Abstract
Biofilms, in which cells attach to surfaces and secrete slime (polymeric substances), are central to microbial life. Biofilms are often thought to require high levels of cooperation because extracellular polymeric substances are a shared resource produced by one cell that can be used by others. Here we examine this hypothesis by using a detailed individual-based simulation of a biofilm to investigate the outcome of evolutionary competitions between strains that differ in their level of polymer production. Our model includes a biochemical description of the carbon fluxes for growth and polymer production, and it explicitly calculates diffusion–reaction effects and the resulting solute gradients in the biofilm. An emergent property of these simple but realistic mechanistic assumptions is a strong evolutionary advantage to extracellular polymer production. Polymer secretion is altruistic to cells above a focal cell: it pushes later generations in their lineage up and out into better oxygen conditions, but it harms others; polymer production suffocates neighboring nonpolymer producers. This property, analogous to vertical growth in plants, suggests that polymer secretion provides a strong competitive advantage to cell lineages within mixed-genotype biofilms: global cooperation is not required. Our model fundamentally changes how biofilms are expected to respond to changing social conditions; the presence of multiple strains in a biofilm should promote rather than inhibit polymer secretion.
Keywords: sociobiology, bacteria, individual-based modeling
It is now a common perception that the vast majority of bacterial life in nature is found in surface-bound communities called biofilms rather than in isolated planktonic cells (1, 2). The importance of biofilms is underlined by their role in many chronic diseases, antibiotic resistance, biofouling, and waste-water treatment (3, 4). However, it is only recently that the consequences of the proximity of individuals in these communities has been approached from a social-evolution perspective (5–11). Biofilms are densely packed systems in which cells share many secreted molecules, including enzymes, iron-scavenging siderophores, and extracellular polymers. This sharing of resources suggests that they are open to the evolution of “cheater” strains that use the resources of others but do not contribute to the group pool (6, 12–14). In particular, when many unrelated strains mix in biofilms, strong evolutionary conflicts are predicted over contribution to shared group resources (6, 7).
A defining feature of many biofilms is the matrix or slime that surrounds the cells (1, 2). These extracellular polymeric substances (EPS) are present in many biofilms where they embed bacteria. Despite the ubiquitous nature of polymer production, the exact evolutionary benefits it provides are unknown. However, polymers are typically viewed as a shared resource that provides a benefit to the biofilm by maintaining its structure (15, 16); protecting it from aggressive agents such as dehydration, ultraviolet radiation, and predator grazing (5–10, 17); and facilitating extracellular enzymatic activity and signaling (6). In the first model of its kind, Kreft (5) analyzed the success of strains with different rates of substrate uptake in mixed biofilms. He showed that slow-growth, high-yield strains could be exploited by high-growth, low-yield strains, and he concluded that cooperation and altruism were necessary for optimal biofilm formation. Like slow substrate uptake, polymer production reduces the available energy for growth. What then maintains polymer production in the face of selection for competition and rapid growth?
Perfect cooperation in biofilms is predicted when they contain a single strain (high genetic relatedness) (5–7). Genetically identical cells, as occurs in most multicellular organisms, do not have evolutionary conflicts of interest, and they are predicted to behave simply as is optimal for the group (18–20). We currently know very little of within-species genetic diversity in biofilms. However, given the huge diversity of species that can occur within biofilms (21–23), it seems unlikely that all or indeed most biofilms will have the clonal structure necessary to generate perfect cooperation throughout. We, therefore, investigated whether cooperation was necessary for polymer production in an individual-based simulation.
Our model investigates the outcome of evolutionary competitions between strains that differ in their level of extracellular polymer production [see supporting information (SI) Fig. 6]. Fig. 1 represents the carbon fluxes involved from the uptake of a substrate (e.g., glucose) to the formation of the end-product biomass (X) and EPS. A central parameter is the investment in polymer production, f, or more specifically, the carbon dedicated to polymer production:
where the r terms capture the rate of EPS and biomass production (for a full list of terms, see Table 1). Further assumptions are needed to characterize the system fully. Substrate uptake is assumed to be the rate-limiting step, and it is described by double-saturation kinetics (Table 2)
The concentration of all intermediaries is assumed to be at steady state. The yield of carbon that goes into organic matter production, i.e., X and EPS, is constant, and the remaining carbon is released as CO2, hence,
where Y is the yield coefficient for use of carbon. The oxygen consumption is related to CO2 consumption by a second yield coefficient, YO,
Having defined the system, mass conservation for carbon produces the following equations for the rate of biomass and EPS,
and
Biofilms consist of densely packed aggregates inside of which fluid flow is reduced and where most of the solute transport to and from the bacteria occurs through diffusion. The dimensions of typical biofilms are large enough that the diffusion of solutes through the matrix is slow compared with the bioconversions occurring, creating well known solute gradients (24). In aerobic biofilms, for example, gradients in oxygen concentration are formed, and consequently cells at the biofilm surface experience more favorable conditions than cells in the interior where conditions are anoxic (25). Importantly, it might be expected that polymer production, which alters the structure and density of the biofilm, will have important effects on the solute gradients and resulting concentrations of resources experienced by cells.
Fig. 1.
Simplified representation of the pathway for growth on glucose of an EPS-producing strain (EPS+) (a) and a corresponding nonproducer (EPS−) (b). Triple dots represent pathway intermediaries whose concentration is assumed to be stationary. In this pathway, glucose uptake is the rate-limiting step, and it has the same expression for both EPS+ and EPS− strains. EPS− has a higher intrinsic growth rate by directing its carbon flux to biomass, as opposed to dedicating part of the flux to EPS synthesis as in the case of EPS+.
Table 1.
Notation summary
| Symbol | Description | Dimensions |
|---|---|---|
| ρEPS | Density of EPS | MCL−3 |
| ρX | Density of biomass | MCL−3 |
| f | Ratio of EPS produced per biomass formed (based on carbon mass) | |
| IS1→S2 | Fitness at invasion of S1 (rare mutant) into strain S2 | |
| KS | Half-saturation constant for substrate (glucose) concentration | MCL−3 |
| KO2 | Half-saturation constant for oxygen concentration | MOL−3 |
| NEPS−,t | No. of individuals of EPS− at time t | |
| NEPS+,t | No. of individuals of EPS+ at time t | |
| [O2] | Concentration of oxygen | MOL−3 |
| qS, max | Maximum specific rate of substrate uptake | T−1 |
| rCO2 | Rate of CO2 production | MCL−3T−1 |
| rEPS | Rate of EPS production | MCL−3T−1 |
| rS | Rate of substrate (glucose) uptake | MCL−3T−1 |
| rX | Rate of biomass formation | MCL−3T−1 |
| [S] | Concentration of substrate (glucose) | MCL−3 |
| wEPS− | Fitness of EPS− strain | |
| wEPS+ | Fitness of EPS+ strain | |
| X | Concentration of biomass | MCL−3 |
| XEPS− | Concentration of EPS− biomass | MCL−3 |
| XEPS+ | Concentration of EPS+ biomass | MCL−3 |
| Y | Yield of carbon from substrate that is used to produce biomass or EPS | |
| YO | Yield of oxygen consumed per CO2 produced | MOMC−1 |
Masses of substrate (glucose), CO2, biomass, and EPS are represented as mass of carbon (MC)O. MO represents mass of oxygen, L represents length, and T represents time.
Table 2.
Stoichiometric table for bioprocesses included in the model
| Reaction | Solutes |
Particulates |
Rate expression | ||||
|---|---|---|---|---|---|---|---|
| S | O2 | CO2 | XEPS− | EPS | XEPS− | ||
| Growth of EPS+ | −1 | −YO(1 − Y) | (1 − Y) | ||||
| Growth of EPS− | −1 | −YO(1 − Y) | (1 − Y) | Y | |||
We investigated the fitness effects of extracellular polymer production in biofilms for different levels of investment in polymer (f) and polymer density (ρX/ρEPS, ratio of cell to polymer density). The complex interplay of physical and biological processes in this system lends it to explicit mechanistic models. Therefore, we apply individual-based modeling to carry out simulations where the success of polymer-producing traits is evaluated. The model used is based on software developed for multispecies models of biofilms (26) that applies the individual-based concept to biofilms (27) and allows any number of bacterial and chemical species to be defined. The numerical methods and other algorithms implemented in the model framework have been extensively described in refs. 26–28 and applied for several applications in environmental biotechnology (28–31), biofilm control (32), and ecology (5). The principles and numerics are explained in the SI Text and SI Figs. 6 and 7. Parameters in the model are based on empirically estimated values (Table 3). Following Kreft (5), the first set of simulations considers direct competition in an initially mixed biofilm with equal starting numbers of two strains: a nonpolymer producer (EPS−) and a polymer-producing strain (EPS+). All else being equal, the former grows faster because it invests all of its carbon in biomass (Fig. 1b). This first set of simulations serves as a first evaluation of the range of values of f and of ρX/ρEPS for which polymer production might provide an advantage within a particular biofilm.
Table 3.
Parameters used in simulations
| Symbol | Description | Value | Notes/ref. |
|---|---|---|---|
| qS,max | Maximum specific substrate uptake rate | 1.02 h−1 | Calculated from values from (48)* |
| KO | Half-saturation constant for oxygen | 1.18 × 10−3 gO2/liter | (48)* |
| Y | Substrate yield | 0.44 | (48)* |
| YO | Yield of oxygen consumed per CO2 produced | 2.66 gO/gC | From the stoichiometry of CO2 production |
| DO2 | Diffusivity of oxygen in water and in biofilm* | 8.33 × 106 μm2h−1 | (30)† |
| CO,env | Environment concentration of oxygen (concentration in the bulk liquid) | 8 × 10−3 go/liter except where noted | Value typically used for saturation concentration of oxygen dissolved in water at 20°C |
| ρX | Biomass density in the biofilm | 200 gC/liter | From typical values enumerated in (49) |
*For Pseudomonas aeruginosa growing on glucose.
†Diffusivity of small solute species in the biofilm matrix may be assumed to be the same as in water for modeling purposes because matrix is typically 97% water (50).
A relative advantage within individual biofilms, however, may not constitute an evolutionary advantage if faster relative growth comes at a large cost to total productivity and biofilm size. To account for such effects we also perform an evolutionary analysis that assesses whether a rare mutant that produces polymers can invade a population of nonproducers and vice versa.
Results and Discussion
Simple Competition of EPS+ and EPS−.
Direct-competition simulations were initialized with an equal amount of individuals of each strain placed randomly at the solid substratum. We observed the outcome after 20 days of growth (Fig. 2, SI Movies 1–3, and http://sysbio.harvard.edu/csb/foster/biofilmeps/). Fitness of both EPS+ and EPS− was then computed as the average number of cell divisions occurring during this time:
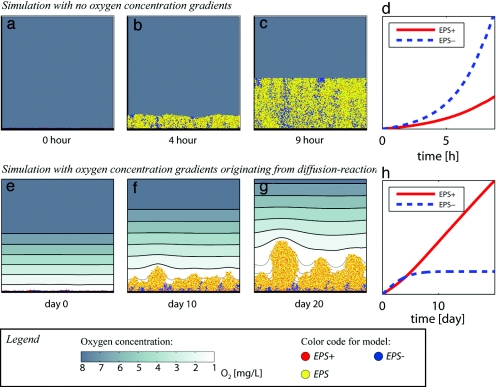
Fig. 3 shows that the outcome of competition depends on the value of EPS production (f) and the density of EPS (ρX/ρEPS ratio). For high EPS densities (ρX/ρEPS = 1) EPS+ producers lose the competition against EPS−, independently of the value of f. However, for low EPS densities (ρX/ρEPS >1), there is a broad range of values of f for which EPS+ wins, despite the higher intrinsic growth rate of EPS−. This observation is illustrated in Fig. 2, which contrasts the effects of EPS production, with and without solute gradients. The upper simulation (a–d) was carried out by shutting off the diffusion–reaction component of our model to simulate the unrealistic case where no solute gradients exist. In these conditions, the competition is purely driven by growth rate, and EPS− wins. The lower simulation (e–h) was carried out with the diffusion–reaction back on. The resulting gradients now give EPS+ an advantage of rapid progression toward the top of the biofilm.
Fig. 2.
Direct competition between an EPS+ and EPS− showing the importance of oxygen gradients in the outcome of the competition. (a–d) Simulation (a–c) and competition outcome (d) in the unrealistic case of absence of oxygen gradients. In this case, competition is decided purely by growth rate, and the fast-growing strain (EPS−) wins. (e–h) Simulation (e–g) and competition outcome (h) in the presence of oxygen gradients, showing the advantage to the EPS+ strain. Note, however, that the EPS+ strains only gain an advantage after a few days of growth so that for transiently formed biofilms, the EPS+ strategy will not be favored. (a–c and e–g) Oxygen concentration is shown in the background, where thick isoconcentration lines are shown for 1 mg/liter steps, and thin gray isoconcentration lines represent order of magnitude changes in the concentration (i.e., 0.1 mg/liter, 0.01 mg/liter, 0.001 mg/liter, etc.). These simulations where carried out at f = 0.55 for the EPS+ strain and ρX/ρEPS = 6. See also SI Movies 1–3.
Fig. 3.
Relative growth rate of an EPS+ in a biofilm with an equal starting frequency of an EPS−. Competition outcome depends on both the rate of EPS production of the producing strain and the density of EPS (ρX/ρEPS). Importantly, the EPS− often loses despite its higher intrinsic growth rate. Each box and whisker set results from 10 replicate simulations. Lines are a curve fitting using results from the 10 replicates. The dashed line represents the fitness of value 1, i.e., the borderline value above which the competition is won by EPS+.
Rare-Mutant Invasion.
The competitive advantage of the EPS+ strain in a biofilm is a proof of principle that polymer production can provide an advantage in mixed biofilms (Fig. 2). However, to evaluate whether polymer production is evolutionarily stable, we performed an invasion analysis that asks when a rare mutant that produces polymer will invade a population of nonproducers. Furthermore, we ask the reverse question to evaluate whether a successful polymer-producing strategy can resist evolutionary reinvasion from nonproducers. We evaluate fitness at invasion from
where, following Eq. 8, wS1 is the fitness of a rare mutant in competition with the strategy S2, which is the majority in the population. Importantly though, 〈wS2〉 is not the fitness of the cells in direct competition with the rare mutant but rather the mean fitness of all S2 individuals in the population. With an assumption of large population sizes, we take this measure to equal the fitness in a biofilm containing only S2 cells, which assumes that cells go through a phase of global competition at the whole-population level (33), i.e., that cells disperse randomly after growth in a biofilm such that the more cells of a strain present in a biofilm, the higher the contribution to subsequent biofilms. Eq. 8 then evaluates whether the rare mutant reproduces more rapidly than an average cell of the majority strategy in the population and therefore its ability to invade. Both the invasiveness of EPS+ over EPS− was computed (EPS+ → EPS−) and vice versa (EPS− → EPS+).
We investigate invasion for different initial frequencies of the rare mutant, which captures the effect of different numbers of strain randomly settling during biofilm initiation. For example, if many different strains are always present, then the initial frequency of a rare mutant will be low: with 10 strains, a rare mutant will start at a frequency of 0.1 in the biofilm. Of course, when a mutant first arises, its proportion in a biofilm will depend on when exactly the mutation occurred. However, this first biofilm will not determine invasion success, so we consider only the subsequent biofilms, in which its initial frequency is defined precisely by the number of strains at biofilm initiation.
The presence of many strains at biofilm initiation favors invasion of polymer-producing strains (Fig. 4). Invasion is also strongly dependent on the polymer density and most favored at low densities (high ρX/ρEPS). Interestingly, for some conditions, we find both that EPS+ can invade an EPS− population and the reverse, which is characteristic of negative frequency-dependent selection (34) and predicts the existence of genetic polymorphism under some conditions, in which both EPS+ and EPS− strains coexist stably in the population.
Fig. 4.
Rare-mutant invasion analysis. (a) Invasion of a rare polymer-producing strain (EPS+) into a population of nonproducers (EPS−). (b) Invasion of a rare EPS− strain into a population of EPS+ strains. The x axis captures the number of strains randomly settling to form biofilms, where increased strains (and reduced within-group relatedness) cause a lower initial frequency of the rare mutant. For example, if 10 strains are present in all biofilms, the initial frequency of a rare mutant will be 0.1. All simulations were carried out at f = 0.5. Each box and whisker set results from 20 replicate simulations.
Another way to view the number of strains in a biofilm is in terms of mean genetic relatedness (6, 7, 19). For many strains, relatedness to the average cell in the biofilm will be low, whereas with few strains, relatedness will be high (6). More precisely, if 10 random strains from the population found a biofilm, then mean relatedness of any cell to other cells in the biofilm at initiation will be 0.1 (1/10 of cells will be clonemates of relatedness 1, and 9/10 will be nonclonemates with relatedness 0). Importantly, however, clonal growth in the biofilm means that although relatedness is low across the whole biofilm, it can be locally high (6, 35). These effects of scale are important in the outcome of the simulation, and they are discussed in the next section.
Why Do Polymer Producers Win?
Polymer-producing cells invade most easily when many strains found a biofilm (Fig. 4); that is, polymer producers perform best with low relatedness at biofilm initiation (6, 7, 19). Indeed, fitness is lower in polymer producers than nonproducers in single-strain biofilms (rightmost bar of Fig. 4a), which suggests that polymer production is selected specifically because it provides a selfish competitive advantage to strains in mixed biofilms. To understand better the fitness effects of polymer production, we modeled a single polymer-producing cell in a field of nonproducers (Fig. 5), which was used to calculate the fitness effects of the EPS+ strategy on the lineage of the focal cell and on lineages of EPS− cells at increasing distance from the focal cell.
Fig. 5.
Polymer production by a founding cell is altruistic to the cells above but spiteful to adjacent cells (38, 39). Simulations were initiated with a single polymer producer (EPS+) surrounded by nonproducers (EPS−) (a) Plot of fitness of founding cells after 20 days of biofilm growth. The lineage of the focal cell (rightmost bar) benefits greatly from polymer production, whereas neighboring cells are suffocated and have reduced fitness. Fitness is calculated as the total number of cells that result from a focal cell in the mixed biofilm, and it is shown here as relative to a cell in a biofilm of pure nonpolymer producers (EPS−; dotted line). (b) Polymer producers are typically very successful in the mixed biofilms. (c) In a minority of cases, the polymer producers are smothered by nonproducers, which explains the high variance in the fitness of the focal polymer-producing cell (a). (d) Polymer production in biofilm has a strong analogy with growth in plants, where competition for light favors vertical growth.
The advantage to polymer production in the focal cell came from differential effects on cells lying above versus laterally to the founding focal cell. For cells lying above the focal, polymer production is altruistic and favored by kin selection (6); that is, the focal EPS cell pays a cost by dividing more slowly than the EPS− cells at the beginning of the simulation (Fig. 2h), but it gains a fitness benefit through relatives because the polymer pushes its descendents up and out into oxygen-rich conditions where there is also reduced competition for space (Fig. 5). This behavior is altruistic because by producing polymer, the focal cell is lowering its rate of division to help other cells to divide. Note that this form of altruism occurs through cells helping descendent cells in the same lineage; that is, it is altruism to the extent that actions that reduce an individual's personal reproduction to help existing offspring reproduce are also considered altruistic (36, 37).
Strong local competition can prevent altruism among relatives (33). However, the upward expansion of the biofilm reduces local vertical and horizontal competition for space within a lineage, and it favors their cooperation. There is strong lateral competition at biofilm initiation, however, which can be seen by the reduced fitness of cells lying horizontal to the polymer-producing lineage (Fig. 5). For these cells, polymer production can be considered spiteful: the focal cell lowers its reproductive rate to produce polymer that reduces the fitness of the lateral neighboring cells (38, 39). The extracellular polymer, therefore, provides a competitive advantage to a lineage by allowing its cells to rise up and over other cells and suffocate them. This suffocation effect also explains the frequency-dependent nature of the advantage, whereby polymer producers invade most easily in biofilms containing many nonproducers (Fig. 4). As the frequency of polymer producers increases in a biofilm, they have less opportunity to overgrow other lineages (increased competition for oxygen and space), which reduces the benefit of polymer production (compare Figs. 2g and 5b).
Conclusions
Cooperative and altruistic adaptations present a significant problem for evolutionary biology because theory predicts that selection for selfishness and cheating should often undermine group behaviors (6, 18, 40, 41). Microbial biofilms are widely viewed as an example of this problem (5–8, 10, 42, 43) because the production of EPS by one cell that can be used by another would seem to be an exploitable resource. By using a simple model of mixed-strain biofilms that incorporates realistic diffusion–reaction effects, here we show that a strong advantage to polymer production arises as an emergent property (44). Secretion of extracellular polymers by a cell allows it altruistically to push descendents into a more oxygen-rich environment. At the same time, it provides a strong competitive advantage at the scale of the cell lineage by suffocating neighboring nonproducers. The effect of polymer production, therefore, has a strong analogy in plants competing for light, where vertical growth and increased foliage area selfishly increase access to light at the expense of competitors (45). The polymer-producing cells that initiate a biofilm are analogous to a tree trunk that allows descendent cells to raise up and outward into the best growth conditions and overgrow the rest (Fig. 5).
Although phrased in terms of oxygen gradients, comparable results can be expected whenever the biofilm forms a barrier to a limiting resource, and an analogous process may also occur in the initiation of biofilms that float on the surface of liquids (14). Our simulations suggest that cells must secrete low-density polymers to gain an advantage. We are unaware of any empirical estimates of extracellular polymer density; but for the values assumed in previous simulations (26, 31, 46, 47), polymer producers would readily invade (ρX/ρEPS = 6; Fig. 4). Another important point is that the advantage to polymer production increases with biofilm age (Fig. 2h). Our model, therefore, predicts that polymer production is most likely to be favored when biofilms last for several days. Finally, polymer production is not expected to increase indefinitely. Modeling the evolution of polymer production as a continuous trait predicts that it will stabilize at an intermediate equilibrium level (see SI Text and SI Fig. 8).
We do not wish to suggest that extracellular polymer production in biofilms will always be selected through within-biofilm competition. Clearly, under some conditions, the polymers may provide global benefits that help both the lineage that produces it and others in the biofilm, such as protection against antimicrobials or desiccation. Nevertheless, our model shows that one should not uncritically assume that biofilms with their matrix of secreted polymers are purely cooperative. These polymers may often provide competitive benefits to lineages by allowing them to suffocate unrelated cells of their own or other species (26).
Why is it important to know whether polymer production requires global cooperation within biofilms? The fitness effects of a trait define how it responds to changing social conditions over ecological and evolutionary time (6). For example, the predicted effect of mixing strains together is reversed between cooperative and competitive traits (7, 48, 49). The standard assumption that extracellular polymers are purely cooperative predicts that the presence of multiple strains will decrease the matrix and, potentially, antimicrobial resistance (7). Our model predicts the opposite: that mixed-strain biofilms will tend to have increased polymer production. General support for this prediction comes from the recent discovery that mixing genetically different strains of the bacterium Pseudomonas aeruginosa results in more robust biofilms (E. Martinez-Garcia, R. Kolter, and K.R.F., unpublished data). The “cooperative” behavior of biofilm-forming bacteria may not be so cooperative after all.
Methods
The individual-based approach used for simulations is a multiagent description of biofilm dynamics where agents (cells) behave independently according to rules mimicking the behavior of a bacterial cell, including growth, division, and polymer production and excretion (26–28). The beginning of each simulation starts with cells adhered to a surface. These cells then grow in a two-dimensional computational space where movement occurs through shoving of neighboring cells, which do not overlap. The dynamics of the biofilm community is emergent from the interactions at the cell scale. Cell growth and polymer production rates are strain-specific following the equations defined in Table 2. The effects of local nutrient concentrations are included in the simulation, which are computed by solving partial differential equations describing diffusion–reaction at each step of the simulation. For an extended description of the model, see SI Text.
Supplementary Material
Acknowledgments
We thank Angus Buckling, Ashlee Earl, Esteban Martinez Garcia, Joan Strassmann, Stuart West, and two anonymous referees for thoughtful comments. This work was supported by National Institute of General Medical Sciences Center of Excellence Grant 5P50 GM 068763-01 (to K.R.F.).
Abbreviations
- EPS
extracellular polymeric substances
- EPS+
EPS-producing strain
- EPS−
non-EPS-producing strain.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607651104/DC1.
References
- 1.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappinscott HM. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 2.Kolter R, Greenberg EP. Nature. 2006;441:300–302. doi: 10.1038/441300a. [DOI] [PubMed] [Google Scholar]
- 3.Fux CA, Costerton JW, Stewart PS, Stoodley P. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 4.Heijnen JJ, van Loosdrecht MCM, Mulder R, Weltevrede R, Mulder A. Wat Sci Technol. 1993;27:253–261. [Google Scholar]
- 5.Kreft JU. Microbiology. 2004;150:2751–2760. doi: 10.1099/mic.0.26829-0. [DOI] [PubMed] [Google Scholar]
- 6.West SA, Griffin AS, Gardner A, Diggle SP. Nat Rev Microbiol. 2006;4:597–607. doi: 10.1038/nrmicro1461. [DOI] [PubMed] [Google Scholar]
- 7.Foster KR. Science. 2005;308:1269–1270. doi: 10.1126/science.1108158. [DOI] [PubMed] [Google Scholar]
- 8.Crespi BJ. Trends Ecol Evol. 2001;16:178–183. doi: 10.1016/s0169-5347(01)02115-2. [DOI] [PubMed] [Google Scholar]
- 9.Keller L, Surette MG. Nat Rev Microbiol. 2006;4:249–258. doi: 10.1038/nrmicro1383. [DOI] [PubMed] [Google Scholar]
- 10.Velicer GJ. Trends Microbiol. 2003;11:330–337. doi: 10.1016/s0966-842x(03)00152-5. [DOI] [PubMed] [Google Scholar]
- 11.Foster KR, Parkinson K, Thompson CRL. Trends Genet. 2007 doi: 10.1016/j.tig.2006.12.003. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffin AS, West SA, Buckling A. Nature. 2004;430:1024–1027. doi: 10.1038/nature02744. [DOI] [PubMed] [Google Scholar]
- 13.Fiegna F, Yu YT, Kadam SV, Velicer GJ. Nature. 2006;441:310–314. doi: 10.1038/nature04677. [DOI] [PubMed] [Google Scholar]
- 14.Rainey PB, Rainey K. Nature. 2003;425:72–74. doi: 10.1038/nature01906. [DOI] [PubMed] [Google Scholar]
- 15.Boyd A, Chakrabarty AM. Appl Environ Microbiol. 1994;60:2355–2359. doi: 10.1128/aem.60.7.2355-2359.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hughes KA, Sutherland IW, Jones MV. Microbiology. 1998;144:3039–3047. doi: 10.1099/00221287-144-11-3039. [DOI] [PubMed] [Google Scholar]
- 17.Hall-Stoodley L, Costerton JW, Stoodley P. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 18.Foster KR. J Evol Biol. 2004;17:1058–1072. doi: 10.1111/j.1420-9101.2004.00747.x. [DOI] [PubMed] [Google Scholar]
- 19.Hamilton WD. J Theor Biol. 1964;7:1–16. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- 20.Frank SA. The Foundations of Social Evolution. Princeton: Princeton Univ Press; 1998. [Google Scholar]
- 21.Kolenbrander PE, Andersen RN, Blehert DS, Egland PG, Foster JS, Palmer RJ. Microbiol Mol Biol Rev. 2002;66:486–505. doi: 10.1128/MMBR.66.3.486-505.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolenbrander PE, Egland PG, Diaz PI, Palmer RJ. Trends Microbiol. 2005;13:11–15. doi: 10.1016/j.tim.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Palmer RJ, Jr, Kazmerzak K, Hansen MC, Kolenbrander PE. Infect Immun. 2001;69:5794–5804. doi: 10.1128/IAI.69.9.5794-5804.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Characklis WG, Turakhia MH, Zelver N. In: Biofilms. Characklis WG, Marshall KC, editors. New York: Wiley Interscience; 1990. pp. 316–340. [Google Scholar]
- 25.De Beer D, Stoodley P, Roe F, Lewadowski Z. Biotech Bioeng. 1994;43:1131–1138. doi: 10.1002/bit.260431118. [DOI] [PubMed] [Google Scholar]
- 26.Xavier JB, Picioreanu C, van Loosdrecht MCM. Environ Microbiol. 2005;7:1085–1103. doi: 10.1111/j.1462-2920.2005.00787.x. [DOI] [PubMed] [Google Scholar]
- 27.Kreft JU, Picioreanu C, Wimpenny JWT, van Loosdrecht MCM. Microbiology. 2001;147:2897–2912. doi: 10.1099/00221287-147-11-2897. [DOI] [PubMed] [Google Scholar]
- 28.Picioreanu C, Kreft JU, van Loosdrecht MCM. Appl Environ Microbiol. 2004;70:3024–3040. doi: 10.1128/AEM.70.5.3024-3040.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Picioreanu C, Batstone DJ, van Loosdrecht MCM. Wat Sci Technol. 2005;52:501–507. [PubMed] [Google Scholar]
- 30.Xavier JB, Picioreanu C, van Loosdrecht MCM. Biofilms. 2004;1:377–391. [Google Scholar]
- 31.Xavier JB, Picioreanu C, van Loosdrecht MCM. Biotechnol Bioeng. 2005;91:651–669. doi: 10.1002/bit.20544. [DOI] [PubMed] [Google Scholar]
- 32.Xavier JB, Picioreanu C, Rani SA, van Loosdrecht MC, Stewart PS. Microbiology. 2005;151:3817–3832. doi: 10.1099/mic.0.28165-0. [DOI] [PubMed] [Google Scholar]
- 33.West SA, Pen I, Griffin AS. Science. 2002;296:72–75. doi: 10.1126/science.1065507. [DOI] [PubMed] [Google Scholar]
- 34.Waxman D, Gavrilets S. J Evol Biol. 2005;18:1139–1154. doi: 10.1111/j.1420-9101.2005.00948.x. [DOI] [PubMed] [Google Scholar]
- 35.West SA, Buckling A. Proc R Soc London Ser B. 2003;270:37–44. doi: 10.1098/rspb.2002.2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehmann L. J Evol Biol. 2006 doi: 10.1111/j.1420-9101.2006.01202.x. [DOI] [Google Scholar]
- 37.Foster KR, Ratnieks FL. Trends Ecol Evol. 2005;20:363–364. doi: 10.1016/j.tree.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 38.Foster KR, Wenseleers T, Ratnieks FLW. Ann Zool Fenn. 2001;38:229–238. [Google Scholar]
- 39.Lehmann L, Bargum K, Reuter M. J Evol Biol. 2006;19:1507–1516. doi: 10.1111/j.1420-9101.2006.01128.x. [DOI] [PubMed] [Google Scholar]
- 40.Sachs JL, Mueller UG, Wilcox TP, Bull JJ. Q Rev Biol. 2004;79:135–160. doi: 10.1086/383541. [DOI] [PubMed] [Google Scholar]
- 41.Keller L, editor. Levels of Selection in Evolution. Princeton: Princeton Univ Press; 1999. [Google Scholar]
- 42.Klausen M, Gjermansen M, Kreft JU, Tolker-Nielsen T. FEMS Microbiol Lett. 2006;261:1–11. doi: 10.1111/j.1574-6968.2006.00280.x. [DOI] [PubMed] [Google Scholar]
- 43.Kreft JU, Bonhoeffer S. Microbiology. 2005;151:637–641. doi: 10.1099/mic.0.27415-0. [DOI] [PubMed] [Google Scholar]
- 44.Anderson C. Biol Bull. 2002;202:247–255. doi: 10.2307/1543475. [DOI] [PubMed] [Google Scholar]
- 45.Goodnight CJ, Schwartz JM, Stevens L. Am Nat. 1992;140:743–761. [Google Scholar]
- 46.Horn H, Neu TR, Wulkow M. Wat Sci Technol. 2001;43:121–127. [PubMed] [Google Scholar]
- 47.Alpkvist E, Picioreanu C, van Loosdrecht MC, Heyden A. Biotechnol Bioeng. 2006;94:961–979. doi: 10.1002/bit.20917. [DOI] [PubMed] [Google Scholar]
- 48.Brown SP, Hochberg ME, Grenfell BT. Trends Microbiol. 2002;10:401–405. doi: 10.1016/s0966-842x(02)02413-7. [DOI] [PubMed] [Google Scholar]
- 49.Brown SP. Proc R Soc London Ser B. 1999;266:1899–1904. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.