Abstract
Thymine glycol (Tg) is a common product of oxidation and ionizing radiation, including that used for cancer treatment. Although Tg is a poor mutagenic lesion, it has been shown to present a strong block to both repair and replicative DNA polymerases. The 2.65-Å crystal structure of a binary complex of the replicative RB69 DNA polymerase with DNA shows that the templating Tg is intrahelical and forms a regular Watson–Crick base pair with the incorporated A. The C5 methyl group protrudes axially from the ring of the damaged pyrimidine and hinders stacking of the adjacent 5′ template guanine. The position of the displaced 5′ template guanine is such that the next incoming nucleotide cannot be incorporated into the growing primer strand, and it explains why primer extension past the lesion is prohibited even though DNA polymerases can readily incorporate an A across from the Tg lesion.
Keywords: oxidative DNA damage, structure, DNA replication
Thymine glycol (5,6-dihydro-5,6-dihydroxythymine; Tg), the most common oxidation product of thymine, is produced endogenously as a consequence of aerobic metabolism or via exogenous factors such as chemical oxidants or ionizing radiation (Fig. 1). It is estimated that 400 Tgs are formed per cell per day (1, 2), and the presence of Tg in DNA has been used as a marker for oxidative stress (1, 3). Moreover, Tg is one of the predominant types of base modifications produced by ionizing radiation (4, 5), including that used in cancer therapy.
Fig. 1.
Tg is the product of ionizing radiation and oxidation. γ-irradiation produces equal amounts of the (5R, 6S) and (5S, 6R) cis isomers, whereas oxidation generates preferentially the (5R, 6S) isomer.
Of the oxidatively modified DNA bases retaining an intact ring, Tg is thought to induce the most distortion in the regular structure of DNA. Even though there are DNA repair enzymes specialized in excising Tg from the genome, such as human NEIL1 and NTH1 (6–8), statistically a few of the damaged bases will evade repair, which means that DNA polymerases will encounter these lesions during replication. Although Tg is a poor mutagenic lesion because it generally pairs with A (9), it has been shown to be a very effective block to DNA replication (10–13). When Tg is encountered as a templating base by replicative or repair polymerases, termination of primer extension occurs immediately past the lesion site with A inserted opposite the Tg. Even in the presence of proofreading, extension proceeds no further. This finding contrasts with the situation observed when the lesion is an abasic site, where, in the presence of proofreading, termination sites are observed one base before the lesion (14). Therefore, it is extension past the Tg·adenine pair, rather than insertion across Tg, that constitutes the block to the replicative polymerases. Because Tg is a strong block to repair and replicative DNA polymerases in vitro (10–13), not surprisingly, it is a lethal lesion in vivo (9, 15–18).
Unlike normal DNA bases, Tg is nonplanar because of the loss of aromatic character that accompanies the addition of hydroxyl groups at the 5 and 6 positions of the ring (19). Computational simulations (20, 21) predict that the axial orientation of the methyl group with respect to the pyrimidine ring would create steric hindrance with the template base on the 5′ side of Tg. NMR studies, on the other hand, have suggested that the structural perturbations attributable to the Tg lesion are localized and result in Tg's being extrahelical (22). To date, there is no published structure of any DNA polymerase in complex with DNA containing Tg that can clarify this lesion's ability to impede extension. Here we report the 2.65-Å resolution crystal structure of a postinsertion binary complex of the replicative DNA polymerase from bacteriophage RB69 with primer/template DNA bearing an acyclic AMP (acyAMP) incorporated opposite Tg. The Tg lesion is intrahelical and engages in a regular Watson–Crick base pair with the incorporated A. In contrast to our previously published structure with the abasic site analog furan (23), translocation takes place after incorporation opposite Tg. Because the methyl group of the oxidized thymine protrudes axially from the pyrimidine ring, it prevents the adjacent 5′ template base from stacking against Tg, thereby impeding extension, in agreement with the computational predictions (20, 21) and the biochemical (10–13) and biological (9, 15–18) studies. Our crystal structure allows visualization of what causes a replicative polymerase to stall when it encounters a Tg lesion.
Results
Primer Extension Assays with DNA Containing Tg.
Primer extension assays were performed with RB69 gp43 (the replicative polymerase of the bacteriophage) with DNA containing a Tg lesion in the templating position. The template strand containing the Tg lesion was synthesized by using a commercially available phosphoramidite (Glen Research, Sterling, VA) (24). Although there are four possible diastereomers of Tg by virtue of the chirality of the C5 and C6 atoms (25), the Tg isomer from the phosphoramidite building block is predominantly the naturally occurring (5R, 6S) cis isomer. Previous work has shown a sequence context dependence of Tg bypass for the Klenow fragment of Escherichia coli DNA polymerase I and DNA polymerase α (11, 26). Tg is a strong block for DNA polymerases in vitro and in vivo, except in the 5′-CTgPur-3′ sequence context, where bypass was observed ≈5% of the time (11, 27). We confirm here that, in a 5′-GTgG-3′ sequence context, Tg yields an absolute block to wild-type RB69 gp43 (Fig. 2a) and a strong pause site for the exonuclease-deficient mutant (D222A/D327A) (Fig. 2b).
Fig. 2.
RB69 gp43 stalls after incorporation across Tg site. (a) Wild-type RB69 gp43 is blocked after incorporation across Tg, even after an incubation of 5 min. (b) With an exonuclease-deficient DNA polymerase (D222A/D327A), Tg is a strong pause site.
Crystal Structure of RB69 gp43 in Complex with Tg-Containing DNA.
A stable complex of RB69 gp43 with Tg-containing DNA was obtained by reacting the polymerase with the annealed primer/template and acyclic ATP (acyATP) in the presence of magnesium. Acyclic nucleotides are chain terminators that have been shown to be readily incorporated by family B polymerases (28). The acyATP originally was used for screening for ternary complexes of RB69 gp43; although incorporation of a chain terminator at the 3′ end of the primer was not necessary to obtain a binary complex, acyATP was used because crystals grew more readily in the presence of acyATP as compared with dATP or dideoxyATP. Diffraction data were collected to 2.65-Å resolution at the Advanced Photon Source at Argonne National Laboratory (Argonne, IL) and phased by using the structure of the previously solved binary complex of RB69 gp43 with DNA containing an abasic site analog (23). The structure was refined to Rwork and Rfree values of 22.8% and 28.2%, respectively.
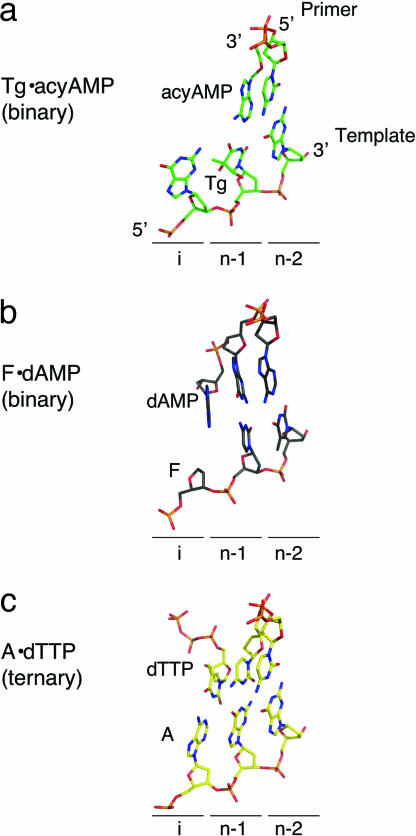
Although the asymmetric unit of the crystal comprises four polymerase–DNA complexes, the description will focus mainly on one of the complexes, molecule B, which has the clearest electron density as well as the lowest average B factors for both protein (51.2 Å2) and DNA (55 Å2). In all four independent complexes, the DNA is located in the polymerase active site rather than the exonuclease active site, and the acyAMP is incorporated opposite the templating Tg lesion, which remains intrahelical. Its adenine base engages in a standard Watson–Crick base pair with the oxidized thymine [distances for Tg (O4)–Adenine (N6) and Tg (N3)–Adenine (N1) are 2.9 and 3.0 Å, respectively] (Fig. 3a). The Tg·acyAMP base pair also maintains the same water-mediated minor groove interaction with the conserved Y567 as a normal G·C base pair (Fig. 3b). The crystal structure reveals that translocation took place after incorporation opposite the lesion (compare Fig. 4 a to c). This finding contrasts with the RB69 gp43 binary complex where dAMP was incorporated opposite furan (denoted in complex by “F”), an abasic site analog (23) (F·dAMP complex), but translocation did not occur (compare Fig. 4 b to c).
Fig. 3.
Tg base-pairs with A and maintains minor groove interactions. (a) Tg·acyAMP base pair with overlaid 2.65-Å simulated annealing omit map contoured at 5 σ. Hydrogen bonds are indicated by black dotted lines. (b) The Tg·acyAMP structure (green) was superimposed onto an RB69 gp43 complex obtained with undamaged DNA (29) (yellow; PDB ID code 1IG9). The Tg·acyAMP base pair (green) maintains the same water-mediated minor groove interaction with Y567 as the normal G·C base pair (yellow), and the universal hydrogen-bond acceptors O2 and N3 (33) occupy the same position in the two superimposed base pairs. The water molecules are shown as red spheres.
Fig. 4.
Translocation occurred after incorporation of A opposite Tg. The letter “i” indicates the position of the insertion site, and “n-1” and “n-2” are the postinsertion sites after one or two cycles of incorporation and translocation. (a) The Tg·acyAMP base pair is located in the postinsertion site (n-1). (b) The F·dAMP base pair, which did not translocate, is in the insertion site (i). (c) The complex with undamaged DNA is shown for comparison. The A·dTTP base pair is in the insertion site.
Tg Bonds to, Rather Than Stacks with, the Adjacent 5′ Template Base.
The methyl group of the oxidized thymine protrudes nearly perpendicularly from the pyrimidine ring, which prevents the adjacent 5′ template guanine from stacking against it. Unexpectedly, the structure further reveals that the minor groove side of the 5′-guanine contacts the major groove side of the oxidized thymine via two hydrogen bonds (Fig. 5). The induced displacement of the guanine base is such that it is no longer in position to serve as a templating base for incorporation of the next incoming nucleotide (Fig. 5.)
Fig. 5.
Interaction of Tg with surrounding bases. Superposition of the Tg·acyAMP complex (green) with an incipient base pair (A·dTTP) from an RB69 gp43 complex with normal DNA (29) (yellow; PDB ID code 1IG9). The adenine (yellow) overlaid on the Tg·acyAMP structure (green) illustrates that the methyl group of Tg would sterically clash with the six-member ring of any purine base in the insertion site (i). The displaced 5′-guanine (green) rotates out of the way and is stabilized by two hydrogen bonds with Tg. Tg also interacts with the 3′-guanine via a water-mediated interaction. Water molecules are shown as red spheres.
Position of the β-Hairpin Loop.
A β-hairpin located in the exonuclease domain (residues 248–265 in RB69) and conserved in polymerases of the B family has been proposed to play a role in strand separation (29) and in active site switching (23, 29, 30) [supporting information (SI) Fig. 6]. In the F·dAMP structure, we observed a unique conformation of the β-hairpin loop, where it swung down toward the polymerase active site and contacted the single-stranded 5′ template (23) (Fig. 6a). The β-hairpin is seen to be in a similar position in the Tg·acyAMP complex, although the two bases at the 5′ end of the template do not contact the β-hairpin loop. G2, the guanine located 5′ to the lesion, hydrogen-bonds with the major groove side of Tg, and C1, the first base in the template strand, stacks against W574 (Fig. 6b).
Discussion
The RB69 replicase harbors two activities, primer extension in the polymerase active site and proofreading in the exonuclease active site. When the polymerase encounters a DNA lesion that slows primer extension long enough, the equilibrium between the two activities is shifted in favor of the proofreading reaction and the primer strand switches to the exonuclease active site. In the previously described F·dAMP binary complex, the asymmetric unit comprised four polymerase–DNA complexes, with two complexes holding DNA in the polymerase active site and two with DNA in the exonuclease active site (23). The F·dAMP pair generated distortions in the DNA within the confines of the polymerase active site such that the F·dAMP pair was sensed by the polymerase as a mispair. This crystal structure trapped what are likely to be intermediate steps of the primer strand switching to the exonuclease active site (23). The crystals obtained with the Tg-containing template crystallize in the same space group (P21) as the F·dAMP crystal does, with very similar cell parameters. In the Tg·acyAMP complex, however, all four polymerase–DNA complexes have DNA in the polymerase active site. The Tg lesion is intrahelical in all four complexes, stacking against the 3′ template base and base-pairing with acyAMP. An NMR structure of a duplex DNA containing Tg suggested that the Tg lesion was largely extrahelical (22), but the differences are likely to be attributable to the fact that the NMR structure was obtained in the absence of a protein. The location of the lesion also differs: it is found within the DNA duplex in the NMR model as opposed to the end of the duplex DNA in the Tg·acyAMP crystal structure. The fact that slippage is not observed at Tg sites, even with the specialized Y family polymerases (31, 32), also concurs with our observation that Tg is intrahelical within the confines of a polymerase active site.
In the structure with the abasic site analog, distortions in the DNA in the vicinity of the active site resulted in the loss of interactions with Y567 and K706, two residues that are posited to check for proper base-pairing via interactions with O2 and N3, the universal hydrogen-bond acceptors in the minor groove of DNA (33). In the event of a mispair, one or both of the bases project toward the major groove, the minor groove interactions are lost, and the DNA is destabilized, which promotes switching of the primer strand to the exonuclease active site. In the structure with Tg, the DNA maintains the minor groove interactions with Y567 and K706, and the Tg·acyAMP base pair thus appears to be a regular Watson–Crick base pair (Fig. 3b). Therefore, in all four polymerase–DNA complexes in the crystal, the DNA is found in the polymerase active site.
The other notable difference with the F·dAMP complex is that, in the Tg·acyAMP crystals, translocation took place after incorporation of A across Tg (Fig. 4). Because Tg·acyAMP is a regular Watson–Crick base pair and translocation occurred after nucleotidyl transfer what, then, is responsible for arresting the polymerase while it attempts to replicate past the Tg lesion? As mentioned earlier, Tg is not planar, because of the presence of the additional hydroxyl groups at C5 and C6. Although in silico studies predicted that the methyl group at C5 would prevent stacking with the base 5′ to the lesion (20, 21), our crystal structure allowed us to visualize the lesion within the confines of a polymerase active site. The methyl group displaces the adjacent 5′ templating guanine base in such a way that it is misaligned and too far to base-pair with an incipient dCTP (the average distance between hydrogen-bonding atoms is 7–8 Å). In addition, the 5′-guanine base is stabilized in its misplaced position by two hydrogen bonds between the NH2 and N3 of guanine on the minor groove side and the C4 and C5 hydroxyl groups of Tg on the major groove side (Fig. 5). Therefore, even though the DNA translocated after incorporation across Tg, the next incoming dNTP cannot be incorporated into the growing primer strand.
The nature of the base 5′ to Tg is known to influence lesion bypass. Replicative DNA polymerases usually are blocked at Tg sites, except in a 5′-CTgPur-3′ sequence context, which allows bypass (11, 26). A superposition of the Tg structure with an A·dTTP base pair in the insertion site (29) indicates that the methyl group would sterically clash with the six-member ring of a 5′-purine (Fig. 5). In our complex, which has a 5′-GTgG-3′ sequence for the template strand, the 5′-guanine is displaced by the methyl group in such a way that the base rotates ≈40° toward the major groove. This finding correlates with both the in vitro data showing that there is no bypass of Tg except in the 5′-CTgPur-3′ sequence context (11, 26) and our failed attempts to crystallize a ternary complex with incoming dCTP in the 5′-GTgG-3′ sequence context. Because of its smaller size, a cytosine at the 5′ template position would be less hindered by the protruding methyl group than a purine would and would be able to rotate slightly to accommodate the methyl group. This fact does not, however, explain why C 5′ to the lesion is more readily bypassed than T in the same position. A structure with each of the two pyrimidines in the 5′ slot should shed light on the differences in their propensity to allow Tg bypass.
The exonuclease domain of RB69 contains a β-hairpin structure, which was shown to participate in strand separation and active site switching (23, 29, 30). In the Tg·acyAMP complex, the β-hairpin swings down toward the polymerase active site. This conformation appears to correlate with complexes where the polymerase has encountered a templating lesion, either furan [F·dAMP complex (23)] or Tg (this complex), and incorporated a nucleotide opposite the damaged base. In the F·dAMP complex, the two 5′ terminal bases are sandwiched between the β-hairpin and F359 (SI Fig. 6). The interactions of the β-hairpin with the 5′ end of the template have been posited to effect active site switching by stabilizing the template strand in the polymerase active site while the primer strand moves to the proofreading site (23). We do not see this snug interaction of the β-hairpin with the two 5′ terminal bases of the template in the Tg complex because, after translocation, G2 binds to, and is nearly coplanar with, the Tg lesion (Fig. 5), leaving just one base, C1, to stack against W574 (SI Fig. 6).
The Tg·acyAMP complex presented here was obtained with the (5R, 6S) stereoisomer. Recent data have shown that RB69 gp43 is blocked by both cis stereoisomers of Tg, (5R, 6S) and (5S, 6R), whereas several DNA polymerases of the A family are blocked by (5R, 6S) Tg but are able to bypass the (5S, 6R) isomer (34). In the (5S, 6R) isomer, the configuration at the C5 position is such that the methyl group would protrude axially toward the adjacent 3′ base. Differences in the active site configurations of replicative polymerases might explain why the A-family polymerases can extend the primer strand despite the protruding methyl group at C5. Further understanding of the precise elements that govern blockage or bypass by Tg will require examining the active sites of replicative DNA polymerases of the A and B families, with the two cis Tg stereoisomers and in different sequence contexts.
In summary, our observations of the Tg·acyAMP complex crystal are consistent with the known effects of Tg on DNA replication, i.e., dAMP is incorporated opposite the lesion, translocation occurs after nucleotidyl transfer, but generally no extension is possible beyond the lesion. Our crystal structure showed that the methyl group of the Tg lesion displaces the 5′ template guanine, which rotates out of the way toward the major groove. The displaced guanine, stabilized by two hydrogen-bond interactions with Tg, adopts a conformation that prohibits incorporation of the next incoming nucleotide, and elongation therefore is impeded.
Materials and Methods
Polyethylene glycol 2000 monomethyl ether (PEG 2000 MME) was purchased from Hampton Research (Aliso Viejo, CA), and all other chemicals were obtained from Fisher Scientific (Waltham, MA) or Sigma (St. Louis, MO). Acyclic adenosine triphosphate was a gift from Andy Gardner (New England BioLabs, Ipswich, MA). DNA primer (5′-GCGGCTGTCATTCC-3′) and template (5′-CG-Tg-GGAATGACAGCCGCG-3′) were synthesized by Midland Certified using a (5R, 6S) phosphoramidite (Glen Research) and gel-purified. Primers used in extension assay were 5′-labeled with tetrachlorofluorescein.
Protein Expression and Purification.
The selenomethionyl variant of an exonuclease-deficient (D222A/D327A) RB69 DNA polymerase was expressed by using the methionine pathway inhibition method (35) and purified according to a previously published protocol (23).
Primer Extension Assays.
In a reaction volume of 50 μl, 1,600 nM enzyme [wild type or exonuclease-deficient (D222A/D327A)] was preincubated with 400 nM duplex DNA containing a Tg lesion in a buffer of 25 mM Tris-acetate (pH 7.5), 50 mM NaCl, 0.5 mM EDTA, and 10 mM β-mercaptoethanol. Reactions were started by addition of 50 μl of a solution containing 20 mM Mg acetate and 8 mM of each dNTP in the same buffer. The final concentrations in the reaction were 800 nM enzyme, 200 nM DNA, 10 mM Mg2+, and 4 mM dNTPs. At the indicated times, 10-μl aliquots were quenched with 10 μl of formamide. Extended primers were separated from unextended primers on 16% polyacrylamide gels containing 8 M urea. DNA bands were visualized on a Bio-Rad (Hercules, CA) Molecular Imager FX at the Alexis 532 setting to excite the tetrachlorofluorescein label.
Crystallization.
The selenomethionyl-polymerase variant (0.1 mM) was mixed in equimolar ratio with annealed primer/template DNA and 2 mM of acyATP (the selenomethionyl variant was used because we observed that it produced slightly larger crystals than its natural counterpart). Hanging drops were made by mixing 0.5 μl of reaction mixture and 0.5 μl of reservoir solution [5% (vol/vol) PEG 2000 MME, 150 mM MgSO4, 100 mM Na acetate, 100 mM Hepes (pH 7.0), 2 mM β-mercaptoethanol, and 6% (vol/vol) glycerol] and equilibrated against 1 ml of reservoir solution at 20°C. Small crystals (100 × 60 × 60 μm3) grew in ≈1 week. Crystals typically underwent three cycles of macroseeding to improve their size and diffraction quality. The resulting crystals (200 × 100 × 100 μm3) belong to space group P21 with unit cell parameters a = 132.61 Å, b = 122.63 Å, c = 168.70 Å, and β = 96.31° and contain four molecules per asymmetric unit. Crystals were cryoprotected by increasing the concentration of PEG 2000 MME and glycerol to 10% and 17%, respectively, and flash-cooled into liquid nitrogen.
Data Collection.
X-ray data were collected at beamline 23 ID-D of the Advanced Photon Source at Argonne National Laboratory by using a MAR m300 CCD detector. One complete 2.65-Å selenomethionyl data set was collected at 100 K at λ = 0.97857 Å. The data set was processed and scaled by using HKL 2000 (36).
Structure Determination and Refinement.
Our previously determined structure of a binary F·dAMP complex (23) (PDB ID code 1RV2) devoid of all nonprotein atoms was used for rigid body refinement of the Tg·acyAMP complex. The structure refinement consisted of cycles of model building with COOT (37), followed by positional refinement, simulated annealing with torsion angle molecular dynamics, and individual B factor refinement with CNS (38). The Rfree value was calculated by using 10% of the reflections that were set aside during refinement. Water molecules were added during the last rounds of refinement. Experimental single-wavelength anomalous diffraction phases calculated from the selenium sites did not seem to increase the map quality and therefore were not used. Maps, however, were substantially improved after using Maxenstein (M.A.R., unpublished program), a program that selects structural fragments from an ensemble of models generated by simulated annealing that best fit the simulated annealing omit maps. The final model includes the following protein residues: molecule A, 1–902; molecule B, 1–902, where residues 501–514 are missing; molecule C, 1–900; and molecule D, 1–897, where residues 254–260 are missing. The quality of the protein model was assessed with PROCHECK (39). All non-glycine residues are found in the allowed regions of the Ramachandran plot, with the exception of T622 (in all four molecules), a residue that has a distorted geometry because of its proximity to active site aspartates D621 and D623 (23). The coordinates for cis (5R, 6S) Tg were obtained from ref. 19. Statistics for data collection and refinement are shown in Table 1. Polymerase structures were superimposed by using the Cα of residues of the palm subdomain (residues 383–468 and 573–729). Figs. 3–6 were drawn with PyMOL (DeLano Scientific, San Carlos, CA).
Table 1.
Data collection and refinement statistics
| Statistic | Tg·acyAMP complex* |
|---|---|
| Data collection | |
| Space group | P21 |
| Cell dimensions | |
| a, b, c, Å | 132.61, 122.63, 168.70 |
| α, β, γ, ° | 90.0, 96.31, 90.0 |
| Resolution, Å | 50–2.65 (2.74–2.65)† |
| Rmerge, % | 11.2 (47.9)† |
| I/σI | 11.0 (2.1)† |
| Completeness, % | 97.7 (84.6)† |
| Redundancy | 4.5 (2.8)† |
| Refinement | |
| Resolution, Å | 50–2.65 |
| No. of reflections | 680,936 (155,892)‡ |
| Rwork/Rfree, % | 22.82/28.18 |
| No. of atoms | 31,942 |
| Protein | 28,693 |
| DNA | 2,637 |
| Water | 612 |
| B factors, Å2 | |
| Protein (molecule A, B, C, D) | 58.5, 51.2, 52.6, 101.2 |
| DNA (molecule A, B, C, D) | 113.0, 54.6, 64.7, 89.2 |
| Water (all molecules) | 43.4 |
| rms deviations | |
| Bond lengths, Å | 0.0069 |
| Bond angles, ° | 1.290 |
*Data collected from a single crystal.
†Data for highest-resolution shell is shown in parentheses.
‡Number of all reflections (number of unique reflections is shown in parentheses).
Supplementary Material
Acknowledgments
We thank Stephen Corcoran, Dr. Ward Smith, and Dr. Ruslan Sanishvili for help with data collection at the General Medicine and Cancer Institutes Collaborative Access Team (GM/CA-CAT) beamline at Advanced Photon Source at Argonne National Laboratory. GM/CA-CAT has been funded in whole or in part with federal funds from National Cancer Institute Grant Y1-CO-1020 and National Institute of General Medical Science Grant Y1-GM-1104. This work was supported by National Institutes of Health Grant CA52040 (to S.S.W.).
Abbreviations
- Tg
thymine glycol
- acyAMP
acyclic AMP
- acyATP
acyclic ATP.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2DY4).
This article contains supporting information online at www.pnas.org/cgi/content/full/0606648104/DC1.
References
- 1.Cathcart R, Schwiers E, Saul RL, Ames BN. Proc Natl Acad Sci USA. 1984;81:5633–5637. doi: 10.1073/pnas.81.18.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saul RL, Ames BN. In: Mechanisms of DNA Damage and Repair. Simic MG, Grossman L, Upton AC, editors. New York: Plenum; 1986. pp. 529–535. [Google Scholar]
- 3.Adelman R, Saul RL, Ames BN. Proc Natl Acad Sci USA. 1988;85:2706–2708. doi: 10.1073/pnas.85.8.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frenkel K, Goldstein MS, Teebor GW. Biochemistry. 1981;20:7566–7571. doi: 10.1021/bi00529a035. [DOI] [PubMed] [Google Scholar]
- 5.Breimer LH, Lindahl T. Biochemistry. 1985;24:4018–4022. doi: 10.1021/bi00336a032. [DOI] [PubMed] [Google Scholar]
- 6.Aspinwall R, Rothwell DG, Roldan-Arjona T, Anselmino C, Ward CJ, Cheadle JP, Sampson JR, Lindahl T, Harris PC, Hickson ID. Proc Natl Acad Sci USA. 1997;94:109–114. doi: 10.1073/pnas.94.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bandaru V, Sunkara S, Wallace SS, Bond JP. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 8.Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Proc Natl Acad Sci USA. 2002;99:3523–3528. doi: 10.1073/pnas.062053799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayes RC, Petrullo LA, Huang HM, Wallace SS, LeClerc JE. J Mol Biol. 1988;201:239–246. doi: 10.1016/0022-2836(88)90135-0. [DOI] [PubMed] [Google Scholar]
- 10.Clark JM, Beardsley GP. Nucleic Acids Res. 1986;14:737–749. doi: 10.1093/nar/14.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayes RC, LeClerc JE. Nucleic Acids Res. 1986;14:1045–1061. doi: 10.1093/nar/14.2.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ide H, Kow YW, Wallace SS. Nucleic Acids Res. 1985;13:8035–8052. doi: 10.1093/nar/13.22.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNulty JM, Jerkovic B, Bolton PH, Basu AK. Chem Res Toxicol. 1998;11:666–673. doi: 10.1021/tx970225w. [DOI] [PubMed] [Google Scholar]
- 14.Sagher D, Strauss B. Nucleic Acids Res. 1985;13:4285–4298. doi: 10.1093/nar/13.12.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laspia MF, Wallace SS. J Bacteriol. 1988;170:3359–3366. doi: 10.1128/jb.170.8.3359-3366.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kow YW, Faundez G, Melamede RJ, Wallace SS. Radiat Res. 1991;126:357–366. [PubMed] [Google Scholar]
- 17.Achey PM, Wright CF. Radiat Res. 1983;93:609–612. [PubMed] [Google Scholar]
- 18.Moran E, Wallace SS. Mutat Res. 1985;146:229–241. doi: 10.1016/0167-8817(85)90063-x. [DOI] [PubMed] [Google Scholar]
- 19.Flippen JL. Acta Crystallogr B. 1973;29:1756–1762. [Google Scholar]
- 20.Clark JM, Pattabiraman N, Jarvis W, Beardsley GP. Biochemistry. 1987;26:5404–5409. doi: 10.1021/bi00391a028. [DOI] [PubMed] [Google Scholar]
- 21.Miaskiewicz K, Miller J, Ornstein R, Osman R. Biopolymers. 1995;35:113–124. doi: 10.1002/bip.360350112. [DOI] [PubMed] [Google Scholar]
- 22.Kung HC, Bolton PH. J Biol Chem. 1997;272:9227–9236. doi: 10.1074/jbc.272.14.9227. [DOI] [PubMed] [Google Scholar]
- 23.Hogg M, Wallace SS, Doublié S. EMBO J. 2004;23:1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iwai S. Angew Chem Int Ed. 2000;39:3874–3876. doi: 10.1002/1521-3773(20001103)39:21<3874::AID-ANIE3874>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 25.Shimizu T, Manabe K, Yoshikawa S, Kawasaki Y, Iwai S. Nucleic Acids Res. 2006;34:313–321. doi: 10.1093/nar/gkj443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark JM, Beardsley GP. Biochemistry. 1989;28:775–779. doi: 10.1021/bi00428a054. [DOI] [PubMed] [Google Scholar]
- 27.Evans J, Maccabee M, Hatahet Z, Courcelle J, Bockrath R, Ide H, Wallace S. Mutat Res. 1993;299:147–156. doi: 10.1016/0165-1218(93)90092-r. [DOI] [PubMed] [Google Scholar]
- 28.Gardner AF, Jack WE. Nucleic Acids Res. 2002;30:605–613. doi: 10.1093/nar/30.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Franklin MC, Wang J, Steitz TA. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 30.Reha-Krantz LJ, Marquez LA, Elisseeva E, Baker RP, Bloom LB, Dunford HB, Goodman MF. J Biol Chem. 1998;273:22969–22976. doi: 10.1074/jbc.273.36.22969. [DOI] [PubMed] [Google Scholar]
- 31.Fischhaber PL, Gerlach VL, Feaver WJ, Hatahet Z, Wallace SS, Friedberg EC. J Biol Chem. 2002;277:37604–37611. doi: 10.1074/jbc.M206027200. [DOI] [PubMed] [Google Scholar]
- 32.Kusumoto R, Masutani C, Iwai S, Hanaoka F. Biochemistry. 2002;41:6090–6099. doi: 10.1021/bi025549k. [DOI] [PubMed] [Google Scholar]
- 33.Seeman NC, Rosenberg JM, Rich A. Proc Natl Acad Sci USA. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takata K, Shimizu T, Iwai S, Wood RD. J Biol Chem. 2006;281:23445–23455. doi: 10.1074/jbc.M604317200. [DOI] [PubMed] [Google Scholar]
- 35.Doublié S. Methods Enzymol. 1997;276:523–530. [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.