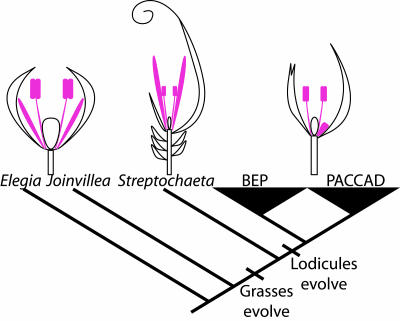
B class genes in angiosperms are expressed consistently in stamens (third whorl), and it is thought that specification of the male reproductive organs in flowers is derived from a role in specification of male cone identity in Gymnosperms (38–42). In the core eudicots, B class genes also have a role in specifying the sterile organs of the second floral whorl, the petal. Similarly, in grasses, these same genes specify the second whorl lodicule. We have isolated B class genes from a basal grass without lodicules and from outgroups to the grasses, and we find that B class genes are consistently expressed in stamens and the organ whorl just outside the stamens in these species. In most grasses, the whorl outside the stamens is composed of lodicules, whereas in the basal grass Streptochaeta, the whorl is composed of bracts X–XII, and in outgroups to the grasses like Joinvillea and Elegia, the whorl is the inner tepals. In Asparagus officinalis, another nongrass monocot with nonshowy tepals, the AP3 and PI orthologs are expressed similarly in stamens and inner tepals (43, 44). Taken together, these data strongly suggest that bracts X–XII of Streptochaeta and lodicules of other grasses evolved by modification of the inner tepal or second whorl of a typical monocot flower (Fig. 4).
Fig. 4.
Evolution of lodicules as indicated by B class gene expression. A schematic of the grass family phylogeny with the position of the outgroups examined in this study as described by the Grass Phylogeny Working Group. Lodicules evolve in the grasses after the divergence of the basal grass Streptochaeta. B class genes are consistently expressed in stamens and the organ whorl just outside the stamens. This whorl just outside the stamens is the inner tepals of Elegia and Joinvillea, the bracts X–XII of Streptochaeta, and the lodicules of the grasses. Both position and B class gene expression indicate lodicules are modifications of the inner tepals, with bracts X–XII of Streptochaeta being an intermediate step in this process. BEP clade, Bambusoideae, Erhartoideae, and Pooideae; PACCAD clade, Panicoideae, Arundinoideae, Centothecoideae, Chloridoideae, Aristidoideae, and Danthonioideae.
It is important to interpret our expression results in light of the loss of function phenotype for AP3 orthologs in the grasses. Expression of B class genes in monocot second whorl organs is not of itself sufficient evidence that these genes control second whorl identity. However, the si1 and spw1 mutants clearly show that the AP3 ortholog in grasses is necessary for lodicule identity. If B class genes were recruited independently to specify lodicule identity in the grasses, then we would not expect B class expression in the second whorl of Streptochaeta and the grass outgroups. To explain our observations, we then would have to hypothesize that B class expressing second whorl organs in Streptochaeta and the outgroups were lost over evolutionary time and were replaced later in grass evolution with lodicules in the same position and whose identity came under control of the same regulatory genes. We feel this is unlikely. The much simpler hypothesis, that B class genes specify a second whorl identity in many angiosperms, that the details of second whorl morphology are variable, and that lodicules are modified second whorl organs, is entirely consistent with the data. Further confirmation must await a B class loss of function mutant for a nongrass monocot.
B Class Genes and Second Whorl Identity as Opposed to Petal Identity.
Our results suggest that B class genes may have two separable roles. The first is a role in establishing a differentiated second whorl organ identity, whereas the second role is to promote petaloid cell identities. B class genes are expressed in the second whorl of grasses and outgroups even though this whorl is not brightly colored or showy. Likewise, Asparagus has two whorls of sepaloid tepals, but B class genes still are expressed in the second whorl.
Petal Evolution and B Class Genes.
We find compelling evidence that B class genes are important for establishing second whorl organ identity in the monocots, suggesting that eudicot petals and inner tepals in the monocots inherited a common mechanism for their specification involving B class MADS-box genes. The independent evolution of petals as described by morphologists simply could be the result of this B class petal program shifting to new organ whorls (45, 46). Expression data, functional genetic data (5, 6, 23), and morphological data taken together suggest that lodicules represent one of multiple possible sorts of second whorl organs, others being petals or second whorl tepals. Although we present evidence here that B class genes have a conserved role in establishing second whorl identity in both grass and nongrass monocots, the fascinating question remains how lodicules evolved their distinct morphology. Identifying the genetic targets of B class MADS-box proteins will help illuminate the evolutionary modifications distinguishing second whorl organs.
Materials and Methods
Plant Material.
Inflorescence primordia were collected from Streptochaeta angustifolia; plants were grown from seed in a growth room at 22°C under constant light conditions. Joinvillea ascendens tissue was collected from plants at the National Tropical Botanical Garden in Kalaheo, HI. Elegia elephas tissue was collected from plants growing in the private collection of Monique and Lambert Devoe of San Diego, CA. cDNA of Pharus virescens was collected from plants growing in a greenhouse of the Missouri Botanical Gardens (St. Louis, MO).
Isolation of B Class Genes.
cDNA was synthesized from RNA isolated from young flowers by using the SuperScript First-Strand cDNA Synthesis kit (Invitrogen, Carlsbad, CA). A polyT primer with a 5′ adapter sequence was used in the cDNA synthesis step (5′-CCGGATCCTCTAGAGCGGCCGCTTT-TTTTTTTTTTTTTT-3′). PCR of B class genes from grass species and outgroups was performed with a degenerate MADS-box sequence forward primer (5′-ATGGGBMGNGGVARKATHGAGA-3′) and the polyT adapter primer. These PCR products were subcloned into pGEM-Teasy (Promega, Madison, WI) or TOPO-TA (Invitrogen) and sequenced. Isolation of AP3 and PI orthologs from some species required a second round of PCR with internal primers: Grass PIrev (5′-YTSCTGBARRTTGGGRTG-3′), and Grass AP3rev (5′-YYARCCSAGGCGSAGGTCGTG-3′). After isolation of a partial sequence, complete cDNA coding sequence was obtained by using 5′ and/or 3′ RACE. DNA sequences were submitted to GenBank with the following accession numbers: PvPI1, DQ662243; PvPI2, DQ662242; SaPI1, DQ662244; SaPI2, DQ662241; JaPI, DQ662245; EePI, DQ662246; SaAP3, DQ662237; JaAP3, DQ662238; EeAP3a, DQ66662239; and EeAP3b, DQ662240.
Phylogenetic Analysis.
Initial DNA sequence alignment was performed with ClustalX, followed by manual adjustments with MacClade4. ModelTest then was used to determine the optimal model of evolution. Based on results from Akaike Information Criterion (AIC), the GTR + G model was selected and Bayesian phylogenetic analysis was performed by using MrBayes v3.1 with 2 million generations, a sample frequency of 100, and a burnin value of 5,000 (25%) for both the AP3 and PI data sets. Maximum Likelihood bootstrap values were determined from a total of 100 replicates, also with the GTR + G model. Published sequences used in the analysis had the following accession numbers: OsMADS2, L37526; ZMM16, AJ292959; HvPI1, BU996044; WPI2, AB107992; OsMADS4, L37527; ZMM18, AJ292960; ZMM29, AJ292961; HvPI2, AY541066; WPI1, AB107991; AhPI, AY621156; SPW1, AF454259; Si1, AF181479; TaAP3, AB107993; HvAP3, AY541065; and AhAP3, AY621154.
Scanning Electron Microscopy.
Developing inflorescences were dissected and fixed in freshly prepared FAA (3.7% formaldehyde, 50% ethanol, 5% acetic acid) containing 0.1% Triton X-100. Samples were dehydrated through an ethanol series and dried with a critical point drier. Dried samples were mounted and dissected when necessary to reveal internal floral organs, then sputter coated with gold palladium and viewed with a Quanta 600 environmental scanning electron microscope.
RNA in Situ Hybridization.
Freshly collected samples (except for Joinvillea, which was collected 24–48 h before fixation) were fixed overnight at 4°C in FAA, then embedded, sectioned, hybridized, washed, and exposed as described in ref. 47; www.its.caltech.edu/∼plantlab/protocols/insitu.htm. Probes were created by PCR amplification of the IKC domains and 3′ UTRs of cDNAs and subcloning this fragment upstream of the T7 promoter of pGEM-Teasy (Promega) or pBluescript (Stratagene, La Jolla, CA). Antisense, digoxygenen-labeled UTP probe was synthesized by using T7 polymerase with either a PCR-amplified DNA template or a linearized plasmid.
Acknowledgments
We thank Dr. Paul Cox and Dr. David Laurence of the National Tropical Botanical Gardens for providing Joinvillea material. Dr. George Chuck (Plant Gene Expression Center, Albany, CA) provided initial Streptochaeta samples for in situ hybridizations. Monique and Lambert Devoe kindly provided Elegia samples from their private collection. Evelyn York of the SIO Analytical Facility provided technical assistance with SEM. This research was supported by a grant from the National Science Foundation and through a Graduate Fellowship from the ARCS Foundation (to C.J.W.) and the National Institutes of Health (to M.Z.).
Footnotes
References
- 1.Coen ES, Meyerowitz EM. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- 2.Weigel D, Meyerowitz EM. Cell. 1994;78:203–209. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- 3.Soltis DE, Soltis PS, Albert VA, Oppenheimer DG, dePamphilis CW, Ma H, Frohlich MW, Theissen G. Trends Plants Sci. 2002;7:22–31. doi: 10.1016/s1360-1385(01)02098-2. discussion 31–34. [DOI] [PubMed] [Google Scholar]
- 4.Mena M, Ambrose BA, Meeley RB, Briggs SP, Yanofsky MF, Schmidt RJ. Science. 1996;274:1537–1540. doi: 10.1126/science.274.5292.1537. [DOI] [PubMed] [Google Scholar]
- 5.Ambrose BA, Lerner DR, Ciceri P, Padilla CM, Yanofsky MF, Schmidt RJ. Mol Cell. 2000;5:569–579. doi: 10.1016/s1097-2765(00)80450-5. [DOI] [PubMed] [Google Scholar]
- 6.Nagasawa N, Miyoshi M, Sano Y, Satoh H, Hirano H, Sakai H, Nagato Y. Development (Cambridge, UK) 2003;130:705–718. doi: 10.1242/dev.00294. [DOI] [PubMed] [Google Scholar]
- 7.Yamaguchi T, Lee DY, Miyao A, Hirochika H, An G, Hirano HY. Plant Cell. 2006;18:15–28. doi: 10.1105/tpc.105.037200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zanis MJ, Soltis PS, Qiu YL, Zimmer E, Soltis DE. Ann Mo Bot Gard. 2003;90:129–150. [Google Scholar]
- 9.Takhtajan A. Evolutionary Trends in Flowering Plants. New York: Columbia Univ Press; 1991. [Google Scholar]
- 10.Albert VA, Gustafsson MHG, Di Laurenzio L. In: Molecular Systematics of Plants II. Soltis DE, Soltis PS, Doyle JJ, editors. New York: Kluwer; 1998. pp. 349–373. [Google Scholar]
- 11.Piper CV. Science. 1906;23:789–790. doi: 10.1126/science.23.594.789-a. [DOI] [PubMed] [Google Scholar]
- 12.Page VM. Bull Torrey Bot Club. 1951;78:22–37. [Google Scholar]
- 13.Clifford HT. In: Grass Systematics and Evolution. Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME, editors. Washington, DC: Smithsonian Inst; 1987. pp. 21–30. [Google Scholar]
- 14.Jack T, Brockman LL, Meyerowitz EM. Cell. 1992;68:683–697. doi: 10.1016/0092-8674(92)90144-2. [DOI] [PubMed] [Google Scholar]
- 15.Goto K, Meyerowitz EM. Genes Dev. 1994;8:1548–1560. doi: 10.1101/gad.8.13.1548. [DOI] [PubMed] [Google Scholar]
- 16.Sommer H, Beltran JP, Huijser P, Pape H, Lonnig WE, Saedler H, Schwarz-Sommer Z. EMBO J. 1990;9:605–613. doi: 10.1002/j.1460-2075.1990.tb08152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trobner W, Ramirez L, Motte P, Hue I, Huijser P, Lonnig WE, Saedler H, Sommer H, Schwarz-Sommer Z. EMBO J. 1992;11:4693–4704. doi: 10.1002/j.1460-2075.1992.tb05574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stellari GM, Jaramillo MA, Kramer EM. Mol Biol Evol. 2004;21:506–519. doi: 10.1093/molbev/msh044. [DOI] [PubMed] [Google Scholar]
- 19.Kramer EM, Dorit RL, Irish VF. Genetics. 1998;149:765–783. doi: 10.1093/genetics/149.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lamb RS, Irish VF. Proc Natl Acad Sci USA. 2003;100:6558–6563. doi: 10.1073/pnas.0631708100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kramer EM, Irish VF. Nature. 1999;399:144–148. doi: 10.1038/20172. [DOI] [PubMed] [Google Scholar]
- 22.Kramer EM, Irish VF. Int J Plant Sci. 2000;161:S29–S40. [Google Scholar]
- 23.Whipple CJ, Ciceri P, Padilla CM, Ambrose BA, Bandong SL, Schmidt RJ. Development (Cambridge, UK) 2004;131:6083–6091. doi: 10.1242/dev.01523. [DOI] [PubMed] [Google Scholar]
- 24.Irish VF. Genome Biol. 2000;1:REVIEWS1015. doi: 10.1186/gb-2000-1-2-reviews1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irish VF. BioEssays. 2003;25:637–646. doi: 10.1002/bies.10292. [DOI] [PubMed] [Google Scholar]
- 26.Remane A. Die Grundlagen des Natürlichen Systems, der Vergleichenden Anatomie und der Phylogenetik. Leipzig, Germany: Akademische Verlagsgesellschaft; 1952. [Google Scholar]
- 27.Chung YY, Kim SR, Kang HG, Noh YS, Chul PM, Finkela D, An G. Plant Sci. 1995;109:45–56. [Google Scholar]
- 28.Moon YH, Jung JY, Kang HG, An G. Plant Mol Biol. 1999;40:167–177. doi: 10.1023/a:1026429922616. [DOI] [PubMed] [Google Scholar]
- 29.Münster T, Wingen LU, Faigl W, Werth S, Saedler H, Theissen G. Gene. 2001;262:1–13. doi: 10.1016/s0378-1119(00)00556-4. [DOI] [PubMed] [Google Scholar]
- 30.Grass Phylogeny Working Group. Ann Mo Bot Gard. 2001;88:373–457. [Google Scholar]
- 31.Paterson AH, Bowers JE, Chapman BA. Proc Natl Acad Sci USA. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Judziewicz EJ, Soderstrom TR. Smithson Contrib Bot. 1989;68:1–52. [Google Scholar]
- 33.Arber A. Ann Bot. 1929;43:35–53. [Google Scholar]
- 34.Kellogg EA. Plant Physiol. 2001;125:1198–1205. doi: 10.1104/pp.125.3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rudall PJ, Bateman RS. New Phytol. 2004;162:25–44. [Google Scholar]
- 36.Decraene LPR, Linder HP, Smets EF. Plant Syst Evol. 2002;231:225–258. [Google Scholar]
- 37.Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lonnig WE, Saedler H, Sommer H. EMBO J. 1992;11:251–263. doi: 10.1002/j.1460-2075.1992.tb05048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fukui M, Futamura N, Mukai Y, Wang Y, Nagao A, Shinohara K. Plant Cell Physiol. 2001;42:566–575. doi: 10.1093/pcp/pce069. [DOI] [PubMed] [Google Scholar]
- 39.Mouradov A, Hamdorf B, Teasdale RD, Kim JT, Winter KU, Theissen G. Dev Genet. 1999;25:245–252. doi: 10.1002/(SICI)1520-6408(1999)25:3<245::AID-DVG7>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 40.Sundstrom J, Carlsbecker A, Svensson ME, Svenson M, Johanson U, Theissen G, Engstrom P. Dev Genet. 1999;25:253–266. doi: 10.1002/(SICI)1520-6408(1999)25:3<253::AID-DVG8>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 41.Sundstrom J, Engstrom P. Plant J. 2002;31:161–169. doi: 10.1046/j.1365-313x.2002.01343.x. [DOI] [PubMed] [Google Scholar]
- 42.Winter K-U, Becker A, Münster T, Kim JT, Saedler H, Theissen G. Proc Natl Acad Sci USA. 1999;96:7342–7347. doi: 10.1073/pnas.96.13.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park JH, Ishikawa Y, Yoshida R, Kanno A, Kameya T. Plant Mol Biol. 2003;51:867–875. doi: 10.1023/a:1023097202885. [DOI] [PubMed] [Google Scholar]
- 44.Park JH, Ishikawa Y, Ochiai T, Kanno A, Kameya T. Plant Cell Physiol. 2004;45:325–332. doi: 10.1093/pcp/pch040. [DOI] [PubMed] [Google Scholar]
- 45.Baum DA, Whitlock BA. Curr Biol. 1999;9:R525–R527. doi: 10.1016/s0960-9822(99)80327-3. [DOI] [PubMed] [Google Scholar]
- 46.Kramer EM, Jaramillo MA. J Exp Zoolog B Mol Dev Evol. 2005;304:526–535. doi: 10.1002/jez.b.21046. [DOI] [PubMed] [Google Scholar]
- 47.Long JA, Moan EI, Medford JI, Barton MK. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]