Abstract
Notch-mediated induction of Nodal at the vertebrate node is a critical step in initiating left–right (LR) asymmetry. In mice and zebrafish we show that Baf60c, a subunit of the Swi/Snf-like BAF chromatin remodeling complex, is essential for establishment of LR asymmetry. Baf60c knockdown mouse embryos fail to activate Nodal at the node and also have abnormal node morphology with mixing of crown and pit cells. In cell culture, Baf60c is required for Notch-dependent transcriptional activation and functions to stabilize interactions between activated Notch and its DNA-binding partner, RBP-J. Brg1 is also required for these processes, suggesting that BAF complexes are key components of nuclear Notch signaling. We propose a critical role for Baf60c in Notch-dependent transcription and LR asymmetry.
Keywords: chromatin, Swi/Snf, node
The bodies of all vertebrates are asymmetric on the left and right sides, resulting in distinct situs of organs, such as the left-pointing heart. Proper regulation of left–right (LR) asymmetry is necessary for normal organ positioning during embryonic development (1, 2). Strong genetic and cell biologic evidence in mammals suggests that the critical events in breaking symmetry take place at the node, an important organizer structure of the vertebrate embryo. These symmetry-breaking events include the asymmetric movement of fluids across the node (so-called nodal flow) and the Notch pathway-dependent activation of the secreted protein Nodal in cells surrounding the node (1–7). This is followed by a cascade of secreted factors and transcription factors restricted to the left side of the embryo, establishing left-sided identity and organ situs (1, 2).
The Swi/Snf-like BAF chromatin remodeling complexes are important regulators of transcription during development (8). BAF complexes are large multisubunit assemblies that are characterized by polymorphic components; e.g., the core ATPase of the complexes can be either Brahma (Brm) or Brahma-related gene 1 (Brg1) (8–10). Another example of this combinatorial assembly of BAF complex subunit composition is the 60-kDa subunit Baf60. Baf60 is found in most BAF complexes, although it is not essential for the chromatin remodeling function of the complex. It can be represented by Baf60a, Baf60b, or Baf60c, which are encoded by the Smarcd1, Smarcd2, and Smarcd3 genes, respectively (10). Baf60 proteins have been shown to interact with transcription factors, including nuclear receptors, the AP-1 complex, and others, and are thought to bridge interactions between these transcription factors and BAF complexes (11–14). Very little is known about the developmental or physiological roles played by Baf60 proteins. Baf60c was recently shown to be critical for heart development (14).
Here, we show that loss of Baf60c in mouse and zebrafish leads to defects in the establishment of the LR asymmetry cascade. In mouse embryos, this is because of impaired activation of Nodal at the node. We further show that Baf60c is an essential component of nuclear Notch signaling and propose that the integration of nuclear Notch signaling components by BAF complexes is a critical mechanism for transcriptional activation of Nodal in the mouse node, and perhaps for Notch-dependent transcription in general.
Results
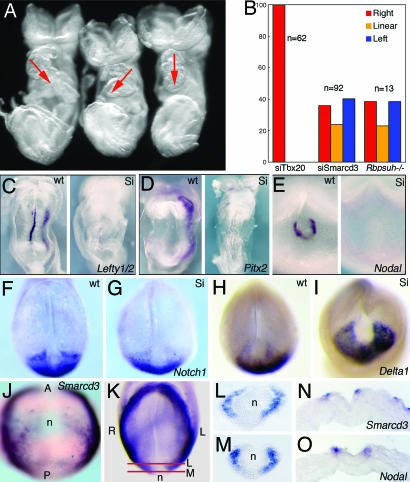
LR Defects in Smarcd3 Knockdown Embryos.
In our analysis of altered cardiogenesis in Smarcd3 short hairpin RNA (shRNA) knockdown embryos (14), we noticed that the situs of the heart and direction of embryonic turning in the most severe knockdown line (A#1) were randomized (Fig. 1 A and B). Lower penetrance of looping defects was also observed for another knockdown line. Expression of key asymmetrically expressed regulators of the LR cascade (Lefty1, Lefty2, and Pitx2) was undetectable in Smarcd3 knockdown embryos (Fig. 1 C and D, n = 5). Nodal expression around the node is essential for lateral plate mesoderm (LPM) expression of LR genes (3–6). Perinodal expression of Nodal was lost in Smarcd3 knockdown embryos (Fig. 1E, n = 5). Expression of Notch1 was reduced in Smarcd3 knockdown embryos (n = 2), whereas Delta1 (Dll1) mRNA was increased (Fig. 1 G and I, n = 2).
Fig. 1.
Baf60c regulates LR asymmetry at the node. (A) Smarcd3 knockdown caused randomized heart looping. Red arrows indicated the direction of heart looping. (B) Histogram shows the percentage of control (siTbx20), Smarcd3 knockdown (siSmarcd3), and Rbpsuh−/− mutant embryos with rightward, linear, and leftward heart morphologies. (C–E) Expression of LR regulators in wild-type (wt) and Smarcd3 knockdown (Si) embryos. Shown are Lefty1/2 (C), Pitx2 (D) at E8.25, and Nodal (E) at E7.75. (F–I) Decreased expression of Notch1 and increased expression of Dll1 in siSmarcd3 embryos. (J–O) Smarcd3 expression in perinodal cells and coexpression with Nodal by in situ hybridization on consecutive sections at E7.75 (O). Red lines in K show the plane of sections shown in L and M.
We previously reported that Smarcd3 was cardiac-specific during development (14). Careful examination of Smarcd3 mRNA distribution in the mouse embryos showed clear expression in Nodal-expressing perinodal cells in mouse embryos (Fig. 1 J–O). Bilateral expression of Smarcd3 expanded beyond the node into the LPM, similar to that of Notch1 (Fig. 1J).
We tested the specificity of the Smarcd3 knockdown in cultured embryos with localized Lipofectamine-mediated shRNA Smarcd3 knockdown [supporting information (SI) Fig. 6 B and C, n = 4] (15). Localized application of Smarcd3 shRNA-expressing plasmids resulted in loss of Nodal in transfected cells (SI Fig. 6 B and C). We further tested the specificity of the knockdown by examining heart situs and Nodal expression in Smarcd3 knockdown embryos expressing a Baf60b-IRES-EGFP transgene under control of the CMV enhancer/β-actin promoter (CAGGS-Baf60b-IRES-EGFP) (14). Although CAGGS-Baf60b-IRES-EGFP is expressed at higher levels in the heart (14), it also expresses homogeneously at lower levels throughout the embryo (SI Fig. 6 E and H) (16). Nodal expression was largely rescued in Smarcd3 knockdown CAGGS-Baf60b-IRES-EGFP embryos (SI Fig. 6 F and I, n = 5), as was cardiac looping [100% leftward looping at embryonic day 9.5 (E9.5); n = 10]. Furthermore, ES cell–tetraploid complementation method or heart defects did not affect the LR pathway, as ES cell-derived embryos lacking the cardiac transcription factor Tbx20 (17) had normal heart situs (Fig. 1B).
We tested whether Baf60c is important in the LPM for receiving Nodal signals from the node by misexpressing Nodal in the right LPM (SI Fig. 6 J–L, n = 6). This resulted in bilateral Nodal expression in wild-type embryos and right-sided expression in Smarcd3 knockdown embryos; Nodal expressed from the transfected expression construct could be detected as strong punctate staining in a few cells, whereas Nodal-induced endogenous Nodal was fainter and broader throughout the LPM. Because endogenous Nodal could be induced by Nodal misexpression in the right LPM of wild-type and Smarcd3 knockdown embryos (n = 6), we conclude that Baf60c is not important for the response of the LPM to Nodal (SI Fig. 6 J–L).
Our results together indicate that Baf60c function at the mouse node, similar to that of Notch signaling, is critical for the initiation of Nodal expression and thus the progression of the LR asymmetry cascade.
Defects in Node Morphology in Smarcd3 Knockdown Embryos.
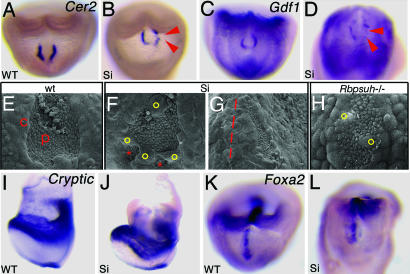
Other genes important for LR determination at the node (Cer2 and Gdf1) (18, 19) were detected in Smarcd3 knockdown embryos (Fig. 2 A–D, n = 3), although Cer2- and Gdf1-expressing crown cells did not properly align around the node, reflecting abnormal node morphology. Indeed, scanning electron microscopy showed abnormal node morphologies in Smarcd3 mutants (Fig. 2 F and G). Abnormal crown cells were seen (Fig. 2F, yellow circles), and node pit cells were separated into noncohesive groups (Fig. 2F, red asterisks). In ≈30% of mutant embryos, the node itself was shifted to the left (Fig. 2G). Monociliated pit cells were apparent in Smarcd3 knockdown embryos, and by video microscopy the cilia in Smarcd3 knockdown embryos (n = 4) were motile, but their motion was restricted, and flow across the node was severely impaired (data not shown). The midline markers Foxa2 (n = 2) and Shh (n = 4) were expressed in Smarcd3 knockdown embryos (Fig. 2 I–L). We believe that these defects in node morphology and flow are secondary to altered crown cell morphology, as has been proposed for mice lacking the Notch ligand Dll1, which also have LR defects (20). We observed similar heart looping and node morphology defects in mouse embryos derived from ES cells homozygous for a deletion of Rbpsuh (Figs. 1B and 2H) (6), which encodes RBP-J, the nuclear effector of Notch signaling (21, 22). We do not believe that the loss of Nodal-expressing cells would be the main cause of disruption in LR asymmetry, because the loss of Nodal mRNA is much broader than the disruptions in crown cell integrity as demonstrated by loss of other perinodal markers. Thus, Baf60c regulates perinodal Nodal expression and node morphology, similar to the Notch signaling pathway.
Fig. 2.
Abnormal node morphology in Smarcd3 knockdown embryos. (A–D) Expression of Cer2 and Gdf1 in wild-type and Smarcd3 knockdown (Si) embryos. Red arrowheads show decreased or missing expression. (E–K) Scanning electron microscopy shows high magnification of the node in wild type (E), abnormal node morphology in Smarcd3 knockdown (F and G), and Rbpsuh−/− (H) embryos at E7.75. c, crown cells; p, pit cells. Smarcd3 knockdown results in abnormal node morphology, with separated pit cells (F, red asterisks) and abnormally migrated crown cells (yellow circles). Some Smarcd3 mutants also have abnormal leftward shifted nodes in addition (G, red line shows embryonic midline). Abnormal crown cells were seen in Rbpsuh−/− embryos (H, yellow circles). (I–L) Expression of Cryptic and Foxa2 in wild-type and Smarcd3 knockdown (Si) embryos.
Misexpression of Smarcd3 Expands the Field of Nodal Expression.
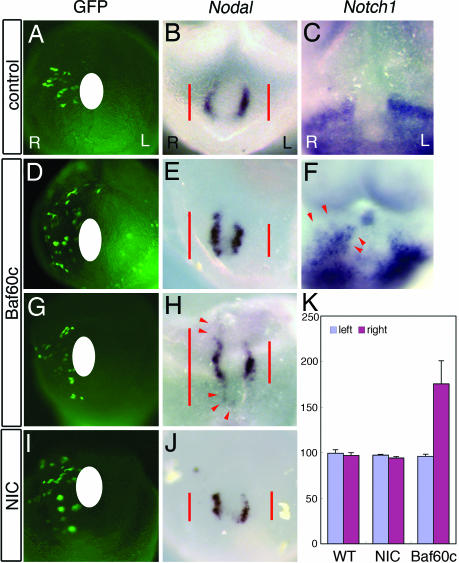
Lipofectamine-mediated misexpression of Baf60c to the right of the node of cultured mouse embryos resulted in expanded Nodal induction along the anteroposterior axis (Fig. 3 E and H, red arrowheads, n = 4) and slightly expanded Notch1 expression (Fig. 3F, red arrowheads, n = 2), whereas activated Notch1 (NIC) misexpression resulted in enhanced Nodal induction at the node without expansion of its expression domain (Fig. 3J, n = 5). This indicates that Baf60c is a limiting factor in Notch-mediated induction of Nodal. These in vivo misexpression studies strongly suggested that Baf60c acts as a critical Nodal inducer at the node and is thus a key factor in the establishment of LR asymmetry.
Fig. 3.
Baf60c is sufficient to induce Nodal. Cultured embryos transfected with an EGFP expression construct and Baf60c, activated Notch (NIC), or Nodal expression constructs, or EGFP vector alone (control) were cultured for 15 h. (A, D, G, and I) EGFP fluorescence, showing transfected cells. (A–C) Control studies showed normal/bilateral nodal (B) at the node and Notch1 (C) expression around the node. (D–H) Smarcd3-transfected embryos: ectopic Nodal induction along the midline was seen (E and H), and expansion of Notch1 expression was also observed (F, red arrowhead). (I and J) Overexpression of NIC enhanced Nodal expression. (K) Histogram shows the length (in millimeters) of Nodal expression on the node between the left side (light purple, control) and the right side (light blue, injected experiments, n = 5).
Zebrafish Smarcd3 Regulates LR Asymmetry.
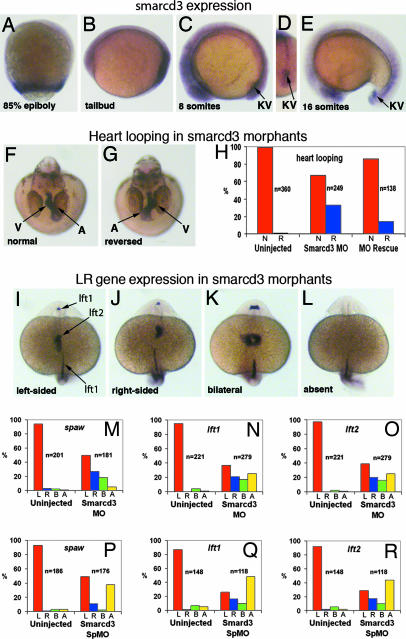
To address conservation of Baf60c function during vertebrate LR asymmetry determination, we used zebrafish (Danio rerio) embryos, because early LR pathways are generally conserved between mammals and fish (1, 2, 7). A candidate for the zebrafish orthologue of Smarcd3 was identified in the region of zebrafish chromosome 7 syntenic to the mouse Smarcd3 locus. Zebrafish smarcd3 is first expressed in a band of three to four cell diameters at the blastoderm margin beginning at later shield/70% epibody (Fig. 4A). At the end of gastrulation (bud stage) smarcd3 is highly expressed in a band of six to eight cell diameters that surrounds the location of the dorsal forerunner cells, the precursors of Kupffer's vesicle (KV), the ciliated organ of asymmetry (analogous to the mouse node) in the zebrafish (Fig. 4B). During early somitogenesis (4–10 s), when LR patterning is being established, smarcd3 is strongly expressed in the notochord and in cells surrounding the KV (Fig. 4 C and D). At later stages smarcd3 is restricted to the KV, eye, midbrain, and forebrain (Fig. 4E). Morpholino (MO) knockdown of Smarcd3 resulted in 36% of embryos with reverse looping heart morphologies (n = 249; vs. 1.3% for control MO, n = 360) (Fig. 4 F–H); this phenotype was partly rescued (14% reverse looping, n = 138) by coinjection of smarcd3 mRNA (Fig. 4H). Similarly, knockdown of Smarcd3 using a splice-blocking MO (Smarcd3 SpMO) resulted in 27% of embryos with reversed heart looping (n = 159; vs. 1% for uninjected controls, n = 173). Expression of left-side-specific genes [lefty1 (lft1), lefty2 (lft2), and southpaw (spaw)] (23, 24) was also altered (Fig. 4 I–R). In smarcd3 morphants, expression of lft1, lft2, and spaw was abnormally right-sided or bilateral, and in ≈30–50% of embryos (10% for spaw using Smarcd3 MO) their expression was completely lost (Fig. 4 M–R), similar to the response in Smarcd3 shRNA knockdown mouse embryos. Midline markers including lft1 (Fig. 4 I–L), ptc1, shh, and ntl (data not shown) were intact.
Fig. 4.
Smarcd3 is required for LR axis specification in zebrafish. (A–E) Expression of smarcd3 (A–C and E are lateral views, anterior to left, and D is a caudal view, dorsal to top). (A) During gastrulation. (B) Expression in the developing tailbud. (C and D) Early somitogenesis. (E) Mid-somitogenesis. Knockdown of smarcd3 randomizes direction of heart looping in embryos 40 h after fertilization (F–H). A, atrium; V, ventricle. In H, red bars show normal looping and blue bars show reversed looping. Heart looping is partially rescued by smarcd3 RNA. (I–L) Altered expression of lefty1 (lft1) in the diencephalon and lefty2 (lft2) in the heart field of 22- to 24-somite-stage embryos. (I) Normal left-sided expression. (J) Right-sided expression. (K) Bilateral expression. (L) Absence of expression. Expression of lft1 in the midline is unaltered. (M–R) Quantification of alterations in asymmetric gene expression patterns in smarcd3 morphants. (M and P) southpaw (spaw) expression in the LPM (19- to 21-somite stage). (N and Q) lft1 expression. (O and R) lft2 expression. L, left-sided; R, right-sided; B, bilateral; A, absent.
Baf60c Functionally and Physically Interacts with Nuclear Notch Components.
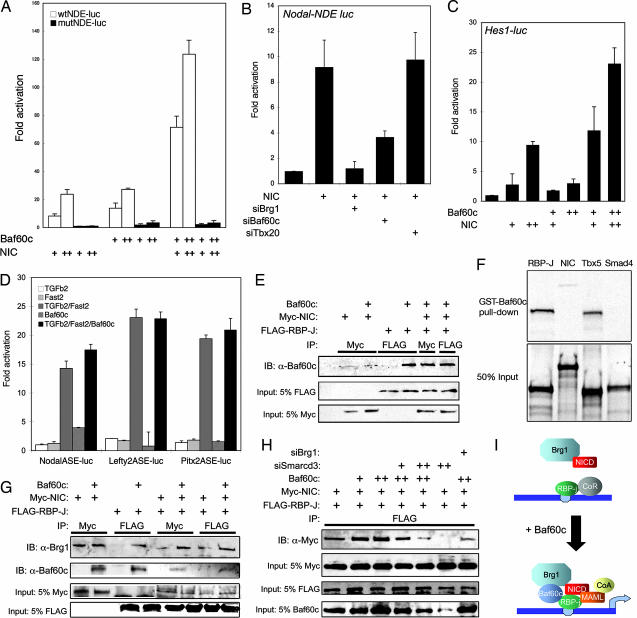
The Nodal node-dependent enhancer (NDE) relies on two RBP-J binding elements for its activity in the node (5, 6). We examined the role of Baf60c in the transcriptional activation of the Nodal-NDE by transient luciferase reporter assay in 10T1/2 cells. Nodal-NDE-luciferase was activated dose-dependently by NIC as well as Baf60c (Fig. 5A), and the combination of Baf60c and NIC synergistically activated Nodal-NDE-luciferase. Activation by NIC or Baf60c depended on intact RBP-J binding sites, suggesting that Baf60c and NIC interact to directly activate the NDE. Depletion of endogenous Baf60c or Brg1 by shRNAs greatly reduced the response of Nodal-NDE-luciferase to NIC (Fig. 5B), indicating a critical requirement for Baf60c and the BAF complex. Partial inhibition by Baf60c RNAi may indicate compensation by other Baf60 proteins. Hes1-luciferase, a well characterized target of Notch that also relies on RBP-J sites (25), was also synergistically activated by Baf60c and NIC (Fig. 5C). LPM expression of Nodal, Lefty1/2, and Pitx2 (which are absent in Smarcd3 knockdown embryos) depends on asymmetry enhancers (ASE) that are activated by TGFβ/FoxH1 (26, 27). TGFβ/FoxH1-dependent activation of Nodal-ASE, Lefty2-ASE, and Pitx2-ASE luciferase constructs was not affected by Baf60c (Fig. 5D). This finding supports the Nodal misexpression data indicating that Baf60c is not involved in mediating TGFβ-mediated induction of genes in the LPM. These results therefore reveal that Baf60c is a critical mediator of Notch signaling and that RBP-J or its DNA binding element is necessary for Baf60c function.
Fig. 5.
Baf60c potentiates transcription by promoting interaction within nuclear Notch complexes and BAF complexes. (A) Baf60c activates the Nodal-NDE-luciferase reporter construct and synergizes with activated Notch (NIC). Baf60c or NIC do not activate Nodal-mutNDE-luciferase (black bars), which has mutated RBP-J binding elements. +, 250 ng of expression construct; ++, 500 ng of expression construct. (B) Depletion of Brg1 (siBrg1) or Baf60c (siBaf60c) by RNAi abrogates the NIC-dependent activation of Nodal-NDE-luciferase. siTbx20 is a control for nonspecific effects of RNAi. (C) Baf60c potentiates transcriptional activation of Hes1-luciferase with NIC. (D) Baf60c does not enhance Nodal-ASE, Lefty2-ASE, or Pitx2-ASE luciferase reporters, which are stimulated by activated TGFβ signaling and Fast2. (E) Coimmunoprecipitation of Flag-RBP-J or Myc-NIC, alone or with a Baf60c expression construct, followed by Baf60c immunodetection shows that Baf60c interacts weakly with NIC and strongly with RBP-J. (F) GST pull-down of 35S-labeled RBP-J, NIC, Tbx5, or Smad4. GST-Baf60c could pull down RBP-J and Tbx5 but not NIC or Smad4. Fifty percent input is shown below the pull-down. (G) Coimmunoprecipitation of Flag-RBP-J or Myc-NIC, cotransfected with Baf60c, followed by immunodetection of Brg1 shows that Brg1 can interact with NIC but not with RBP-J. The Brg1/RBP-J interaction depends on Baf60c, and Brg1/NIC/RBP-J interaction is strengthened by Baf60c. (H) Interaction of NIC with RBP-J is enhanced by Baf60c. Depletion of endogenous Baf60c (siSmarcd3) or Brg1 (siBrg1) destabilized the interaction of RBP-J with NIC in 10T1/2 cells. (I) Model for the integration of Notch signaling by Baf60c.
Immunoprecipitation experiments in transfected cells revealed a strong interaction of Baf60c with RBP-J and a much weaker interaction with NIC (Fig. 5E). This finding was confirmed by GST pull-down assays (Fig. 5F). We tested whether Baf60c potentiated interactions between NIC or RBP-J and Brg1 by coimmunoprecipitation: Brg1 interacted with NIC whether or not Baf60c was coexpressed; however, Baf60c was necessary for an association of Brg1 and RBP-J (Fig. 5G). Furthermore, Baf60c could enhance an interaction between Brg1 and NIC/RBP-J in this system. Interestingly, the interaction between NIC and RBP-J in10T1/2 cells was also strongly enhanced by Baf60c, and, most importantly, depletion of endogenous Baf60c destabilized the NIC/RBP-J interaction, as did depletion of Brg1 (Fig. 5H). We conclude from these results that Baf60c is a critical nuclear factor in Notch signaling that promotes interactions between activated Notch and its nuclear partner RBP-J and enhances the interaction of the NIC/RBP-J complex with the BAF complexes. We propose that Baf60c interacts preferentially with RBP-J, thus creating a bridge between RBP-J and the BAF complex. Furthermore, because NIC interacts with Brg1 (and thus presumably the BAF complex), we propose that four interactions are important for stabilization of the nuclear Notch signaling complex: (i) NIC with RBP-J, (ii) NIC with Brg1, (iii) Baf60c with Brg1, and (iv) Baf60c with RBP-J (Fig. 5I). The aggregate effect of these interdependent interactions is the formation of a stable complex at RBP-J binding sites that includes the BAF chromatin remodeling complex.
Discussion
Baf60c Regulate LR Asymmetry at the Node.
We have shown that embryos with a specific knockdown of Smarcd3, encoding Baf60c, have defects in the establishment of the LR asymmetry pathway due to impaired induction of Nodal at the mouse node. Nodal expression in perinodal cells is essential for the initiation of the downstream cascade of gene expression that establishes the left side of the embryo as distinct from the right side, including the expression of Nodal itself in the LPM (3, 4). Our data show that the role of Baf60c in regulating LR asymmetry is conserved between mouse and zebrafish, suggesting a similar mode of action. However, whereas in mouse we consistently observed a loss of left-sided gene expression, in zebrafish we also noted bilateral and reversed gene expression. Clear differences exist between species in the regulation of the LR pathway from the node or its orthologous structures (1, 2). Recently, a self-enhancement/lateral inhibition model based on Nodal and Lefty diffusion has been proposed to explain the robust establishment of left-sided gene expression (28). This model predicts that slight differences in left-sided expression or diffusion of Nodal from the node would disrupt the normal LR pattern; it is possible that in the Smarcd3 zebrafish morphants spaw expression surrounding the KV is partly reduced, thus still leading to disrupted LR patterning via an imbalance in the self-enhancement/lateral inhibition system, albeit in a pattern that differs from the mouse embryos in which Nodal expression was eliminated. Regardless of the precise mechanism, it is certain that Baf60c has important roles in regulating the LR pathway in both species. Interestingly, a portion of the Smarcd3 knockdown hearts did not loop at all, reminiscent of hearts in which the left-sided regulator of organ sidedness Pitx2 has been misexpressed (29). This may be an indicator of a linkage of Baf60c and cell-autonomous organ orientation.
We also found that Smarcd3 knockdown mouse embryos had defects in node morphology, including defective arrangement of perinodal crown cells and nodal pit cells. Zebrafish smarcd3 knockdown did not result in any anomalies in the KV in terms of morphology or acetylated tubulin staining of cilia (data not shown). The abnormalities in node morphology may contribute to the defective LR patterning in Smarcd3 knockdown embryos, but the primary defect appears to be the absence of Nodal expression in perinodal cells.
Baf60c and the BAF Complex Functionally Interact with Notch Signaling.
Notch signaling at the mouse node is a primary regulator of Nodal transcription via two conserved RBP-J binding sites (5, 6). Notch signaling also regulates Nodal at the node in chick embryos (30) and has been implicated in LR asymmetry in zebrafish as well (6, 31). In the mouse it is likely that the function of Baf60c in node morphogenesis is also partly related to Notch signaling, because we found similar defects in Rbpsuh−/− embryos, and similar abnormal nodes are seen in mice lacking the Notch ligand Delta-1 (5, 20). The clear involvement of Notch signaling in Nodal regulation and node morphogenesis prompted us to examine a possible functional relationship between Baf60c and Notch. Our combined biochemical data strongly suggest that the BAF complex interacts with NIC/RBP-J and that this interaction is promoted by Baf60c. Interestingly, the stability of the interaction between NIC and RBP-J was critically dependent on the presence of endogenous Baf60c, suggesting not only that there was an interaction between BAF complexes and NIC/RBP-J, but that the recruitment of the BAF complex is absolutely critical for the stability of the NIC/RBP-J interaction. Thus, we propose that Baf60c and the BAF complex are integral mediators of Notch signaling in the nucleus via stabilization of NIC/RBP-J interaction and possibly remodeling of chromatin at sites bound by NIC/RBP-J.
The finding that stabilization of NIC and RBP-j interactions is mediated by Baf60c and the BAF complex is perhaps unexpected considering that, to date, no requirement for a stabilizing protein in nuclear Notch signaling has been demonstrated. Biochemical data demonstrate strong interactions between NIC and RBP-j in vitro, and they are readily isolated together from cells by coimmunoprecipitation (22, 25, 32–34). However, these data collectively do not exclude a requirement for a stabilizing effect of other proteins, because in vitro data solely demonstrate that two proteins can interact sufficiently well under experimental conditions, and immunopurification from cells by default includes other potential cellular components. In the case of the BAF complex, compelling evidence in other organisms suggests that this function may be a general feature of Notch signaling and may extend to invertebrates: in Caenorhabditis elegans, ZK1128.5, encoding a Baf60c ortholog, genetically interacts with Notch signaling (35), and in a screen for genetic interactions with Drosophila Brahma, encoding the Brg1/Brm homolog, genes encoding Notch signaling components were the predominant class of mutations identified (36). Taken together these results suggest that Baf60c and the BAF complex is a conserved key cellular nuclear component of Notch signaling in several contexts, including the establishment of LR asymmetry in mammals. Our findings may have important implications for interaction of the Notch pathway and Baf60c in cardiogenesis, because Baf60c is a critical factor for heart morphogenesis (14), and Notch signaling is important for cardiac differentiation in Xenopus (37), in ES cells (38), and as a causative factor in human congenital heart defects (39). Our findings overall suggest a mechanism for transcriptional potentiation of critical signaling pathways in development.
Methods
Mouse in Vivo RNAi and Transgenesis.
In vivo RNA interference for Smarcd3 and tetraploid aggregations was performed as previously described (14). Embryo transfection was performed on E7–E7.5 embryos by using Lipofectamine 2000 as previously described (15). Rbpsuh−/− ES cells were generated from Rbpsuh+/− ES cells (40) selected in medium containing high levels of G418. Nodal flow was assessed in mouse embryos as described in ref. 41.
MO Knockdown of Smarcd3 Function.
Translation-blocking (0.6 ng) and splice-blocking (4 ng) MO antisense oligonucleotides (Gene Tools, Philomath, OR) were used to knock down the function of smarcd3: smarcd3 MO is complementary to a region immediately upstream of the translation start site 5′-TTCCCTCCGCTTCTCCTGCCTTTTG-3′. smarcd3 SpMO is complementary to the splice donor site of exon 3, 5′-TCAGATCTCTTACTCACCCTTTGTG-3′. The MO phenotype was rescued by coinjection of smarcd3 MO with 75 pg of in vitro synthesized smarcd3 RNA containing the ORF of Smarcd3, with the 5′ UTR truncated to remove 18 bp of sequence recognized by the MO.
Transactivation Assays and Immunoprecipitation.
Transactivation assays and coimmunoprecipitation experiments were performed as previously described (14). Nodal, Pitx2, and Lefty2 enhancers (5, 26, 27) were subcloned into pGL3 to generate luciferase reporter constructs. Anti-Brg1 (Upstate Biotechnology, Lake Placid, NY), anti-FLAG (Sigma, St. Louis, MO), anti-HA (Sigma), and anti-myc (Santa Cruz Biotechnology, Santa Cruz, CA) antisera were commercially obtained; anti-Baf60c antiserum (12) was kindly provided by J. Auwerx (Institut de Génétique et de Biologie Moléculaire et Cellulaire, Illkirch, France).
GST Pull-Down Assay.
GST and GST-Baf60c were expressed in BL21 Escherichia coli. GST protein was purified from cell lysates with glutathione Sepharose 4B beads (Amersham). Proteins were translated in vitro and labeled with [35S]methionine by using a reticulocyte lysate system (Promega). Labeled proteins were incubated with GST or GST-Baf60c beads overnight at 4°C. The beads were washed, mixed with SDS loading buffer, and heated to 100°C. The bound proteins were analyzed by SDS/PAGE. The gel was dried and exposed to autoradiograph film overnight.
Supplementary Material
Acknowledgments
We thank S. McMaster (Mount Sinai Hospital Research Institute, University of Toronto, Toronto, ON, Canada) for tetraploid aggregations; D. Holmyard for scanning electron microscopy; J. N. Campbell and B. L. McMahan for technical assistance; T. Honjo (Kyoto University, Kyoto, Japan) for Rbpsuh+/− ES cells; and A. Israel (Institute Pasteur, Paris, France), R. Kopan (Washington University, St. Louis, MO), and J. Wrana (Mount Sinai Hospital Research Institute) for expression and reporter constructs. This research was funded by the Canadian Institutes of Health Research (B.G.B. and J.R.), the Heart and Stroke Foundation of Ontario (B.G.B.), March of Dimes Birth Defects Foundation Grant 1-FY05-117 (to B.G.B.), an Emmy Noether fellowship of the Deutsche Forschungsgemeinschaft (to H.L.), and a Human Frontiers Science Program long-term fellowship (to J.K.T.). J.R. is a Canadian Institutes of Health Research Distinguished Scientist. B.G.B. held a Canada Research Chair in Developmental Cardiology.
Abbreviations
- LR
left–right
- LPM
lateral plate mesoderm
- shRNA
short hairpin RNA
- KV
Kupffer's vesicle
- MO
morpholino
- NDE
node-dependent enhancer
- ASE
asymmetry enhancer
- En
embryonic day n
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0608118104/DC1.
References
- 1.Shiratori H, Hamada H. Development (Cambridge, UK) 2006;133:2095–2104. doi: 10.1242/dev.02384. [DOI] [PubMed] [Google Scholar]
- 2.Raya A, Belmonte JC. Nat Rev Genet. 2006;7:283–293. doi: 10.1038/nrg1830. [DOI] [PubMed] [Google Scholar]
- 3.Brennan J, Norris DP, Robertson EJ. Genes Dev. 2002;16:2339–2344. doi: 10.1101/gad.1016202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saijoh Y, Oki S, Ohishi S, Hamada H. Dev Biol. 2003;256:160–172. doi: 10.1016/s0012-1606(02)00121-5. [DOI] [PubMed] [Google Scholar]
- 5.Krebs LT, Iwai N, Nonaka S, Welsh IC, Lan Y, Jiang R, Saijoh Y, O'Brien TP, Hamada H, Gridley T. Genes Dev. 2003;17:1207–1212. doi: 10.1101/gad.1084703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raya A, Kawakami Y, Rodriguez-Esteban C, Buscher D, Koth CM, Itoh T, Morita M, Raya RM, Dubova I, Bessa JG, et al. Genes Dev. 2003;17:1213–1218. doi: 10.1101/gad.1084403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Development (Cambridge, UK) 2005;132:1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- 8.de la Serna IL, Ohkawa Y, Imbalzano AN. Nat Rev Genet. 2006;7:461–473. doi: 10.1038/nrg1882. [DOI] [PubMed] [Google Scholar]
- 9.Olave IA, Reck-Peterson SL, Crabtree GR. Annu Rev Biochem. 2002;71:755–781. doi: 10.1146/annurev.biochem.71.110601.135507. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, Crabtree GR. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 11.Hsiao PW, Fryer CJ, Trotter KW, Wang W, Archer TK. Mol Cell Biol. 2003;23:6210–6220. doi: 10.1128/MCB.23.17.6210-6220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Debril MB, Gelman L, Fayard E, Annicotte JS, Rocchi S, Auwerx J. J Biol Chem. 2004;279:16677–16686. doi: 10.1074/jbc.M312288200. [DOI] [PubMed] [Google Scholar]
- 13.Ito T, Yamauchi M, Nishina M, Yamamichi N, Mizutani T, Ui M, Murakami M, Iba H. J Biol Chem. 2001;276:2852–2857. doi: 10.1074/jbc.M009633200. [DOI] [PubMed] [Google Scholar]
- 14.Lickert H, Takeuchi JK, von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 15.Yamamoto M, Saijoh Y, Perea-Gomez A, Shawlot W, Behringer RR, Ang SL, Hamada H, Meno C. Nature. 2004;428:387–392. doi: 10.1038/nature02418. [DOI] [PubMed] [Google Scholar]
- 16.Rhee JM, Pirity MK, Lackan CS, Long JZ, Kondoh G, Takeda J, Hadjantonakis AK. Genesis. 2006;44:202–218. doi: 10.1002/dvg.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takeuchi JK, Mileikovskaia M, Koshiba-Takeuchi K, Heidt AB, Mori AD, Arruda EP, Gertsenstein M, Georges R, Davidson L, Mo R, et al. Development (Cambridge, UK) 2005;132:2463–2474. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- 18.Marques S, Borges AC, Silva AC, Freitas S, Cordenonsi M, Belo JA. Genes Dev. 2004;18:2342–2347. doi: 10.1101/gad.306504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rankin CT, Bunton T, Lawler AM, Lee SJ. Nat Genet. 2000;24:262–265. doi: 10.1038/73472. [DOI] [PubMed] [Google Scholar]
- 20.Przemeck GK, Heinzmann U, Beckers J, Hrabe de Angelis M. Development (Cambridge, UK) 2003;130:3–13. doi: 10.1242/dev.00176. [DOI] [PubMed] [Google Scholar]
- 21.Lai EC. Development (Cambridge, UK) 2004;131:965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- 22.Lubman OY, Korolev SV, Kopan R. Mol Cell. 2004;13:619–626. doi: 10.1016/s1097-2765(04)00120-0. [DOI] [PubMed] [Google Scholar]
- 23.Bisgrove BW, Essner JJ, Yost HJ. Development (Cambridge, UK) 1999;126:3253–3262. doi: 10.1242/dev.126.14.3253. [DOI] [PubMed] [Google Scholar]
- 24.Long S, Ahmad N, Rebagliati M. Development (Cambridge, UK) 2003;130:2303–2316. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- 25.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 26.Saijoh Y, Adachi H, Sakuma R, Yeo CY, Yashiro K, Watanabe M, Hashiguchi H, Mochida K, Ohishi S, Kawabata M, et al. Mol Cell. 2000;5:35–47. doi: 10.1016/s1097-2765(00)80401-3. [DOI] [PubMed] [Google Scholar]
- 27.Shiratori H, Sakuma R, Watanabe M, Hashiguchi H, Mochida K, Sakai Y, Nishino J, Saijoh Y, Whitman M, Hamada H. Mol Cell. 2001;7:137–149. doi: 10.1016/s1097-2765(01)00162-9. [DOI] [PubMed] [Google Scholar]
- 28.Nakamura T, Mine N, Nakaguchi E, Mochizuki A, Yamamoto M, Yashiro K, Meno C, Hamada H. Dev Cell. 2006;11:495–504. doi: 10.1016/j.devcel.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 29.Logan M, Pagan-Westphal SM, Smith DM, Paganessi L, Tabin CJ. Cell. 1998;94:307–317. doi: 10.1016/s0092-8674(00)81474-9. [DOI] [PubMed] [Google Scholar]
- 30.Raya A, Kawakami Y, Rodriguez-Esteban C, Ibanes M, Rasskin-Gutman D, Rodriguez-Leon J, Buscher D, Feijo JA, Izpisua Belmonte JC. Nature. 2004;427:121–128. doi: 10.1038/nature02190. [DOI] [PubMed] [Google Scholar]
- 31.Kawakami Y, Raya A, Raya RM, Rodriguez-Esteban C, Belmonte JC. Nature. 2005;435:165–171. doi: 10.1038/nature03512. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T. Curr Biol. 1995;5:1416–1423. doi: 10.1016/s0960-9822(95)00279-x. [DOI] [PubMed] [Google Scholar]
- 33.Wilson JJ, Kovall RA. Cell. 2006;124:985–996. doi: 10.1016/j.cell.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 34.Nam Y, Sliz P, Song L, Aster JC, Blacklow SC. Cell. 2006;124:973–983. doi: 10.1016/j.cell.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 35.Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Nat Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- 36.Armstrong J, Sperling A, Deuring R, Manning L, Moseley S, Papoulas O, Piatek C, Doe CQ, Tamkun JW. Genetics. 2005;170:1761–1774. doi: 10.1534/genetics.105.041327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rones MS, McLaughlin KA, Raffin M, Mercola M. Development (Cambridge, UK) 2000;127:3865–3876. doi: 10.1242/dev.127.17.3865. [DOI] [PubMed] [Google Scholar]
- 38.Schroeder T, Fraser ST, Ogawa M, Nishikawa S, Oka C, Bornkamm GW, Nishikawa S, Honjo T, Just U. Proc Natl Acad Sci USA. 2003;100:4018–4023. doi: 10.1073/pnas.0438008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 40.Oka C, Nakano T, Wakeham A, de la Pompa JL, Mori C, Sakai T, Okazaki S, Kawaichi M, Shiota K, Mak TW, Honjo T. Development (Cambridge, UK) 1995;121:3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- 41.Nonaka S, Shiratori H, Saijoh Y, Hamada H. Nature. 2002;418:96–99. doi: 10.1038/nature00849. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.