Abstract
Type Ib diamonds emit bright fluorescence at 550–800 nm from nitrogen-vacancy point defects, (N-V)0 and (N-V)−, produced by high-energy ion beam irradiation and subsequent thermal annealing. The emission, together with noncytotoxicity and easiness of surface functionalization, makes nano-sized diamonds a promising fluorescent probe for single-particle tracking in heterogeneous environments. We present the result of our characterization and application of single fluorescent nanodiamonds as cellular biomarkers. We found that, under the same excitation conditions, the fluorescence of a single 35-nm diamond is significantly brighter than that of a single dye molecule such as Alexa Fluor 546. The latter photobleached in the range of 10 s at a laser power density of 104 W/cm2, whereas the nanodiamond particle showed no sign of photobleaching even after 5 min of continuous excitation. Furthermore, no fluorescence blinking was detected within a time resolution of 1 ms. The photophysical properties of the particles do not deteriorate even after surface functionalization with carboxyl groups, which form covalent bonding with polyl-lysines that interact with DNA molecules through electrostatic forces. The feasibility of using surface-functionalized fluorescent nanodiamonds as single-particle biomarkers is demonstrated with both fixed and live HeLa cells.
Keywords: blinking, photobleaching, single-molecule detection, single-particle tracking, live cell
One of the key avenues to understanding how biological systems function at the molecular level is to probe biomolecules individually and observe how they interact with each other directly in vivo. Laser-induced fluorescence is a technique widely adopted for this purpose owing to its ultrahigh sensitivity and capabilities of performing multiple-probe detection (1–3). However, in applying this technique to imaging and tracking a single molecule or particle in a biological cell, progress is often hampered by the presence of ubiquitous endogenous components such as flavins, nicotinamide adenine dinucleotides, collagens, and porphyrins that produce high fluorescence background signals (4–6). These biomolecules typically absorb light at wavelengths in the range of 300–500 nm and fluoresce at 400–550 nm (Fig. 1). To avoid such interference, a good biological fluorescent probe should absorb light at a wavelength longer than 500 nm and emit light at a wavelength longer than 600 nm, at which the emission has a long penetration depth through cells and tissues (5, 7). Organic dyes and fluorescent proteins are two types of molecules often used to meet such a requirement (1, 8, 9); however, the detrimental photophysical properties of these molecules, such as photobleaching and blinking, inevitably restrict their applications for long-term in vitro or in vivo observations. Fluorescent semiconductor nanocrystals (or quantum dots), on the other hand, have gained considerable attention in recent years because they hold a number of advantageous features including high photobleaching thresholds and broad excitation but narrow emission spectra well suited for multicolor labeling and detection (10–14). Unfortunately, most nanomaterials are toxic, and hence reduction of cytotoxicity and human toxicity through surface modification plays a pivotal role in successful application of quantum dots to in vivo labeling, imaging, and diagnosis (10, 15, 16).
Fig. 1.
Comparison of emission spectra of flavin and FND. Note that the entire emission profile of the 35-nm FNDs (red) is well separated from that of flavin (orange) and also coincides with the high quantum efficiency region (gray) of the back-illuminated CCD camera used in this experiment. The absorption (blue) of flavin at 532 nm is negligible (39).
An additional concern with respect to quantum dots is that their photophysical properties are subject to change, depending on how their surfaces are modified. This limits the scope of their surface modification and functionalization, which often involve complicated surface chemistry. Insulator-based nanoparticles such as nanodiamonds, in contrast, are free of this limitation. We recently demonstrated that diamond crystallites with a nominal size of 100 nm are capable of producing stable fluorescence from color centers after surface treatment with strong oxidative acids (17). The result is not unanticipated because the fluorescence of nanodiamonds originates from point defects embedded in the crystal lattice and has little to do with their surface structures. One of the most noteworthy point defects in diamond is the negatively charged nitrogen-vacancy center, (N-V)−, which is the dominant end product of thermal annealing of irradiation-damaged type Ib diamond containing atomically dispersed nitrogen atoms (18). This defect center absorbs strongly at ≈560 nm and emits fluorescence efficiently at ≈700 nm, which is well separated from the spectral region where endogenous fluorescence occurs (Fig. 1). The material is nontoxic, and, moreover, its fluorescence shows no sign of fading due to photobleaching (17, 19), thereby allowing long-term monitoring of a single diamond nanoparticle in a biological cell.
Here we report our observations of single fluorescent nanodiamonds (FND) with an average size of 35 nm on a glass substrate and also in a HeLa cell using both confocal and wide-field epifluorescence microscopies. In addition to the photobleaching property as examined previously for an ensemble of 100-nm diamond powders (17), fluorescence blinking was also closely characterized for the individual 35-nm FNDs. We found that FNDs are exceptionally photostable under high-power laser excitation (up to an intensity of 1 MW/cm2) and that the brightness of the fluorescence remains almost at the same level even after covalent immobilization of biomolecules such as polypeptides and DNA onto their surfaces. It is demonstrated that FND is a promising material to be used as a fluorescent biomarker for in vitro as well as in vivo studies at the single-particle level.
Results and Discussion
Fig. 2A shows a confocal scanning image of single 35-nm FNDs dispersed on a bare glass substrate. The FWHM of each peak is two to three pixels, corresponding to a physical distance of 400–600 nm, which coincides well with the diffraction limit of the optical microscope. The observation of these diffraction-limited spots suggests that they derived from single isolated FNDs. This suggestion was indeed confirmed independently by obtaining a SEM image of the FNDs on the same glass plate showing that these particles are well separated on the surface [see supporting information (SI) Fig. 7]. The FND particles have an average size of ≈35 nm, consistent with the diameter specified by the vendor.
Fig. 2.
Detection of single FNDs. (A) Confocal scanning image of 35-nm FNDs dispersed on a bare glass substrate. The FWHM of each peak is two to three pixels, corresponding to a physical distance of 400–600 nm. (B) Fluorescence spectra of three different single FNDs. Indicated are two ZPLs at 576 and 638 nm originated from the defect centers (N-V)0 and (N-V)−, respectively.
The identification of single FNDs in Fig. 2A was also supported by dispersed fluorescence studies. As shown in Fig. 2B, each FND emits a characteristic spectrum in the extended red region, which is indicative of single-particle detection. Whereas the observed fluorescence spectra are highly heterogeneous, two sharp zero-phonon lines (ZPL) can be found for all of the 35-nm particles interrogated. The first ZPL peaking at 576 nm is ascribable to the electronic transition of the neutral defect center (N-V)0, and the second ZPL peaking at 638 nm corresponds to the 3A → 3E transition of the negatively charged defect center, (N-V)− (18–21). Both the ZPLs are accompanied with broad phonon sidebands red-shifted by ≈50 nm. A close comparison of the spectra reveals that these three particles mainly differ in the ratio of (N-V)− to (N-V)0 contents.
To test the photostability of FNDs, we monitored the fluorescence intensities of the individual particles over an extended period (Fig. 3A). In accord with our previous finding for an ensemble of 100-nm diamond powders (17), excellent photostability was observed for the single 35-nm diamonds as well. Under the excitation with 532-nm light at a power density of 8 × 103 W/cm2, the fluorescence intensity of the individual FNDs stays essentially the same over a time period of 300 s. No sign of fluorescence blinking was detected within the time resolution of 1 ms (Fig. 3B). In contrast, single dye molecules such as Alexa Fluor 546 covalently linked to dsDNA photobleached within 12 s (Fig. 3A).
Fig. 3.
Characterization of single FNDs. (A) Typical time traces of the fluorescence from a single 100-nm FND (green), a single 35-nm FND (red), and a single Alexa Fluor 546 dye molecule attached to a single dsDNA molecule (blue). Note that no sign of photobleaching was detected within 300 s of the continuous excitation for the FNDs. (B) Time traces of the fluorescence from a single 100-nm FND (green) and a single 35-nm FND (red) acquired with a time resolution of 1 ms, showing a nonblinking behavior. (C) Plot of fluorescence intensity as a function of laser power density over the range of 1 × 102 to 1 × 106 W/cm2 for 100-nm and 35-nm FNDs. The corresponding increase of the fluorescence intensity is 300-fold, and no photobleaching was observed for both samples. (D) Fluorescence lifetime measurements of 100-nm FNDs (green) and Alexa Fluor 546 dye molecules (blue). Fitting the time traces with two exponential decays reveals a fast component of 1.7 ns (4%) and a slow component of 17 ns (96%). The latter is four times longer than that (≈4 ns) of Alexa Fluor 546.
For the 35-nm FNDs, an analysis of the fluorescence intensity for a large number (≈30) of measurements revealed that the brightness of the single nanodiamonds varies ≈5-fold among the individuals. Such a variation is likely to result from heterogeneities in size and quantity, as well as the fluorescence quality of the N-V defect centers embedded in different nanocrystallites. Regardless of the difference in absolute intensity, both the 35-nm and 100-nm FNDs show similar photostability (compare time traces in Fig. 3A), which implies that the photostability of nanodiamonds is size-independent. Additionally, the intensity ratio between these two samples is ≈30-fold, in accord with their volume ratio of ≈23:1. Although the fluorescence intensity does not scale quite linearly with the laser energy, this intensity ratio stays almost the same as the laser power density increases from 1 × 104 to 1 × 106 W/cm2 (Fig. 3C). All of the observations are consistent with the suggestion that the fluorescence signals detected in this experiment derive primarily from the N-V defect centers distributed evenly in the nanodiamond matrices.
In Fig. 3D we also show the result of the fluorescence lifetime measurements for 100-nm FNDs. The fluorescence decay was obtained by coadding the time traces of 30 individual particles. The major component of the decay has a lifetime of 17 ns, which is comparable to that (11.6 ns) measured for bulk diamonds (22) but is substantially longer than that of dye molecules (≈4 ns for Alexa Fluor 546 in Fig. 3D) and cell autofluorescence. Such a distinct difference in fluorescence lifetime is expected to be useful for isolation of FND emission from these (and other) background signals using various time-gating methods (23, 24), with which the contrast of single-nanodiamond imaging in biological cells can be further enhanced.
It is instructive to compare the fluorescence brightness of nanodiamonds with quantum dots. From a measurement of ≈100 particles for each sample, we estimated that the average fluorescence intensity of the 35-nm FNDs is approximately the same as that of quantum dots such as CdSe/ZnS that emit light in the similar wavelength range (see SI Fig. 8). Further information about the fluorescence brightness of FND can be deduced from a close comparison of our result with the single-defect measurements (21, 25), which indicates that the number of defect centers in the individual 35-nm diamonds prepared in this experiment is ≈100. Note that this number of defect centers is ≈10-fold less than expected if all of the nitrogen impurities in the diamond lattice are converted to N-V centers, given a nitrogen concentration of 300 ppm (or ≈3 × 107 nitrogen atoms per cubic micrometer) for type Ib diamond (26). This discrepancy suggests that if we can properly optimize our experimental conditions, 10-nm diamonds (or “diamond dots”) with fluorescence brightness comparable to that of quantum dots can be produced. Such fluorescence brightness is sufficient for single-particle tracking in cells (9), particularly in the extended red region where the autofluorescence background is much reduced.
In a step to demonstrate that nanodiamond is a promising biomarker candidate for in vivo imaging and diagnosis, 35-nm FNDs were incubated together with HeLa cells cultured in DMEM at 37°C on a chamber slide. For the HeLa cell, intense cell autofluorescence was observed at 510–560 nm when exposed to blue light at 476 nm. Switching the laser excitation wavelength to 532 nm and collecting the emission at 650–720 nm where the FND fluorescence resides greatly reduces the fluorescence background signals (see SI Fig. 9). Fig. 4A shows an overlay of the bright-field and epifluorescence images of a single HeLa cell after the FND uptake. Translocation of the FNDs through the cell's membrane was confirmed by obtaining vertical cross-section images (see SI Fig. 10) of the cell with the wide-field epifluorescence microscope. Although many FNDs are found to form aggregates in the cell, some isolated nanodiamonds can be detected clearly in the cytoplasm (Fig. 4B). These particles are identified as single nanodiamonds because the spot sizes of their images are diffraction-limited (Fig. 4C) and their fluorescence intensities are comparable to that of the single FNDs spin-coated on the coverglass plate. It is of interest to note that the FNDs uptaken are mainly distributed in the cytoplasm of the HeLa cell and that they are photostable even after continuous excitation of the sample for 20 min at a laser power density of 100 W/cm2 (Fig. 4C Inset). Similar to the earlier finding, neither photobleaching nor blinking of the fluorescence was observed for these FNDs within the limits of our detection sensitivity and time window.
Fig. 4.
Observation of single FNDs in a HeLa cell. (A) Bright-field and epifluorescence images of a HeLa cell after uptake of 35-nm FNDs. Most of the uptaken FNDs are seen to distribute in the cytoplasm. (B) Epifluorescence fluorescence image of a single HeLa cell after the FND uptake. An enlarged view of the fluorescence spots (denoted by “1” and “2”) with diffraction-limited sizes (FWHM ≈ 500 nm) is shown in Inset. The separation between these two particles is ≈1 μm. (C) Intensity profile of the fluorescence image along the line drawn in B Inset. (C Inset) Integrated fluorescence intensity (after subtraction of the signals from cell autofluorescence and background fluorescence from the microscope slides) as a function of time for particle “1.” The signal integration time was 0.1 s. No sign of photobleaching was detected after continuous excitation of the particle for 20 min.
As pointed out in our previous work (27–29) and described in Materials and Methods, the surface of nanodiamonds can be easily functionalized with carboxyl groups and their derivatives for specific or nonspecific binding with nucleic acids and proteins. Such a unique characteristic opens many opportunities for both in vitro and in vivo applications of FNDs. One such example consists of coating carboxylated FNDs with polyl-lysines (PLs) to facilitate binding of the particles nonspecifically with DNA through electrostatic interactions (27, 29, 30). In this experiment, a single T4 DNA molecule fluorescently labeled with TOTO-1 dye molecules was stretched on an amine-terminated glass substrate with a channel-combing method (31). How the positively charged FND particles interact with the negatively charged DNA molecules was observed directly by a wide-field epifluorescence microscope equipped with a dual-view system.
Two channels were monitored simultaneously to reveal the microscopic details of the interaction between PL-coated FND and DNA: (i) the shorter-wavelength channel detects 545- to 605-nm emission from both FND and the TOTO-1 dyes intercalated within the T4 DNA molecule, and (ii) the longer wavelength channel detects 675- to 685-nm emission from FND only. As shown in Fig. 5A, the T4 DNA was combed to have a V-shape configuration and stretched to a length of ≈60 μm, close to its full contour length of 75 μm (32). An overlay of the images from these two detection channels reveals that the T4 DNA molecule was actually wrapped around the PL-coated FND particle. To quantify the fluorescence decay rates of FND and TOTO-1 separately, the fluorescence intensity (after background subtraction) of each channel was integrated, normalized, and plotted as a function of time. The fluorescence intensity of the TOTO-1 dyes was seen to decrease to one-third of its initial value (owing to photobleaching) after continuous excitation of the sample at 514 nm for 40 s. In contrast, the fluorescence intensity of the FND stayed essentially the same during the same time period of excitation (Fig. 5B).
Fig. 5.
Observation of a single PL-coated FND particle bound with a single T4 DNA molecule on an amine-terminated glass substrate. (A) Dual-view fluorescence images of a single DNA/FND complex. An overlay (Right) of the images from the shorter (545–605 nm) and longer (675–685 nm) wavelength channels reveals that the T4 DNA molecule was wrapped around the 100-nm FND particle and stretched to a V-shape configuration. (B) Fluorescence decays of the 100-nm FND particle (red) and the TOTO-1 dyes (green) intercalated in the T4 DNA molecule.
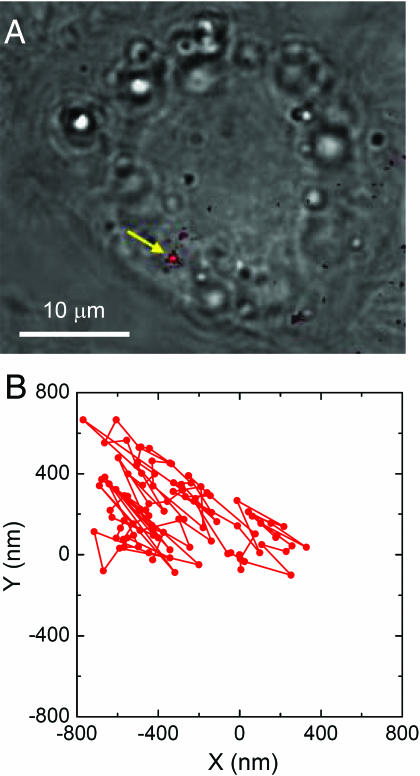
Finally, we demonstrate that it is possible to conduct single-particle tracking for the 35-nm FND in the cytoplasm of a live HeLa cell. Fig. 6B shows the trajectory of the diffusion motion of the particle appearing as a bright red spot indicated in Fig. 6A. Starting at the coordinate (0, 0), each data point was obtained by fitting the intensity profile of the red spot with a two-dimensional Gaussian function to deduce its center and go beyond the optical diffraction limit (33). The particle's motion is Brownian and is confined within a small area of 1 × 1 μm2 near the nucleus over our observation time window of 13.9 s. Note that in Fig. 6B the trajectory is plotted as a projection on the x-y plane of the glass slide. Three-dimensional confocal tracking (34) of the single FND is practical if either the bright fluorescence or the strong scattered laser light (35) from the diamond nanoparticle is used as a feedback signal to locate its position in the live cell.
Fig. 6.
Tracking of a single FND in a live HeLa cell. (A) Overlay of bright-field and epifluorescence images of a live HeLa cell after uptake of 35-nm FNDs. (B) A 100-step (139 ms per step) trajectory of a single FND, indicated by the yellow arrow in A, moving in the cytoplasm of the HeLa cell. A video showing the diffusion motion of this particle is given in SI Movie 1.
Conclusions
We have characterized in detail the photophysical properties of FND with an average size of 35 and 100 nm at the single-particle level. The material possesses several unique features including facile surface modification, long-term photostability, and no fluorescence blinking. The latter two properties are size-independent and do not show any sign of deterioration even after surface treatment with strong oxidative acids. The easiness of surface functionalization endows nanodiamonds with a significant advantage over other nanomaterials for specific or nonspecific binding with proteins and nucleic acids. Centered in the extended red region (≈700 nm), the fluorescence of FND can be detected with minimal interference from cell autofluorescence. We have demonstrated in this work that a combination of these unique chemical and photophysical properties ensures long-term (short-term as well) single-particle tracking and fluorescent imaging of FNDs as cellular biomarkers.
For future applications where smaller-sized particles are required, it is possible to employ standard separation methods to extract FNDs with a size in the range of 10 nm from the current sample. Each particle of this size is expected to contain up to 30 nitrogen-vacancy defect centers after optimization of our sample preparation conditions and have the photostability similar to that described in this article. A natural extension of the present application to in vivo studies includes the use of FND as a drug or gene carrier, as a device for tumor targeting, and as a fluorescent probe for two-photon confocal microscopy (36). Developed originally for surface finishing industry, the diamond nanoparticle, interestingly, is now finding new and far-reaching applications in modern biomedical science and biotechnologies.
Materials and Methods
Production of FNDs.
Synthetic type Ib diamond powders with a nominal size of 35 nm (MSY, Microdiamant) and 100 nm (μm+ MDA, Element Six) were purified in concentrated H2SO4-HNO3 solution (3:1, vol/vol) at 90°C for 30 min. After separation by centrifugation and rinsing extensively with deionized water (Millipore), the diamond powders were resuspended in water to a concentration of 0.1 g/ml as a stock solution. A thin diamond film (≈0.5-cm2 area × ≈50-μm thickness) was prepared by depositing 50 μl of the diamond suspension on a 0.5-mm-thick silicon wafer and air-dried. The dry diamond film was irradiated by a 3-MeV proton beam from a tandem accelerator (9SDH-2; National Electrostatics) at a dose of 1 × 1016 ions per square centimeter. Formation of nitrogen-vacancy defect centers was facilitated by annealing the proton beam-treated nanodiamonds in vacuum at 700°C for 2 h. To remove graphitic surface structures induced by thermal annealing, the freshly prepared FNDs were additionally cleaned in concentrated H2SO4-HNO3 (9:1, vol/vol) solution at 75°C for 3 days, followed by separation with centrifugation and extensive rinsing with deionized water. Both of the diamond samples were treated following the same protocols.
Surface Modification of FNDs.
FNDs (100 nm) were surface-functionalized with carboxyl groups by strong oxidative acid treatment in concentrated H2SO4-HNO3 (9:1, vol/vol) at 75°C for 3 days, subsequently in 0.1 M NaOH aqueous solution at 90°C for 2 h, and finally in 0.1 M HCl aqueous solution at 90°C for 2 h. The resulting carboxylated/oxidized FNDs were separated by centrifugation, rinsed extensively, and resuspended in deionized water. To functionalize the surfaces of FNDs with amino groups, 8 mg of N-(3-dimethylaminopropyl)-N′-ethyl-carbodiimide hydrochloride (Sigma) and 6 mg of N-hydroxysuccinimide (Sigma) were dissolved together in 5 ml of the diamond suspension containing 6 mg of FND, followed by addition of 3 mg of PLs (MW ≈ 30,000; Sigma) to the suspension to form covalent linkages with the surface carboxylate groups activated by N-(3-dimethylaminopropyl)-N′-ethyl-carbodiimide hydrochloride. After incubation of the mixture at room temperature for 24 h, the amine-terminated FNDs were thoroughly washed with deionized water and separated by centrifugation before use.
Observation of Single FNDs, Single Dye Molecules, and Single Quantum Dots on Coverglass Plates.
Coverglass plates (Marienfeld Laboratory Glassware) were cleaned by immersing them in concentrated H2SO4-HNO3 (vol/vol, 3:1) solution for >24 h, followed by extensive rinse with deionized water. The FNDs well dispersed in a diluted suspension were then spin-coated on the coverglass plate at 14 × g. To observe the fluorescence from a single dye molecule, Alexa Fluor 546 covalently linked to a dsDNA molecule was used. The dye-labeled dsDNA molecule (Integrated DNA Technologies) was prepared by heating a T50 buffer (10 mM Tris·HCl, pH 8/50 mM NaCl) containing both 33-bp ssDNA (5′ end-labeled with Alexa Fluor 546) and 27-bp ssDNA to 90°C for 10 min, after which the solution was slowly cooled to room temperature. The solution was finally diluted with the same buffer to a concentration of 40–400 pM before spin-coating of the annealed dsDNA molecules on the coverglass plate at 57 × g. In the comparative study of fluorescence brightness between FNDs and quantum dots, CdSe/ZnS particles (Evident Technologies) of a core diameter of ≈4 nm were used as the sample. The particles (emission peak at 600 nm and fluorescence quantum yield of 30–50%) were first diluted in toluene with polymethyl methacrylate (3% vol/vol) and then spin-coated on the coverglass plate for single-particle detection.
The glass plate spin-coated with the sample was mounted on a modified confocal optical microscope (E600; Nikon) for inspection (37). Excitation of the sample was made through a ×100 objective (Plan Fluor, N.A. 1.3 oil; Nikon) by using a solid-state laser (JL-LD532-GTE; Jetlaser) operated at 532 nm. Epifluorescence passing through a 565-nm long-pass filter (E565lp; Chroma Tech) was collected and detected by an avalanche photodiode (SPCM-AQR-15; PerkinElmer). Fluorescence images were first obtained by raster scanning with a piezo-driven nanopositioning and scanning system (E-710.4CL and P-734.2CL; Physik Instrument). After obtaining the images of single FNDs, each particle was moved consecutively to the focal point of the objective to record the time evolution of the fluorescence intensity. The corresponding spectra were acquired by using a monochromator (SP300i; Acton Research) equipped with a liquid-nitrogen-cooled CCD camera (LN/CCD-1100-PB; Princeton Instruments). For fluorescence lifetime measurements, a frequency-doubled picosecond Nd:YAG laser (IC-532-30; High Q Laser) served as the excitation source. The corresponding fluorescence decays were measured with a time-correlated single-photon counting module (SPC-600; Becker & Hickl).
Cell Culture, FND Uptake, and Observation of Single FNDs in HeLa Cells.
HeLa cells (105 cells per milliliter) were cultured in DMEM (SH30243.02; HyClone) supplemented with 10% FBS and 1% penicillin/streptomycin. Suspensions of the 35-nm FNDs were first diluted with DMEM without serum to a final concentration of 0.8 μg/ml. After sonication for 30 min to ensure complete dispersion of the FNDs, an aliquot (typically 0.1 ml) of the suspension was added to the wells of a chamber coverglass plate (Lab-Tek II; Nunc) containing the HeLa cells precultured for 24 h. The chamber slide was then incubated at 37°C in a 5% CO2 incubator for 5 h for FND uptake. The FNDs not uptaken were removed by washing the sample three times with warm Dulbecco's phosphate buffer saline, after which the cells were fixed with 4% paraformaldehyde (Sigma). The excess paraformaldehyde was washed away with deionized water. In the experiment of observing the diffusion motion of a single diamond in a live HeLa cell, the sample was treated and inspected following the same procedures described above but with no paraformaldehyde treatment.
The fluorescence images of FNDs in HeLa cells were acquired by using a wide-field epifluorescence microscope (IX70; Olympus) equipped with a Nd:YAG laser (DPSS 532; Coherent) operating at 532 nm (power density ≈ 100 W/cm2). The resulting emission was collected by a ×100, N.A. 1.35 oil objective (UPLFL ×100; Olympus) and selected by a 565-nm long-pass filter (E565lp; Chroma Tech) for detection. Images were recorded with an electron-multiplying CCD (DV887DCS-BV; Andor) having a pixel size of 16 μm and an exposure time of 0.1 s.
Observation of Single DNA/FND Complexes on a Glass Plate.
T4 DNA (165.6 kbp; Wako) was fluorescently labeled with TOTO-1 (T3600; Molecular Probes) at a molar ratio of 4 bp per dye molecule. The length of the T4 DNA molecule extended from 56 μm to ≈75 μm after labeling (32). To prepare the DNA/FND complex, 3 μg of PL-coated FNDs were suspended in 200 μl of 0.5× Tris/borate/EDTA buffer (15581-044; Invitrogen) and mixed with T4 DNA at a molar ratio of eight DNA molecules per FND particle. After incubation at room temperature for 10 min, the sample mixture was diluted to a concentration suitable for single-particle detection. 2-Mercaptoethanol (Sigma) was added afterward to the sample solution with a volume ratio of ≈3% to avoid rapid photobleaching of TOTO-1.
A microchannel-combing method (31) was used to stretch and fix DNA on a coverglass plate surface-modified with amino groups. The microchannel was constructed with three components: a coverglass plate pretreated with 3-aminopropyltriethoxysilane (APTES; Fluka), a standard microscope slide containing two 1-mm holes separated by 16 mm, and a 3M double-side tape cut with a channel pattern with a dimension of 20 mm × 3 mm (38). To prepare the microchannel, a clean coverglass plate was first soaked in 6 mM APTES (pH ≈ 3) at 60°C for 18 h, after which it was rinsed thoroughly with distilled water. The double-side tape was then sandwiched in between the microscope slide and the APTES-coated coverglass plate to form a microchannel with a height of ≈100 μm. After filling the microchannel with the sample solution, atmospheric air was slowly pushed into the channel through a pipette firmly attached to one of the holes on the microscope slide. The DNA molecule was consequently stretched and fixed on the coverglass plate by the moving air–water interface.
Inside the channel, FND and the fluorescently labeled DNA molecule were coexcited by the 514-nm light from an Ar ion laser (Innova 90; Coherent) and inspected by a fluorescence microscope (IX70; Olympus) equipped with a dual-view system (Dual-View; Optical Insights). This dual-view system allowed simultaneous capture of two images using a single electron-multiplying CCD (DV887DCS-BV; Andor) operating at an exposure time of 0.1 s. The wavelength coverage of each channel is 545–605 nm and 675–685 nm, as defined separately by two bandpass filters (HQ575/60m and HQ680/10m; Chroma Tech).
Supplementary Material
Acknowledgments
We thank J. H. Hsu (Department of Materials Science and Optoelectonics Engineering, National Sun Yat-sen University, Kaohsiung, Taiwan) for proving the quantum dot samples. This research was supported by Academia Sinica and National Science Council Grants NSC 93-3112-B-001-020-Y and NSC 94-2120-M-002-009 (National Nanoscience and Nanotechnology Project).
Abbreviations
- FND
fluorescent nanodiamond
- ZPL
zero-phonon line
- PL
polyl-lysine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0605409104/DC1.
References
- 1.Michalet X, Kapanidis AN, Laurence T, Pinaud F, Doose S, Pflughoefft M, Weiss S. Annu Rev Biophys Biomol Struct. 2003;32:161–182. doi: 10.1146/annurev.biophys.32.110601.142525. [DOI] [PubMed] [Google Scholar]
- 2.Weijer CJ. Science. 2003;300:96–100. doi: 10.1126/science.1082830. [DOI] [PubMed] [Google Scholar]
- 3.Weiss S. Science. 1999;283:1676–1683. doi: 10.1126/science.283.5408.1676. [DOI] [PubMed] [Google Scholar]
- 4.Billinton N, Knight AW. Anal Biochem. 2001;291:175–197. doi: 10.1006/abio.2000.5006. [DOI] [PubMed] [Google Scholar]
- 5.Mansfield JR, Gossage KW, Hoyt CC, Levenson RM. J Biomed Opt. 2005;10 doi: 10.1117/1.2032458. 041207. [DOI] [PubMed] [Google Scholar]
- 6.Troy T, Jekic-McMullen D, Sambucetti L, Rice B. Mol Imaging. 2004;3:9–23. doi: 10.1162/15353500200403196. [DOI] [PubMed] [Google Scholar]
- 7.Lim YT, Kim S, Nakayama A, Stott NE, Bawendi MG, Frangioni JV. Mol Imaging. 2003;2:50–64. doi: 10.1162/15353500200302163. [DOI] [PubMed] [Google Scholar]
- 8.Lippincott-Schwartz J, Patterson GH. Science. 2003;300:87–91. doi: 10.1126/science.1082520. [DOI] [PubMed] [Google Scholar]
- 9.Yu J, Xiao J, Ren XJ, Lao KQ, Xie XS. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- 10.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Science. 2005;307:538–544. doi: 10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medintz IL, Uyeda HT, Goldman ER, Mattoussi H. Nat Mater. 2005;4:435–446. doi: 10.1038/nmat1390. [DOI] [PubMed] [Google Scholar]
- 12.Lacoste TD, Michalet X, Pinaud F, Chemla DS, Alivisatos AP, Weiss S. Proc Natl Acad Sci USA. 2000;97:9461–9466. doi: 10.1073/pnas.170286097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao XH, Cui YY, Levenson RM, Chung LWK, Nie SM. Nat Biotechnol. 2004;22:969–976. doi: 10.1038/nbt994. [DOI] [PubMed] [Google Scholar]
- 14.Zhang CY, Yeh HC, Kuroki MT, Wang TH. Nat Mater. 2005;4:826–831. doi: 10.1038/nmat1508. [DOI] [PubMed] [Google Scholar]
- 15.Derfus AM, Chan WCW, Bhatia SN. Nano Lett. 2004;4:11–18. doi: 10.1021/nl0347334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirchner C, Liedl T, Kudera S, Pellegrino T, Javier AM, Gaub HE, Stolzle S, Fertig N, Parak WJ. Nano Lett. 2005;5:331–338. doi: 10.1021/nl047996m. [DOI] [PubMed] [Google Scholar]
- 17.Yu S-J, Kang M-W, Chang H-C, Chen K-M, Yu Y-C. J Am Chem Soc. 2005;127:17604–17605. doi: 10.1021/ja0567081. [DOI] [PubMed] [Google Scholar]
- 18.Davies G, Hamer MF. Proc R Soc London Ser A. 1976;348:285–298. [Google Scholar]
- 19.Gruber A, Drabenstedt A, Tietz C, Fleury L, Wrachtrup J, von Borczyskowski C. Science. 1997;276:2012–2014. [Google Scholar]
- 20.Jelezko F, Tietz C, Gruber A, Popa I, Nizovtsev A, Kilin S, Wrachtrup J. Single Mol. 2001;2:255–260. [Google Scholar]
- 21.Treussart F, Jacques V, Wu E, Gacoln T, Grangier P, Roch JF. Physica B. 2006;376:926–929. [Google Scholar]
- 22.Collins AT, Thomaz MF, Jorge MIB. J Phys C: Solid State Phys. 1983;16:2177–2181. [Google Scholar]
- 23.Pepperkok R, Squire A, Geley S, Bastiaens PIH. Curr Biol. 1999;9:269–272. doi: 10.1016/s0960-9822(99)80117-1. [DOI] [PubMed] [Google Scholar]
- 24.Dahan M, Laurence T, Pinaud F, Chemla DS, Alivisatos AP, Sauer M, Weiss S. Opt Lett. 2001;26:825–827. doi: 10.1364/ol.26.000825. [DOI] [PubMed] [Google Scholar]
- 25.Kurtsiefer C, Mayer S, Zarda P, Weinfurter H. Phys Rev Lett. 2000;85:290–293. doi: 10.1103/PhysRevLett.85.290. [DOI] [PubMed] [Google Scholar]
- 26.Woods GS. In: Properties and Growth of Diamond. Davies G, editor. London: INSPEC, Institute of Electrical Engineers; 1994. Chap 3.1. [Google Scholar]
- 27.Huang L-CL, Chang H-C. Langmuir. 2004;20:5879–5884. [PubMed] [Google Scholar]
- 28.Kong XL, Huang L-CL, Hsu C-M, Chen W-H, Han C-C, Chang H-C. Anal Chem. 2005;77:259–265. doi: 10.1021/ac048971a. [DOI] [PubMed] [Google Scholar]
- 29.Kong XL, Huang L-CL, Liau S-CV, Han C-C, Chang H-C. Anal Chem. 2005;77:4273–4277. doi: 10.1021/ac050213c. [DOI] [PubMed] [Google Scholar]
- 30.Zinchenko AA, Yoshikawa K, Baigl D. Phys Rev Lett. 2005;95:228101. doi: 10.1103/PhysRevLett.95.228101. [DOI] [PubMed] [Google Scholar]
- 31.Petit CAP, Carbeck JD. Nano Lett. 2003;3:1141–1146. [Google Scholar]
- 32.Perkins TT, Smith DE, Larson RG, Chu S. Science. 1995;268:83–87. doi: 10.1126/science.7701345. [DOI] [PubMed] [Google Scholar]
- 33.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 34.Cang H, Wong CM, Xu CS, Rizvi AH, Yang H. Appl Phys Lett. 2006;88:223901. [Google Scholar]
- 35.Colpin Y, Swan A, Zvyagin AV, Plakhotnik T. Opt Lett. 2006;31:625–627. doi: 10.1364/ol.31.000625. [DOI] [PubMed] [Google Scholar]
- 36.Larson DR, Zipfel WR, Williams RM, Clark SW, Bruchez MP, Wise FW, Webb WW. Science. 2003;300:1434–1436. doi: 10.1126/science.1083780. [DOI] [PubMed] [Google Scholar]
- 37.Wang C-F, White JD, Lim T-L, Hsu J-H, Yang S-C, Fann WS, Peng K-Y, Chen S-A. Phys Rev B. 2003;67 035202. [Google Scholar]
- 38.Ha T. Methods. 2001;25:78–86. doi: 10.1006/meth.2001.1217. [DOI] [PubMed] [Google Scholar]
- 39.Harms GS, Cognet L, Lommerse PH, Blab GA, Schmidt T. Biophys J. 2001;80:2396–2408. doi: 10.1016/S0006-3495(01)76209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.