Abstract
The navigation of axons toward their targets is a highly dynamic and precisely regulated process during nervous system development. The molecular basis of this navigation process is only partly understood. In Caenorhabditis elegans, we isolated the RNAi-hypersensitive strain nre-1(hd20) lin-15b(hd126), which allows us to phenocopy axon guidance defects of known genes by feeding RNAi. We used this mutant strain to systematically screen 4,577 genes on chromosomes I and III for axon guidance phenotypes. We identified 93 genes whose down-regulation led to penetrant ventral cord fasciculation defects or motoneuron commissure outgrowth defects. These genes encode various classes of proteins, ranging from secreted or putative cell surface proteins to transcription factors controlling gene expression. A majority of the genes is evolutionary conserved and previously uncharacterized. In addition, we found axon guidance functions for known genes like pry-1, a component of the Wnt-signaling pathway, and ced-1, a receptor required for the engulfment of neurons undergoing apoptosis during development. Our screen provides insights into molecular pathways operating during the generation of neuronal circuits and provides a basis for a more detailed analysis of gene networks regulating axon navigation.
Keywords: Wnt, neuron, development, axon navigation
Directed outgrowth of neuronal processes like axons and dendrites reflects a complex navigational problem because of the large number of neurons involved in this process. Several conserved signaling systems have been described that influence the direction of outgrowth of navigating axons (for reviews, see refs. 1–3). Despite this progress in the identification of axon guidance signals, their small number does not match the complexity of the guidance process and the large number of neurons involved. The rather limited defects in mutants of known guidance cues indicate that a substantial fraction of axon guidance genes remains to be identified.
The nematode Caenorhabditis elegans is a suitable model for such a purpose, not only because its nervous system is simple and well described, but also because large-scale screens can be performed fast and easily. C. elegans has a comparatively small nervous system, with exactly 302 neurons in the adult hermaphrodite (4, 5). Axons and dendrites are typically unbranched and grow out in a stereotypic fashion. Because it is now possible to label neurons in vivo using fluorescent proteins like GFP (6, 7), large-scale direct visual screens for axon navigation defects have become feasible. In the last few years, RNAi in C. elegans (8) has become a powerful method to identify genes controlling particular biological processes, such as longevity, fat regulation, genome stability, RNAi, or transposon silencing (9). Unfortunately, RNAi, by feeding, does not function efficiently in the nervous system (10), effectively preventing use of this tool for the molecular analysis of axon guidance. Here we report the isolation of a strain in C. elegans that shows enhanced RNAi in the nervous system and the use of this RNAi hypersensitive strain for identification of genes regulating axon guidance.
Results
Isolation of an RNAi-Sensitive C. elegans Strain.
To identify RNAi-hypersensitive mutants suitable for analysis of axon navigation, we performed a genetic screen with a transgenic C. elegans strain expressing GFP at moderate levels in the entire nervous system (unc-119::GFP). In a WT background, this strain showed no reduction of GFP expression in neurons after feeding of bacteria expressing GFP dsRNA (= feeding RNAi) (Fig. 1c). We screened for mutants with decreased neuronal GFP expression after feeding RNAi against GFP and identified a mutant we named nre-1(hd20) for neuronal RNAi efficient. In feeding RNAi experiments with nre-1(hd20), typically only a small and variable number of neurons continued to express GFP at low levels (Fig. 1d). Nre-1(hd20) mutant animals are further characterized by a reduced number of progeny at 20°C (159 ± 18, n = 4) compared with WT (270 ± 18, n = 4). At 25°C, brood size is reduced even further to 7 ± 2 (n = 10) compared with 169 ± 10 in WT (n = 4). Nre-1(hd20) was mapped to the right arm of the X chromosome to a region containing the lin-15b gene, which was recently described to be RNAi hypersensitive (11). Sequencing of the lin-15b locus in our hypersensitive strain revealed a G to A transition in the splice acceptor site of the fifth intron (position 2463 of cosmid ZK678; GenBank accession no. Z79605). Destruction of this splice acceptor most likely leads to splicing out of the sixth exon, causing a frame shift. This effectively truncates the protein and eliminates the DNA-binding domains, suggesting that our strain contains a strong loss-of-function mutation in lin-15b. Our strain, however, has additional phenotypes, such as the sterility at 25°C, that are not shared by lin-15 alleles. In fact, lin-15ab(n765ts) complements this sterility phenotype of nre-1(hd20). We therefore conclude that our RNAi-hypersensitive strain contains two closely linked mutations, one of them in lin-15b and a second in a nearby gene. All of the phenotypes cosegregated in all genetic experiments, and we were never able to separate the different phenotypes in our strain. Most importantly, both mutations contribute to RNAi hypersensitivity, because our nre-1(hd20) lin-15b(hd126) strain is more effective than lin-15b(n744), particularly in RNAi experiments targeting neuronally expressed genes (Table 1). Further, comparison with other RNAi-hypersensitive strains like rrf-3, eri-1, and an eri-1; lin-15b double mutant indicates that our nre-1(hd20) lin-15b(hd126) strain is the only one where known axon guidance genes can be phenocopied with high penetrance (Table 1; RNAi directed against unc-40 or unc-73), making it the strain of choice for an RNAi screen for axon guidance genes.
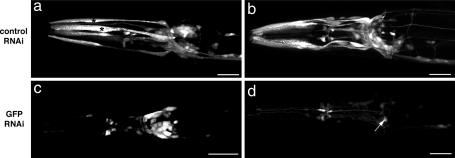
Fig. 1.
Effectiveness of RNAi in neurons in nre-1(hd20) lin-15b(hd126) mutants. (a) unc-119::GFP is expressed in nonneuronal cells (asterisks) and neurons. Feeding of an empty RNAi vector does not affect GFP expression. (c) WT animals fed with a GFP RNAi vector (L4417): GFP expression is decreased in nonneuronal cells but not in neurons. (b) Feeding the control vector to nre-1(hd20) lin-15b(hd126) mutant animals does not affect GFP expression. (d) Feeding the GFP RNAi vector to nre-1(hd20) lin-15b(hd126) mutant animals leads to down-regulation of GFP expression in all cells. Only few neurons are left that weakly express GFP (arrow). Anterior to the left. (Scale bars, 20 μm.)
Table 1.
Comparison of RNAi-hypersensitive strains
| RNAi against* | Percentage of animals with phenotype in |
|||||
|---|---|---|---|---|---|---|
| WT | rrf-3 | eri-1 | lin-15b† | lin-15b; eri-1† | lin-15b nre-1‡ | |
| GFP | 0 ± 0 | 0 ± 0 | 1 ± 1 | 74 ± 7 | 97 ± 3 | 98 ± 3§¶ |
| unc-40 | 0 ± 0 | 0 ± 0 | 5 ± 4 | 9 ± 2 | 29 ± 1 | 73 ± 15§¶ |
| unc-73 | 14 ± 5 | 57 ± 10 | 10 ± 2 | 15 ± 3 | 35 ± 4 | 77 ± 13§¶ |
| dpy-13‖ | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 | 100 ± 0 |
| daf-2 | 7 ± 3 | 6 ± 2 | 8 ± 4 | 3 ± 3 | 18 ± 6 | 31 ± 10§¶ |
| lin-1 | 7 ± 9 | 27 ± 15 | 39 ± 15 | 68 ± 11 | 87 ± 5 | 77 ± 12§¶ |
Numbers are mean ± SD for three independent experiments (n = 56–100 for each experiment with lin-1, daf-2, dpy-13, and GFP; n = 28–65 for unc-40 and unc-73).
*Phenotypes scored: GFP expression in neurons scored with rhls13 [unc-119::GFP] (GFP); ventral cord defasciculation and commissure defects scored with hdls17 (unc-40, unc-73); Dpy (dpy-13); dauer formation (daf-2); multivulva (lin-1).
†lin-15b (n744).
‡lin-15b (hd126).
§Significant with P < 0.01 compared with WT (unpaired t test).
¶Significant with P < 0.01 compared with lin-15b (n744) (unpaired t test).
‖ Strains show different severity of dumpy phenotype: wt < nre-1 lin-15b/lin-15b < rrf-3/eri-1/lin-15b; eri-1.
Large-Scale RNAi Screens for Axon Guidance Genes.
We used nre-1(hd20) lin-15b(hd126) in a large-scale screen for axon guidance genes on chromosomes I and III. A fluorescent reporter was used to label interneurons of the motor circuit, as well as several classes of motoneurons (DA, DB, DD, and VD). This allowed analysis of the majority of axons in the ventral and dorsal cord, as well as outgrowth and navigation of motoneuron commissures that connect ventral and dorsal cord (Fig. 2a and e). Feeding of 4,577 RNAi clones led to the identification of 93 genes with reproducible axon guidance phenotypes. Most prominent were defects in motoneuron commissure navigation, where commissures fail to reach the dorsal cord and extend in various lateral positions instead. Frequently, this was combined with abnormal branching and dorsal cord defaciculation [Table 2(Com); Fig. 2 f–h]. Another common defect was abnormal midline crossing of ventral cord axons [Table 2 (VNC); Fig. 2 b and d]. Seventy-six different RNAi clones led to defects where motor- and/or interneuron axons crossed over into the left ventral cord axon tract either once or multiple times (Table 2). In one case, we found gaps in the dorsal cord or more rarely in the ventral cord (Table 2; Fig. 2c), indicating a failure of the outgrowth process per se rather than a navigation error. To analyze the defects in more detail, we labeled subsets of neurons with different fluorescent proteins, namely the excitatory DA/DB type motoneurons with cyan fluorescent protein and interneurons with DsRed and DD/VD motoneurons with yellow fluorescent protein (YFP). The majority of commissure as well as ventral cord defects are found in DD/VD motoneurons (Table 2). Ventral cord crossover defects in DD/VD motoneuron axons are found in 42 of the 93 RNAi clones, with 25 also sharing interneuron axon crossover defects and 8 both interneuron and DA/DB motoneuron crossover defects. Only three clones led to defects exclusively in interneurons, and no clone was found to interfere only with DA/DB axon guidance in the ventral cord.
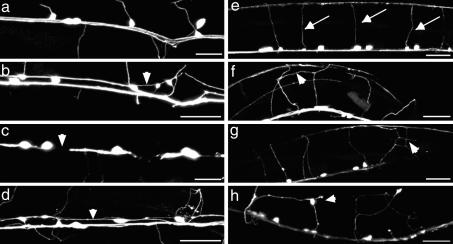
Fig. 2.
Representative axon guidance phenotypes. (b, c, and f) Phenotypes generated by RNAi. (g and h) Phenotypes of genetic mutants. (a) WT: all GFP labeled axons run tightly fasciculated in the right tract. (b) Cdh-4(RNAi): axons cross over into the left axon bundle (arrowhead). (c) Unc-101(RNAi): gaps (arrowhead) indicate a premature stop of axon outgrowth. (d) Pry-1(mu38): ventral cord midline crossing defects (arrowhead). (Scale bar, 20 μm.) Ventral view, anterior to the left. (e) WT: commissures grow circumferentially to the dorsal cord (arrows). (f) F10G8.4(RNAi): premature branching of commissures (arrowheads). (g) ced-1(e1735): abnormal branching of commissures (arrow). (h) Unc-101(m1): lateral extending axons (arrowhead). (Scale bar, 20 μm.) Lateral view, ventral side down.
Table 2.
RNAi clones leading to axon guidance phenotypes
| Gene (Chr) | Size, aa | Description | Conserved* | Com† | Com type | VNC‡ | Axon type | DNC§ | PVQ¶ | Other phenotype‖ |
|---|---|---|---|---|---|---|---|---|---|---|
| Secreted or cell surface (12 genes) | ||||||||||
| unc-40 (I) | 1,415 | Netrin receptor, DCC homolog | Yes | +++ | D | +++ | I | ++ | − | Gro, Unc, Eg |
| unc-71 (III) | 1,042 | ADAM-type metalloprotease | Yes | ++ | D | + | D | +++ | − | |
| ced-1 (I) | 1,111** | EGF domain | Yes | + | D | ++ | D | − | − | |
| B0041.5 (I) | 429 | RhaT domain | Yes(d) | ++ | D | ++ | ++ | ++ | ||
| nas-5 (I) | 360 | Astacin metalloprotease | Yes | ++ | D | +++ | ++ | − | ||
| C24G7.1 (I) | 608 | Sodium channel | Yes | ++ | D | ++ | D | ++ | − | |
| F44E2.4 (III) | 1,283 | LDL receptor domain | No | +++ | A,D | − | − | − | ||
| grl-22 (III) | 162 | Groundhog related | No | ++ | D | + | − | − | ||
| cdh-4 (III) | 4,307 | FAT-like Cadherin | Yes | − | ++ | D,I | + | − | ||
| C09D4.1 (I) | 586** | AraJ domain | Yes | + | D | ++ | def | + | − | Unc |
| T23B3.4 (I) | 395 | Sulfakinin receptor | Yes | + | D | ++ | D,I | + | − | |
| D1044.2† (III) | 1,090 | Nidogen N terminus | Yes(d) | + | D | + | − | − | ||
| Signaling (12 genes) | ||||||||||
| pry-1 (I) | 586 | Axin homolog, Wnt signaling | Yes | ++ | D | ++ | D | + | − | Gro, Unc, Lvr |
| unc-73 (I) | 2,488** | GNEF, Trio homolog | Yes | +++ | D | ++ | A,D,I | ++ | − | Gro, Unc, Egl |
| unc-13 (I) | 2,155** | C2, DUF1041 domains | Yes | + | D | + | D,I | − | ++ | Gro |
| unc-101 (I) | 422 | Clathrin adaptor protein AP1 | Yes | + | D | ++ | − | − | ||
| ZC581.9 (I) | 827 | S/T kinase | Yes | ++ | ++++ | + | − | |||
| F10G8.4 (I) | 395 | Tyrosine phosphatase | Yes(d) | +++ | D | +++ | D,I | ++ | − | Gro |
| smd-1 (I) | 368 | SAM decarboxylase | Yes | ++ | D | +++ | D,I | + | − | |
| F47B3.1 (I) | 363 | Tyrosine phosphatase | Yes | ++ | D | ++ | D,I | ++ | − | |
| pde-6 (I) (Y95B8A.10) | 760 | Phosphodiesterase | Yes | ++ | D | ++ | A,D,I | − | + | |
| F26E4.5 (I) | 474 | SH2 domain, tyrosine kinase | Yes | + | D | ++ | D,I | − | − | |
| ZC155.7 (III) | 329 | Syntaxin | Yes | ++ | D | + | − | − | ||
| Y52D3.1 (III) | 388 | S/T kinase | Yes (d) | + | D | + | − | − | ||
| Cytoskeleton (6 genes) | ||||||||||
| unc-70# (V) | 2,302** | β-Spectrin | Yes | ++++ | D | − | + | ++ | Jnc, Gro | |
| nmy-2 (I) | 2,003 | Myosin heavy chain | Yes | ++ | D | +++ | def | − | − | Gro |
| unc-95 (I) | 350 | LIM domain adaptor protein | No | +++ | D | +++ | D,I | ++ | − | Unc, Egl |
| lem-3 (I) | 732 | Ankyrin repeat, LEM domain | Yes (d) | ++ | D | +++ | D,I | ++ | − | |
| hum-5† (III) | 1,017** | Myosin heavy chain | Yes | ++ | D | − | − | − | ||
| unc-116 (III) | 815 | Kinesin heavy chain | Yes | ++ | D | − | + | − | ||
| Transcription, splicing, protein modification/degradation (15 genes) | ||||||||||
| lin-59 (I) | 1,312 | ASH1/trithorax homolog | Yes (d) | +++ | D | +++ | ++ | − | Gro, Egl | |
| sur-2 (I) | 1,589** | CRSP3 homolog | Yes | ++ | D | +++ | A,D,I | ++ | − | |
| ZK1128.5 (III) | 446 | Swp73/BAF60 homolog | Yes | ++++ | D | ++ | I | +++ | − | |
| rnp-6 (I) | 749** | HALF-PINT, (FIR)/PUF6 homolog | Yes | ++ | D | +++ | D | ++ | ++ | Gro, Unc, Egl |
| hmg-3 (I) | 689 | SSRP1 homolog | Yes | +++ | D | +++ | D,I | ++ | − | Gro, Unc |
| tbx-9 (III) | 292 | T-box transcription factor | Yes | +++ | A,D | − | +++ | − | ||
| F25D7.1 (I) | 245 | Derlin-1 homolog | Yes | + | D | ++++ | − | − | ||
| lin-39 (III) | 253 | HoxD4/Dfd homolog | Yes | ++++ | D | − | − | ++ | ||
| cnd-1 (III) | 192 | NeuroD homolog | Yes | +++ | D | − | + | − | Egl | |
| Y95B8A.7 (I) | 608 | zfr1 homolog | Yes | ++ | D | ++ | D,I | ++ | − | |
| wwp-1 (I) (Y65B4BR.4) | 794** | WWP1 homolog | Yes | ++ | D | ++ | D,I | ++ | − | Unc |
| ZC328.2 (I) | 386 | Zinc finger TF | Yes(d) | + | D | ++ | − | − | ||
| ceh-13 (III) | 202 | Hox1/labial homolog | Yes | + | D | ++ | A,D,I | + | ++ | |
| dpy-27 (III) | 1,469 | Condensin complex member | Yes | ++ | A,D | − | − | − | Dpy | |
| hse-5 (III) | 616 | HS-C5-epimerase | Yes | + | D | − | − | − | ||
| Cell metabolism (17 genes) | ||||||||||
| cua-1 (III) | 1,238 | Copper transporter | Yes | ++++ | A,D | ++++ | def | ++++ | − | Unc, gro, Lvr |
| Y56A3A.21 (III) | 159 | TRAP-delta homolog | Yes | ++++ | D | ++++ | ++ | − | Unc, gro, | |
| Y105E8B.9 (I) | 628 | β-Glucuronidase | Yes | ++ | D | +++ | ++ | ++ | Gro | |
| T05E8.3 (I) | 856 | DEAH-box RNA helicase | Yes | + | +++ | D,I | ++ | − | Gro, Unc | |
| Y105E8A.23 (I) | 944 | RNA polymerase | Yes | + | D | +++ | def | ++ | − | Gro |
| T21G5.5 (I) | 445** | KH1 domain (RNA binding) | Yes | ++ | D | +++ | ++ | + | Unc, Egl | |
| mdt-18 (I) (C55B7.9) | 232 | MED18 homolog | Yes | ++ | D | ++ | D,I | + | ++ | Gro, Unc |
| lpd-3 (I) | 1,599 | Lipid metabolism | Yes | ++ | D | ++ | ++ | − | ||
| rpl-17 (I) | 187** | Ribosomal L17 protein | Yes | ++ | D | ++ | D,I | + | − | Gro, Unc |
| F43G9.3 (I) | 294 | Mitochondrial carrier protein | Yes | ++ | D | +++ | − | − | ||
| E03H4.1 (I) | – | Transposase | - | ++ | D | ++ | A,D,I | + | − | |
| C41D11.2 (I) | 365 | Translation initiation factor 3 | Yes | ++ | D | ++ | def | − | − | |
| mdt-21 (III) | 132 | Peptidase | Yes | ++ | D | + | A,D,I | + | − | Unc, Gro |
| lbp-5 (I) | 136 | H-FABP homolog | Yes | ++ | D | ++ | D | − | − | |
| W03D8.8 (I) | 430 | Acyl-CoA thioesterase | Yes | + | D | ++ | D | + | − | |
| dhs-9 (III) | 319 | Short-chain dehydrogenase | Yes | ++ | D | + | D,I | − | − | |
| Y37D8A.7 (III) | − | Transposase | − | ++ | D | − | − | − | ||
| Unknown function (31 genes) | ||||||||||
| Y106G6H.8 (I) | 112 | DUF423, TM | Yes | +++ | D | +++ | ++ | − | Gro | |
| Y63D3A.9 (I) | 277 | F-box protein | No | ++ | D | +++ | D,I | ++ | − | |
| T24D1.3 (I) | 349 | RING finger | No | ++ | D | ++ | ++ | + | ||
| T07A5.2 (III) | 301 | UNC50 domain | Yes | +++ | D | − | + | − | Unc, Lev | |
| pqn-20 (I) | 1,239 | Poly-Q-containing protein | Yes(d) | + | D | ++ | + | − | ||
| C38C10.3# (III) | 301 | DUF508 | No | ++ | D | − | − | − | − | |
| D2007.2 (III) | 195 | MSP domain | No | ++ | D | + | − | − | ||
| T28A8.3 (III) | 621 | SPK domain | No | + | D | + | D,I | − | ++ | |
| F59A2.6 (III) | 1,133 | GRIP domain | Yes | + | D | − | − | − | ||
| Y47G6A.29 (I) | 2460 | No known domains | Yes(d) | ++ | D | +++ | A,D,I | ++ | − | |
| F46F11.9 (I) | 1,282** | No known domains | Yes(d) | ++ | D | ++ | +++ | − | Gro, Unc, Egl | |
| C40H1.3 (III) | 504 | No known domains | Yes(d) | + | D | − | − | − | ||
| K04H4.2 (III) | 967** | No known domains | Yes(d) | + | D | − | − | − | Gro | |
| Y34D9A.3 (I) | 639 | No known domains | No | +++ | D | +++ | D,I | ++ | − | Unc |
| Y48G1C.8 (I) | 828 | No known domains | No | + | D | +++ | def | +++ | − | Gro, Unc |
| F29D11.2 (I) | 1,153 | No known domains | No | ++ | A,D | +++ | A,D,I | ++ | − | Unc, Dpy |
| F26B1.1 (I) | 304 | No known domains | No | ++ | D | +++ | D | ++ | − | |
| C17F3.1 (I) | 88 | No known domains | No | +++ | D | ++ | D,I | ++ | ||
| Y55D5A. 1 (III) | 285** | No known domains | No | ++ | D | ++ | +++ | − | Egl | |
| F48C1.4 (I) | 104 | No known domains | No | ++ | D | ++ | D | + | − | Dpy, Lvr |
| Y106G6A.5 (I) | 210 | No known domains | No | ++ | D | +++ | D,I | − | − | |
| Y43F4B. 3 (III) | 216 | No known domains | No | +++ | D | + | − | − | ||
| Y106G6A.2 (I) | 428 | No known domains | No | + | D | +++ | D,I | − | − | |
| ZK973.8 (I) | 392 | No known domains | No | + | D | +++ | def | − | − | |
| Y18D10A.21 (I) | 286 | No known domains | No | − | +++ | − | − | |||
| Y37H9A.1 (I) | 514** | No known domains | No | + | D | ++ | I | − | − | |
| T15D6.9 (I) | 407 | No known domains | No | + | D | ++ | D,I | ++ | − | |
| Y106G6A. 4 (I) | 175 | No known domains | No | ++ | D | ++ | D,I | + | − | |
| F54D8.6 (III) | 783 | No known domains | No | + | D | − | + | − | ||
| T04C9.2 (III) | 116 | No known domains | No | + | D | + | − | − | ||
| F54H12.2 (III) | 419 | No known domains | No | + | D | − | D,I | − | − | |
Chr, chromosome; Com, commissure.
*Yes indicates BLAST scores higher than 1e-10 for >70% of the protein length. Yes(d) indicates BLAST scores higher than 1e-10 for >30% but <70% of the protein length. No indicates BLAST scores lower than 1e-10 and/or <30% protein length. Similar or homologous proteins found in other animals (BLAST search results evaluated).
†More than two commissures not reaching the dorsal cord (1% defects in WT); Comm type A, DA/DB motoneuron commissures have defects; Comm type D, DD/VD motoneuron commissures are affected. RNAi clone contained wrong insert (see Materials and Methods).
‡Midline crossing defects in the ventral nerve cord (VNC) (6% defects in WT); axon type A, DA/DB motoneuron axons in the ventral cord are affected; Axon type D, DD/VD motoneuron axons; axon type I, interneuron axons; def, overall defasciculation of the ventral cord.
§Dorsal nerve cord (DNC) defasciculation (6% defects in WT).
¶PVPR/PVQL guidance defects (midline crossing or outgrowth in the right axon tract 4% defects in WT). +, Defects <25% but significantly higher than in WT (P < 0.01, χ2 test). ++, 25–49% animals with defects. +++, 50–74% animals with defects. ++++, 75–100% animals with defects.
‖Additional phenotypes observed: Dpy, dumpy; Egl, egg-laying defect; Gro, growth defect; Lev, levamisole resistant; Lvr, larval arrest; Unc, uncoordinated movement.
**Longest splice variant.
To determine whether the identified genes govern axon guidance in other neuron populations, we repeated the RNAi feeding experiments with the positive clones isolated in the first screen and scored PVP and PVQ axons in the ventral cord using a different reporter strain. A total of 12 genes showed defects in PVPR and PVQL axon guidance with a penetrance ranging from 20% to 43% (Table 2, PVQ).
The axon guidance genes identified fall into distinct functional categories. A number of genes encode putative secreted or cell surface proteins like cdh-4, a member of the cadherin family of adhesion molecules (Table 2, Secreted or cell surface). Cdh-4(RNAi) leads to midline crossing defects within the ventral cord and defasciculation of the dorsal cord, suggesting that cdh-4 plays a role in the adhesion of axons extending together in the same axon tract. Ced-1(RNAi) also leads to midline crossing and commissure navigation defects. The second category of genes contains signaling molecules that may connect signal–receptor complexes to effectors acting on the cytoskeleton (Table 2, Signaling). RNAi against pry-1, a component of the Wnt-signaling pathway, leads to defects in dorsal and ventral cord fasciculation and commissure guidance defects. RNAi against unc-101, a clathrin adapter protein implicated in trafficking of olfactory receptors (12), leads to gaps in the ventral cord and commissure guidance defects. Other genes in this category include kinases and phosphatases, which currently cannot be placed in any of the known signaling pathways. A smaller group (Table 2, Cytoskeleton) consists of proteins directly associated with the cytoskeleton, like myosin and kinesin family members as well as unc-70/spectrin. Another group of genes encodes transcription factors and regulators of protein degradation (Table 2, Transcription, splicing, protein modification/degradation). Among them is cnd-1, a NeuroD homolog that determines motoneuron cell fate as well as aspects of terminal differentiation like directed axon outgrowth (13) and homeobox and T-box containing transcription factors. Targeted protein degradation after ubiquitinylation is another way of controlling protein activity. We identified wwp-1, an E3 ubiquitin ligase, leading to defects in all analyzed axon trajectories. As in previous RNAi screens, a number of genes involved in general cellular metabolic processes were identified as well (Table 2, Cell metabolism). Here the axon guidance phenotypes most likely are a secondary consequence of a disturbed cell metabolism. In many cases, these RNAi clones produced additional phenotypes like slow growth or even larval or embryonic arrest, indicating that this category includes genes with a lethal phenotype that might not be completely penetrant in the RNAi experiment. Finally, a large fraction of the genes encodes proteins with no recognizable domains, making a functional classification difficult (Table 2, Unknown function). Several are conserved genes like Y106G6H.8, which encodes a small membrane protein, possibly a member of a novel receptor family involved in axon guidance.
Pry-1, ced-1, and unc-101 Mutants Show Axon Guidance Defects.
Genetic mutations are not available for most of the genes identified in our screen. However, we were able to test the axon guidance function of several genes identified in the screen by analyzing existing genetic mutants. In lin-39 mutant animals, we could not observe any axon guidance defect, indicating that our collection contains some “false positives.” By contrast, pry-1, ced-1, and unc-101 mutants show axon outgrowth and/or navigation defects like the corresponding RNAi experiments (Fig. 2 d, g–i and Table 3). Pry-1(mu38) mutants showed the strongest defects, with branched commissures and ventral cord midline crossing defects (Fig. 2d). Ced-1(e1735) mutants exhibit only commissure defects (Fig. 2g), and unc-101(m1) mutants are characterized by branched lateral outgrowing commissures and prominent gaps, predominantly in the dorsal cord (Fig. 2 h and i). This shows that the RNAi phenotypes can be confirmed by the analysis of genetic mutants, which are more suitable for a detailed study of gene function.
Table 3.
Axon guidance defects in unc-101, ced-1, and pry-1 mutants
| Genotype | Com* | VNC† | DNC‡ | n |
|---|---|---|---|---|
| WT | 6 | 2 | 0 | 111 |
| unc-101 (m1) | 26§ | 8 | 30§ | 106 |
| ced-1 (e1735) | 23§ | 6 | 4 | 129 |
| pry-1 (mu38) | 66§ | 42* | 48§ | 65 |
Animals were examined with an extrachromosomal transgene hdEx191[glr-1::YFP, unc-129::YFP, unc-47::YFP, rol-6(su1006)] labeling motor- and interneurons.
*More than one commissure not reaching the dorsal cord.
†Midline crossing defects in the ventral nerve cord (VNC).
‡Dorsal nerve cord (DNC) defasciculation or gaps in the dorsal cord (unc-101 only).
§Significant with P < 0.01 (χ2 test). Com, commissure.
Discussion
Nre-1(hd20) lin-15b(hd126) Is a Potent RNAi-Sensitive Mutant Combination.
Mutations affecting the RNAi response of cells have been identified. Mutations in genes abolishing the RNAi response have been instrumental in the identification and characterization of the molecular machinery mediating the effects of dsRNA in cells (14, 15). Delivering the dsRNA by feeding (the method of choice for high-throughput screens) does not work efficiently for neuronal genes. Mutations with increased sensitivity to dsRNA, like rrf-3 (16) or eri-1 (17), have been isolated but have not really solved this problem. Recently, it was shown that mutations in retinoblastoma pathway components, like lin-15b or lin-35/Rb, can enhance RNAi (11, 18). This pathway seems to act in parallel to the eri-1/rrf-3 pathway, because a combination of mutations in those pathways is even more RNAi-hypersensitive than the single mutants (11, 18). A combination of eri-1 and lin-15b was recently used successfully in an RNAi screen to identify genes affecting synaptic transmission (19). Although the eri-1;lin-15b mutant combination also allows to phenocopy axon guidance defects to a certain extent, we find that the nre-1(hd20) lin-15(hd126) mutant combination described here provides a better genetic background to screen for genes involved in axon guidance. One of the mutations in our strain, nre-1(hd20), shares phenotypes with eri-1 and rrf-3, like the temperature-sensitive decrease in brood size. Nre-1 might be involved in RNA metabolism like eri-1 [a 3′ exonuclease (17)] and rrf-3 [a RNA-dependent RNA polymerase (16)], possibly interfering with the down-regulation of the RNAi response.
Genes Controlling Axon Navigation.
Our screen identified most of the known axon guidance genes in C. elegans on the investigated chromosomes (i.e., unc-40, unc-71, unc-73, and hse-5). A few genes like lin-11 or unc-119 were missed, indicating that even in our highly sensitized background, it is not possible to identify the complete set of genes needed for axon navigation. We also identified a few genes (i.e., lin-39) as having a role in axon navigation, where gene mutations do not seem to confirm this function. One possible explanation for the discrepancy is a dependence of the RNAi effect on axon guidance on the particular genetic background used in the screen. Lin-39 and lin-15b, which is mutated in the strain used in the RNAi screen, are both transcription factors and share a function in vulva cell fate specification (20). The RNAi experiments could point to a synergistic function of these genes in axon guidance.
The 93 genes affecting interneuron and/or motoneuron navigation fall into distinct functional categories, showing that our screening approach was able to identify genes at all levels of axon guidance regulation. One class of genes encodes putative secreted or cell surface proteins. One of those proteins (D1044.2) has structural similarity to nidogen (NID-1), a component of the basement membrane known to be important for the correct positioning of axon bundles in C. elegans (21). Another gene in this category is the cadherin CDH-4, one of two fat-like cadherins encoded in the C. elegans genome (22). No function for cdh-4 has been described. We have isolated mutants, confirming the function for cdh-4 in axon guidance (C.S. and H.H., unpublished results). Fat-like cadherins are expressed in the developing nervous system in vertebrates, and mice lacking mFat1 exhibit defects in forebrain and eye development (23, 24), suggesting an evolutionary conserved function for fat-like cadherins in nervous system development.
Midline crossing defects of interneuron and/or motoneuron axons in the ventral cord were a prominent phenotype in our RNAi screen. PVP and PVQ axons, which also run in the ventral cord, were rarely affected, leading to two conclusions. First, the majority of the genes identified has a specific role in the navigation of certain classes of axons rather than a general role in axon guidance affecting every neuron (as would a global signal). Second, different axons extending in the same axon bundle apparently use different combinations of signals to navigate and stay on course. This adds an additional level of complexity and hints at how neuronal circuits can be formed in a robust way: by using not a few globally acting signals but separate sets of signals for different groups of axons, even for those that travel along the same path. This confirms earlier studies, which suggested that different classes of neurons with axons in the ventral cord use different combinations of guidance signals to navigate (25). Conversely, particular guidance signals in the ventral cord apparently are shared by neurons belonging to the motor circuit. Ventral cord defects in DA/DB motoneurons are always, and defects in glr-1::GFP expressing interneurons are predominantly, found in combination with VD/DD defects, suggesting that the affected genes belong to an axon guidance pathway shared by these neurons.
Functions for ced-1, unc-101, and pry-1 in Axon Navigation.
To validate the screen results, we analyzed mutants of ced-1, pry-1, and unc-101 and found axon guidance defects similar to those in the RNAi screen, suggesting that these genes have previously unrecognized functions in neuronal circuit formation. Ced-1 is an EGF-domain-containing receptor implicated in cell–cell recognition between apoptotic and hypodermal cells destined to engulf them (26). Ced-1 has been shown to be expressed in many neurons in the head and ventral cord (26). Our results show that the receptor has a second function in circumferential guidance of motoneuron commissures. Unc-101 encodes a homolog of the medium chain of the clathrin adaptor complex AP1 (9, 27). Trafficking of olfactory receptors depends on unc-101 (12), suggesting that in unc-101 mutants, altered axon guidance receptor localization could be responsible for the observed defects. Pry-1 encodes the C. elegans axin homolog (28). RNAi against pry-1 leads to severe defects, indicating that Wnts play a role in a variety of axon guidance decisions in C. elegans. Members of the Wnt family have been shown to control axon navigation in Drosophila as well as in mice (reviewed in refs. 29 and 30). An important role for Wnts in axonal navigation along the anterior–posterior axis was shown recently in C. elegans as well (31–33). We show here that a central component of the canonical Wnt signaling pathway, pry-1, is involved in left–right (within the ventral cord) and dorsoventral axon guidance decisions (commissures). It seems most likely that a combination of the above-mentioned Wnts is involved here as well, because RNAi against individual Wnts does not phenocopy the defects seen in pry-1 (data not shown).
In summary, we isolated the RNAi-hypersensitive mutant combination nre-1(hd20) lin-15b(hd126) suitable for RNAi analysis of axon guidance. The large number of axon guidance genes identified in our screen provides a basis for further molecular analysis of directed axon outgrowth at all regulation levels, from extracellular signals to transcriptional control of axon guidance genes.
Materials and Methods
Nematode Strains and GFP Markers.
For the RNAi screen, the following promoters were used to drive YFP expression in a subset of neurons with axons in the ventral cord: interneurons, glr-1(35, 36); D-type motoneurons, unc-47 (37); and DA/DB motoneurons, unc-129 (38). The marker strain hdIs17[glr-1::YFP, unc-47::YFP, unc-129::YFP, rol-6(su1006)] was generated (originally for a different purpose) in a CB4856 background (Hawaii isolate). For the RNAi screen, hdIs17 (three times outcrossed to Bristol N2) was crossed into nre-1(hd20) lin-15b(hd126) to obtain VH715: hdIs17I; hdIs10[unc-129::CFP, glr-1::YFP, unc-47::DsRed, hsp-16::rol-6] V; nre-1(hd20) lin-15b(hd126) X. The following mutations and markers also were used: Bristol N2, rhIs13[unc-119::GFP; dpy-20(+)] V; hdIs26[odr-2::CFP, sra-6::DsRed2] III; hdIs32[glr- 1::DsRed2] III; rrf-3(pk1426) X; ced-1(e1735) I; unc-101(m1) I; pry-1(mn38) I,; eri-1(mg366) IV; lin-15(n744) X; eri-1(mg366) IV; lin-15(n744) X. All strains were cultured at 20°C using standard methods.
Mutant Isolation.
The nre-1(hd20) lin-15b(hd126) mutant strain was isolated after ethyl-methyl-sulfonate (EMS) mutagenesis (50 mM EMS for 4 h) of rhIs13[unc-119::GFP; dpy-20(+)] (panneuronal GFP expression). The F1 and F2 generations of the mutagenized animals were fed with the E. coli strain HT115(DE3) containing the RNAi feeding vector L4417 (Fire vector kit; Addgene, Cambridge, MA). Nre-1(hd20) lin-15b(hd126) was selected out of 20,000 genomes as having significantly reduced GFP expression in neurons. Nre-1(hd20) lin-15b(hd126) was mapped by using SNP after crossing with the strain CB4856 (Hawaii isolate). The sterility phenotype was used to identify nre-1 homozygotes, which were then also tested for RNAi hypersensitivity. Both traits mapped to a region between SNP pkP6117 on cosmid C27C12 (corresponding to map position 21.4) and the end of the right arm of the X chromosome.
RNAi Screening and Analysis of Axonal Defects.
RNAi by feeding was performed as described (39) with the following modifications: RNAi feeding clones were grown for 16 h in LB culture with 50 μg/ml ampicillin and seeded onto agar plates containing 1 mM isopropyl β-d-thiogalactoside and 50 μg/ml carbenicillin. Twenty-four hours later, L3 stage hermaphrodites were placed on the plates and incubated for 5 days at 20°C. Bacterial clones leading to axonal defects were retested twice, and only clones giving significant phenotypes with P < 0.01 (χ2test) in all three experiments were scored as positive. All inserts from positive clones were sequenced to confirm identity of the clone. The following discrepancies were detected: well for clone JA_C16C10.8 contained clone JA_T02C12.1, JA_D1044.1 contained JA_D1044.2, JA_C38C10.2 contained JA_C38C10.3, JA_B0280.4 insert corresponds to unc-70. For phenotypic analysis, worms were washed off the feeding plates with M9 buffer containing 3 mM levamisole and mounted on agar pads. Axon guidance defects were scored with a Zeiss Axioskop Microscope (Zeiss, Oberkochen, Germany) by using a 40× Plan Neofluar objective and a standard YFP filter set. Stacks of confocal images were recorded with a Leica (Deerfield, IL) TCS SP2 microscope and are shown as maximum-intensity projections generated by ImageJ software (http://rsb.info.nih.gov/ij).
Statistical Analysis.
Significances of differences in defects between control strains and mutant strains/RNAi-treated animals were determined by χ2test. All axon guidance defects in Table 2 are significant, with P < 0.01 in comparison with the YFP marker strain in WT background.
Acknowledgments
We thank Andrew Fire (Stanford University, Stanford, CA) for vectors, the Ahringer laboratory (University of Cambridge, Cambridge, U.K.) for the RNAi library (obtained from Geneservice, Cambridge, U.K.), the Kaplan laboratory (Harvard University, Cambridge, MA) for the eri-1(mg366) IV; lin-15(n744) X strain, Suse Zobeley and Ilse Wunderlich for help with cloning and the generation of transgenic strains, and members of our laboratory for critical discussion of the experiments and comments on the manuscript. Some of the strains used in this work were obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN). This work was supported by the Max Planck Society, Deutsche Forschungsgemeinschaft Grants GRK 484 and GRK 791 and Canadian Institutes of Health Research Grant MOP77786 (to H.H.).
Abbreviation
- YFP
yellow fluorescent protein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Araujo SJ, Tear G. Nat Rev Neurosci. 2003;4:910–922. doi: 10.1038/nrn1243. [DOI] [PubMed] [Google Scholar]
- 2.Dickson BJ. Science. 2002;298:1959–1964. doi: 10.1126/science.1072165. [DOI] [PubMed] [Google Scholar]
- 3.Yu TW, Bargmann CI. Nat Neurosci. 2001;4(Suppl):1169–1176. doi: 10.1038/nn748. [DOI] [PubMed] [Google Scholar]
- 4.Sulston JE, Horvitz HR. Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 5.Sulston JE, Schierenberg E, White JG, Thomson JN. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- 6.Chalfie M, Tu Y, Euskirchen G, Ward WW, Prasher DC. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 7.Hutter H. J Microsc. 2004;215:213–218. doi: 10.1111/j.0022-2720.2004.01367.x. [DOI] [PubMed] [Google Scholar]
- 8.Timmons L, Fire A. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- 9.Sugimoto A. Differentiation. 2004;72:81–91. doi: 10.1111/j.1432-0436.2004.07202004.x. [DOI] [PubMed] [Google Scholar]
- 10.Timmons L, Court DL, Fire A. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 11.Wang D, Kennedy S, Conte D, Jr, Kim JK, Gabel HW, Kamath RS, Mello CC, Ruvkun G. Nature. 2005;436:593–597. doi: 10.1038/nature04010. [DOI] [PubMed] [Google Scholar]
- 12.Dwyer ND, Adler CE, Crump JG, L'Etoile ND, Bargmann CI. Neuron. 2001;31:277–287. doi: 10.1016/s0896-6273(01)00361-0. [DOI] [PubMed] [Google Scholar]
- 13.Hallam S, Singer E, Waring D, Jin Y. Development (Cambridge, UK) 2000;127:4239–4252. doi: 10.1242/dev.127.19.4239. [DOI] [PubMed] [Google Scholar]
- 14.Filipowicz W. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 15.Mello CC, Conte D., Jr Nature. 2004;431:338–342. doi: 10.1038/nature02872. [DOI] [PubMed] [Google Scholar]
- 16.Simmer F, Tijsterman M, Parrish S, Koushika SP, Nonet ML, Fire A, Ahringer J, Plasterk RH. Curr Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- 17.Kennedy S, Wang D, Ruvkun G. Nature. 2004;427:645–649. doi: 10.1038/nature02302. [DOI] [PubMed] [Google Scholar]
- 18.Lehner B, Calixto A, Crombie C, Tischler J, Fortunato A, Chalfie M, Fraser AG. Genome Biol. 2006;7:R4. doi: 10.1186/gb-2006-7-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sieburth D, Ch'ng Q, Dybbs M, Tavazoie M, Kennedy S, Wang D, Dupuy D, Rual JF, Hill DE, Vidal M, et al. Nature. 2005;436:510–517. doi: 10.1038/nature03809. [DOI] [PubMed] [Google Scholar]
- 20.Chen Z, Han M. Curr Biol. 2001;11:1874–1879. doi: 10.1016/s0960-9822(01)00596-6. [DOI] [PubMed] [Google Scholar]
- 21.Kim S, Wadsworth WG. Science. 2000;288:150–154. doi: 10.1126/science.288.5463.150. [DOI] [PubMed] [Google Scholar]
- 22.Hill E, Broadbent ID, Chothia C, Pettitt J. J Mol Biol. 2001;305:1011–1024. doi: 10.1006/jmbi.2000.4361. [DOI] [PubMed] [Google Scholar]
- 23.Down M, Power M, Smith SI, Ralston K, Spanevello M, Burns GF, Boyd AW. Gene Expr Patterns. 2005;5:483–490. doi: 10.1016/j.modgep.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 24.Ciani L, Patel A, Allen ND, ffrench-Constant C. Mol Cell Biol. 2003;23:3575–3582. doi: 10.1128/MCB.23.10.3575-3582.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutter H. Development (Cambridge, UK) 2003;130:5307–5318. doi: 10.1242/dev.00727. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Z, Hartwieg E, Horvitz HR. Cell. 2001;104:43–56. doi: 10.1016/s0092-8674(01)00190-8. [DOI] [PubMed] [Google Scholar]
- 27.Lee J, Jongeward GD, Sternberg PW. Genes Dev. 1994;8:60–73. doi: 10.1101/gad.8.1.60. [DOI] [PubMed] [Google Scholar]
- 28.Korswagen HC, Coudreuse DY, Betist MC, van de Water S, Zivkovic D, Clevers HC. Genes Dev. 2002;16:1291–1302. doi: 10.1101/gad.981802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fradkin LG, Garriga G, Salinas PC, Thomas JB, Yu X, Zou Y. J Neurosci. 2005;25:10376–10378. doi: 10.1523/JNEUROSCI.3429-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zou Y. Trends Neurosci. 2004;27:528–532. doi: 10.1016/j.tins.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 31.Pan CL, Howell JE, Clark SG, Hilliard M, Cordes S, Bargmann CI, Garriga G. Dev Cell. 2006;10:367–377. doi: 10.1016/j.devcel.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 32.Hilliard MA, Bargmann CI. Dev Cell. 2006;10:379–390. doi: 10.1016/j.devcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 33.Prasad BC, Clark SG. Development (Cambridge, UK) 2006;133:1757–1766. doi: 10.1242/dev.02357. [DOI] [PubMed] [Google Scholar]
- 34.Maduro M, Pilgrim D. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hart AC, Sims S, Kaplan JM. Nature. 1995;378:82–85. doi: 10.1038/378082a0. [DOI] [PubMed] [Google Scholar]
- 36.Maricq AV, Peckol E, Driscoll M, Bargmann CI. Nature. 1995;378:78–81. doi: 10.1038/378078a0. [DOI] [PubMed] [Google Scholar]
- 37.McIntire SL, Reimer RJ, Schuske K, Edwards RH, Jorgensen EM. Nature. 1997;389:870–876. doi: 10.1038/39908. [DOI] [PubMed] [Google Scholar]
- 38.Colavita A, Krishna S, Zheng H, Padgett RW, Culotti JG. Science. 1998;281:706–709. doi: 10.1126/science.281.5377.706. [DOI] [PubMed] [Google Scholar]
- 39.Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]