Abstract
d-Mannitol is the predominant carbon compound in conidiospores of the filamentous fungus Aspergillus niger and makes up 10 to 15% of the dry weight. A number of physiological functions have been ascribed to mannitol, including serving as a reserve carbon source, as an antioxidant, and to store reducing power. In this study, we cloned and characterized the A. niger mpdA gene, which encodes mannitol 1-phosphate dehydrogenase (MPD), the first enzyme in the mannitol biosynthesis pathway. The mpdA promoter contains putative binding sites for the development-specific transcription factors BRLA and ABAA. Furthermore, increased expression of mpdA in sporulating mycelium suggests that mannitol biosynthesis is, to a certain extent, developmentally regulated in A. niger. Inactivation of mpdA abolished mannitol biosynthesis in growing mycelium and reduced the mannitol level in conidiospores to 30% that in the wild type, indicating that MPD and mannitol 1-phosphate phosphatase form the major metabolic pathway for mannitol biosynthesis in A. niger. The viability of spores after prolonged storage and germination kinetics were normal in an mpdA null mutant, indicating that mannitol does not play an essential role as a reserve carbon source in A. niger conidia. However, conidiospores of a ΔmpdA strain were extremely sensitive to a variety of stress conditions, including high temperature, oxidative stress and, to a lesser extent, freezing and lyophilization. Since mannitol supplied in the medium during sporulation repaired this deficiency, mannitol appears to be essential for the protection of A. niger spores against cell damage under these stress conditions.
Polyols or polyhydroxyalcohols are present in all organisms, from bacteria to animals. In particular, plants and fungi are known to accumulate high levels of polyols intracellularly, up to several hundred millimoles per liter. The filamentous fungus Aspergillus niger produces a number of different polyols, including glycerol, erythritol, and d-mannitol (41). The intracellular concentrations of the individual polyols in A. niger depend on growth conditions and developmental stage, suggesting that polyols have important functions in fungal physiology.
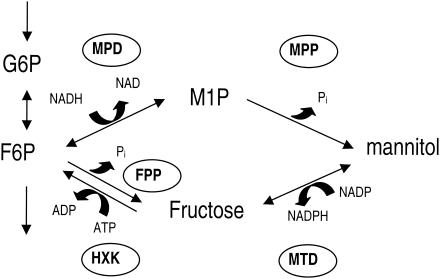
The hexitol d-mannitol is accumulated by vascular plants (37) and many fungal species (21). In A. niger conidiospores, d-mannitol is the predominant carbon-containing compound and makes up 10 to 15% of the dry weight (41). The metabolic pathway for mannitol biosynthesis in ascomycete and deuteromycete fungi most likely consists of two steps (19) (Fig. 1). Fructose 6-phosphate is reduced to mannitol 1-phosphate by NAD+-dependent mannitol 1-phosphate dehydrogenase (MPD), followed by the dephosphorylation of mannitol 1-phosphate, which is catalyzed by mannitol 1-phosphate phosphatase (MPP), yielding mannitol. The presence of MPD and MPP in several fungi, such as A. niger (22), Aspergillus nidulans (34), and Alternaria alternata (18), has been demonstrated, but these enzymes are also responsible for mannitol biosynthesis in protozoan parasites of the genus Eimeria (32). In contrast, the basidiomycete Agaricus bisporus probably produces mannitol from fructose via a reduction step catalyzed by NADP+-dependent mannitol dehydrogenase (MTD) (36). In ascomycete and deuteromycete fungi, however, MTD is believed to be involved in the first step in the catabolism of mannitol and thus operates in the direction opposite that in A. bisporus in vivo. The phosphorylation of fructose by hexokinase completes the mannitol cycle (Fig. 1) originally proposed by Hult and Gatenbeck (18). Genes encoding enzymes with MPD activity have been cloned from Cryptococcus neoformans (38), Eimeria tenella (GenBank accession no. AF055716; see also reference 32), and numerous bacteria.
FIG. 1.
Mannitol metabolism in A. niger. The two possible pathways for mannitol biosynthesis from fructose 6-phosphate, the pathway for mannitol catabolism, and part of the glycolytic pathway are indicated. G6P, glucose 6-phosphate; F6P, fructose 6-phosphate; M1P, mannitol 1-phosphate; HXK, hexokinase; FPP, fructose 6-phosphate phosphatase.
Physiological functions which have been ascribed to mannitol include serving as a reserve carbon source, as an antioxidant, to store reducing power, as a carbon compound which is transported through hyphae, and in redox balancing. The high concentrations of mannitol in conidia of A. niger (41) and several other fungi, such as Aspergillus oryzae (17) and Aspergillus clavatus (8), support a role in the survival of spores. During spore germination in A. niger, mannitol is rapidly metabolized, suggesting that it plays a role in the storage of carbon or reducing power (41). In the avian parasite E. tenella, mannitol is stored in unsporulated oocysts (32). Mannitol functions as the endogenous carbon and energy source to enable sporulation of the oocysts outside of the host (2). Similarly, in A. bisporus fruiting bodies, mannitol contributes up to 50% of the dry weight and is believed to be the main source of carbon and energy in the mushroom after harvest (16).
Mannitol is known to quench reactive oxygen species (ROS) (35), and there is some evidence that fungi use mannitol to prevent oxidative damage. An example is the phytopathogenic fungus A. alternata, which excretes mannitol during plant infection or in the presence of host plant extracts (20). This fungus appears to use mannitol as an antioxidant to suppress ROS-mediated plant defense. Likewise, the human fungal pathogen C. neoformans produces large quantities of mannitol in infected hosts (6). A mutant producing low quantities of mannitol is less virulent in mice and more susceptible to oxidative killing (5, 6). Heterologous expression of bacterial mtlD, encoding MPD, in a yeast gpd1 mutant results in mannitol accumulation and restores the growth of the mutant at high NaCl concentrations (7). Thus, mannitol can replace glycerol as a compatible solute during osmotic stress. Moreover, the mannitol-producing yeast strain is more resistant to oxidative killing, indicating that mannitol serves as an antioxidant.
Hult and Gatenbeck (18) have proposed that the mannitol cycle (Fig. 1) plays a role in NADPH production in the fungus A. alternata. The net result of mannitol cycle operation is transhydrogenase activity (NADH + NADP+ → NAD+ + NADPH) at the expense of ATP. The enzymes of the mannitol cycle are present in many fungi (19), but there is no evidence that mannitol turnover is required for NADPH production in any fungus. Singh et al. (34) did not find any correlation between the activities of the mannitol cycle enzymes and the need for NADPH in A. nidulans.
It is possible that mannitol has distinct functions in different organisms; unfortunately, however, no fungal mutants deficient in mannitol biosynthesis have been available to date in order to demonstrate unequivocally the role of mannitol in fungal physiology. In this report, we describe the cloning of the A. niger mpdA gene, encoding MPD; the disruption of the gene by gene replacement; and the physiological characterization of the ΔmpdA mutant.
MATERIALS AND METHODS
Strains, culture media, and growth conditions.
The A. niger strains used in this study were descendants of strain N400 (CBS 120.49) and are listed in Table 1. Somatic recombination was performed as described by Bos et al. (3). Unless stated otherwise, preparation of conidiospores was done by culturing for 3 to 4 days at 30°C on complete medium plates (27) containing 1% (wt/vol) glucose. Spores were harvested with a solution containing 0.05% (wt/vol) Tween 80 and 0.9% (wt/vol) NaCl. The spore suspension was filtered through sterile glass wool to remove mycelium. Spores were then washed by centrifugation of the spore suspension at 5,000 × g for 5 min and subsequent resuspension of the pellet in 0.05% (wt/vol) Tween 80-0.9% (wt/vol) NaCl at 108 to 1010 spores ml−1. The entire procedure for harvesting spores was performed at 4°C.
TABLE 1.
A. niger strains used in this study
| Strain | Genotype | Origin |
|---|---|---|
| N485 | fwnA1 trpA1 cspA1 | 3 |
| NW129 | goxC17 pyrA1 cspA1 | 29 |
| NW131 | goxC17 cspA1 | 29 |
| NW283 | fwnA1 pyrA13 lysA7 cspA1 creA4 | 30 |
| NW294 | ΔargB goxC17 pyrA6 cspA1 leuA1 ΔmpdA::argB+nicA1 | This study |
| NW302 | ΔargB goxC17 cspA1 leuA1 ΔmpdA::argB+ | This study |
| NW305 | ΔargB goxC17 cspA1 | This study |
| NW306 | ΔargB goxC17 cspA1 ΔmpdA::argB+ | This study |
| NW324 | ΔargB goxC17 pyrA6 cspA1 leuA1 nicA1 | P. A. vanKuyk et al., unpublished data |
Mycelium was cultured at 30°C in minimal medium (MM) containing, per liter, 6 g of NaNO3, 1.5 g of KH2PO4, 0.5 g of KCl, 0.5 g of MgSO4 · 7H2O, 0.03 g of yeast extract, 0.04 ml of a trace metal solution (40), and 20 g of glucose as a carbon source, unless indicated otherwise. Three-liter jacketed stirred tank reactors (Applikon) were used, and the culture pH was 4, unless indicated otherwise. Cultures were inoculated with 109 spores liter−1 and mixed at 500 rpm by using headspace aeration for 6 to 8 h to allow germination. Subsequently, sparging of the cultures with 2.5 liters of air min−1 was started, 0.5 ml of 30% (vol/vol) polypropylene glycol in alcohol was added per liter of medium as an antifoam agent, and pH and oxygen control was started. The culture pH was controlled by automatic addition of either 2 M HCl or 5 M NaOH, while the addition of pure oxygen was used to keep the dissolved oxygen tension above 30% air saturation. To analyze polyols in germinating spores, cultures were inoculated with 1010 spores liter−1. When necessary, the medium was supplemented with 0.2 g of arginine liter−1, 1 mg of nicotinamide liter−1, 0.2 g of tryptophan liter−1, 0.2 g of leucine liter−1, 0.365 g of lysine ml−1, and 1.22 g of uridine liter−1.
For analysis of the expression of mpdA in sporulating mycelium, 100 ml of MM containing 2% (wt/vol) glucose was inoculated with 106 spores ml−1 and incubated at 30°C without shaking. The mycelial mat developing at the liquid surface was harvested and analyzed. For analysis of the effect of a creA-derepressing mutation on mpdA expression, wild-type strain NW131 and creA mutant strain NW283 were precultured in MM containing 2% (wt/vol) fructose, transferred to MM containing 2% (wt/vol) glucose, and cultured for another 4 h before mycelium was harvested for Northern analysis.
Germination kinetics were analyzed by microscopic examination of slides coated with MM containing 1% (wt/vol) glucose and 1.5% (wt/vol) agar and spot inoculated with 2 × 104 conidiospores. The percentage of germinated spores was monitored over time by examining 50 to 100 spores at 30-min intervals. The viability of spores was determined with a suspension containing approximately 108 spores ml−1 in 0.05% (wt/vol) Tween 80- 0.9% (wt/vol) NaCl. Following different treatments (15 min to 2 h of incubation at 50°C, 1 h of incubation in the presence of 1 to 10 mM sodium hypochlorite [NaOCl], a freeze-thaw step, lyophilization, or 0 to 24 days of storage at 4 or 20°C), aliquots of the spore suspension were serially diluted in 0.05% (wt/vol) Tween 80- 0.9% (wt/vol) NaCl and plated on complete medium containing 1% (wt/vol) glucose. The numbers of colonies were counted after 24 h of growth at 30°C.
Complementation of Escherichia coli mtlD mutant JWL239 was performed with M9 minimal medium plates (31) supplemented with 50 mM mannitol, 100 μg of isopropyl-β-d-thiogalactopyranoside (IPTG) ml−1, 50 μg of ampicillin ml−1, thiamine, and leucine.
Extraction and analysis of polyols and mannitol 1-phosphate.
Spores and germinating spores for polyol analysis were harvested by filtration with 0.2-μm-pore-size filters. Polyol extraction and determination were performed as described by Witteveen et al. (42).
Sampling of mycelium and extraction of metabolites for analysis of mannitol 1-phosphate levels were performed as described previously (28). Mannitol 1-phosphate levels were determined with 50 mM triethanolamine- 5 mM MgCl2 (pH 7.6) in the presence of 1 mM NAD+, 1 mM NADP+, 3 U of glucose 6-phosphate dehydrogenase ml−1, 15 U of phosphoglucose isomerase ml−1, and 0.5 U of MPD ml−1. MPD is not commercially available and was therefore purified from an A. niger mpdA multicopy transformant (see below).
Preparation of cell extracts, enzyme assays, and purification of MPD.
Cell extracts were prepared as described previously (29). The extraction buffer for MPD and MTD contained 50 mM potassium phosphate (pH 7.0), 5 mM MgCl2, 5 mM 2-mercaptoethanol, and 0.5 mM EDTA, whereas for MPP and fructose 6-phosphate phosphatase, 50 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES) (pH 7.0) was used instead of potassium phosphate. Enzyme activities were determined at 30°C. MPD activity was determined with 50 mM Tris (pH 7.0) containing 0.2 mM NADH and 2.5 mM fructose 6-phosphate. MTD activity was determined with 100 mM glycine (pH 9.6) containing 0.5 mM NADP+ and 100 mM d-mannitol. MPP and fructose 6-phosphate phosphatase activities were determined with discontinuous assays. The phosphatase reactions were carried out with 25 mM PIPES (pH 7.5)-2.5 mM MgCl2 and 2 mM mannitol 1-phosphate or fructose 6-phosphate. Samples of the reaction mixture (200 μl) were taken at various time points and mixed with 760 μl of fourfold-diluted acid molybdate solution (Sigma). To this mixture, 40 μl of a solution containing 159 mg of Fiske-Subbarow reducer (Sigma) ml−1 was added. After 10 min of incubation at room temperature, phosphate concentrations were determined by reading the absorbance at 660 nm and were compared to values obtained for a phosphate standard series. Protein concentrations in cell extracts were determined with a bicinchoninic acid protein kit (Sigma) according to the supplier's instructions and with bovine serum albumin as a standard.
A. niger MPD was purified from transformant NW129::pIM4906-12, which overproduced the enzyme approximately 80-fold. A cell extract was prepared with extraction buffer, containing 10 mM bis-Tris (pH 6.0), 5 mM MgCl2, 1 mM 2-mercaptoethanol, and 0.5 mM EDTA, at 0°C. The suspension was centrifuged at 10,000 × g for 10 min, and the resulting supernatant was loaded into a 20-ml Procion Blue P-GR Sepharose CL-4B column. Following washing of the column with extraction buffer, MPD was eluted with extraction buffer containing 0.1 mM NADH. The enzyme was then concentrated by anion-exchange chromatography with a 1-ml ResourceQ column (Amersham Biotech) and stored at −70°C.
DNA manipulation and cloning of mpdA.
Standard methods (31) were used for DNA manipulations, such as Southern analysis, subcloning, DNA digestion, and plasmid DNA isolation. Chromosomal DNA was prepared as described previously (10). For Southern analysis, a 1.5-kb EcoRI-BglII mpdA fragment and a 0.7-kb EcoRV-SalI mpdA fragment were used as probes. Sequencing reactions were performed by using a Thermo-Sequenase fluorescence-labeled primer cycle sequencing kit (Amersham Biotech) with universal sequencing primers and a Cy5 Autoread sequencing kit (Amersham Biotech) with specific primers. Sequencing reactions were analyzed on an ALFexpress apparatus (Amersham Biotech). Sequence analysis was done by using a PCGENE package (Intelligenetics). The mpdA sequence was determined from both strands. Transformation of A. niger was performed as described by Kusters-van Someren et al. (24). For cotransformation, the pyrA gene (15) was used as a selective marker.
Complementation cloning of mpdA was performed by transformation of E. coli JWL239 (thi-1 leuB6 supE44 rpsL8 lacZ4 mtlAo mtlAp49 mtlD60 gut+ gatR49), provided by J. W. Lengeler, with an A. niger NW147 cDNA library (39). With one of the mpdA cDNAs being used as a probe, a genomic mpdA clone was obtained by screening of an A. niger N400 library in λEMBL4. A 2.9-kb NsiI-KpnI fragment, containing A. niger mpdA, was cloned in pGEM7, giving pIM4906. A construct used for the disruption of mpdA, pIM4907, was made by replacing a 0.7-kb EcoRV-SalI fragment from pIM4906 with a 2.2-kb HindIII-SalI fragment from pIM2101, which contains A. niger argB (25). pIM4907 was linearized with KpnI before transformation of A. niger to achieve gene replacement.
Northern analysis.
Total RNA was extracted from powdered mycelium by using TRIzol (Life Technologies) according to the supplier's instructions. For denaturation, RNA (5 μg) was incubated with 3.3 μl of 6 M glyoxal, 10 μl of dimethyl sulfoxide, and 2 μl of 0.1 M sodium phosphate buffer (pH 7) in a total volume of 20 μl for 1 h at 50°C. Following electrophoresis in 1.5% agarose gels containing 10 mM sodium phosphate (pH 7), RNA was transferred to nylon membranes (Hybond-N; Amersham Biotech) by capillary blotting in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Hybridization was performed at 42°C with a solution containing 50% (vol/vol) formamide, 0.9 M NaCl, 90 mM trisodium citrate, 0.2% (wt/vol) Ficoll, 0.2% (wt/vol) polyvinylpyrrolidone, 0.2% (wt/vol) bovine serum albumin, 0.1% (wt/vol) sodium dodecyl sulfate (SDS), 10% (wt/vol) dextran sulfate, and 100 μg of single-stranded herring sperm DNA ml−1. The following probes were used: a 1.5-kb EcoRI-BglII mpdA fragment and a 0.7-kbp EcoRI fragment of the 18S rRNA subunit (26) as a loading control. Blots were washed with 30 mM NaCl- 3 mM trisodium citrate- 0.5% (wt/vol) SDS at 65°C.
Nucleotide sequence accession number. The GenBank accession number of the mpdA sequence is AY081178.
RESULTS
Cloning and characterization of A. niger mpdA.
To clone the A. niger gene encoding MPD, mpdA, we made use of the fact that many bacteria require MPD for the catabolism of mannitol. In bacteria, mannitol is transported and concomitantly phosphorylated by the phosphotransferase system, yielding mannitol 1-phosphate, which is converted to fructose 6-phosphate by MPD and then further metabolized. Complementation of E. coli mtlD mutant JWL239, lacking MPD, was performed by transformation with an A. niger cDNA library and selection for transformants capable of growth on mannitol as the sole carbon source. Out of 2 × 105 transformants, 4 were able to grow on mannitol and had, in contrast to JWL239, MPD activity (data not shown). From these transformants, plasmid DNA was isolated and used to transform JWL239. All four plasmids produced a high frequency of mannitol-positive colonies, indicating that the ability to grow on mannitol was plasmid borne. Sequence analysis of one of the four clones showed that the protein encoded by the cDNA had sequence similarity to bacterial MPDs. This mpdA cDNA was used as a probe to isolate a genomic mpdA clone from an A. niger N400 library. A 2.9-kb NsiI-KpnI fragment was cloned in pGEM7, resulting in pIM4906.
Sequencing of the genomic clone (GenBank accession no. AY081178) and comparison to the cDNA sequence led to the identification of an intron-less open reading frame encoding a protein of 388 amino acids with an Mr of 43368. The derived amino acid sequence was most similar to that of bacterial MPDs, and the gene was therefore named mpdA. The percentages of identity across the entire protein with the four most similar bacterial MPDs were 43% for Bacillus halodurans MPD, 40% for Streptococcus mutans and Streptococcus pneumoniae MPDs, and 39% for Bacillus stearothermophilus MPD. A. niger mpdA is not homologous to C. neoformans mpd1 (38) or the E. tenella MPD-encoding gene (GenBank accession no. AF055716). Sequences with strong similarity to A. niger mpdA are present in the genome sequences of Aspergillus fumigatus (75.8% identity) (12; http://www.tigr.org) and Neurospora crassa (60.7% identity) (http://www-genome.wi.mit.edu). Low-stringency hybridization of A. niger genomic DNA with mpdA as a probe did not reveal any additional detectable signals (data not shown), suggesting that there are no genes similar to mpdA in the A. niger genome.
Within a 646-bp sequence upstream of the ATG, the following putative regulatory elements were identified: a TATA-like box at position −200; CCAAT elements at −49 and −486 bp relative to the ATG; and at position −328 an element (GTGGGG) meeting the consensus sequence (SYGGRG) for the binding of CREA, the broad domain regulator for carbon catabolite repression in Aspergillus (23). In addition, we found one putative bristle response element (AGAGGGG) at position −183 and two putative abacus response elements (CATTCT) at positions −238 and −188, which correspond to binding sites for the development-specific transcription factors BRLA (4) and ABAA (1), respectively, in A. nidulans.
By cotransformation of A. niger strain NW129 with pIM4906, containing mpdA, and pGW635, containing the pyrA gene, transformants with up to 80-fold overproduction of MPD activity were obtained, indicating that the cloned mpdA gene was functional. MPD was purified from transformant NW129::pIM4906-12 by affinity chromatography with Procion Blue P-GR Sepharose CL-4B. The apparent molecular mass determined by SDS-polyacrylamide gel electrophoresis was 45 kDa (Fig. 2), which corresponds well to the value calculated from the deduced amino acid sequence.
FIG. 2.
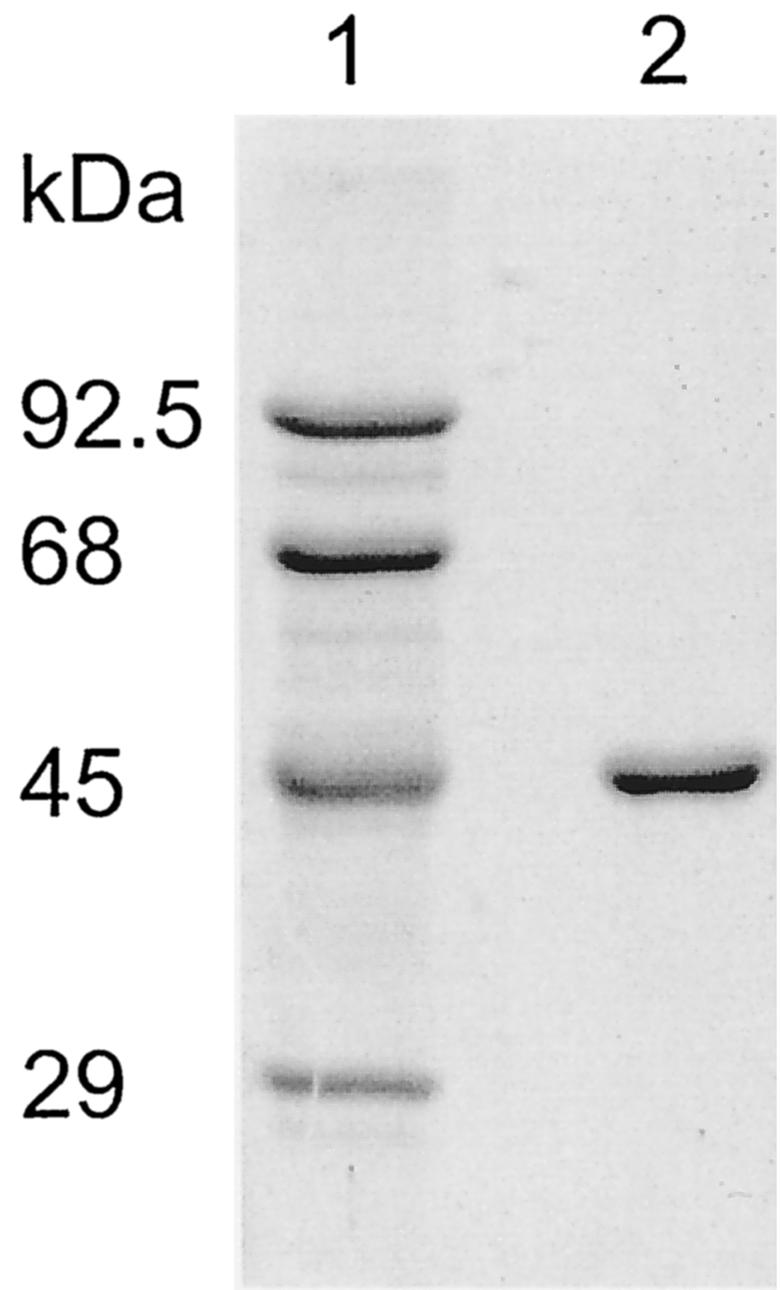
SDS-polyacrylamide gel electrophoresis of purified A. niger MPD. Lane 1, marker proteins with indicated molecular masses; lane 2, purified MPD.
Transcriptional regulation of mpdA.
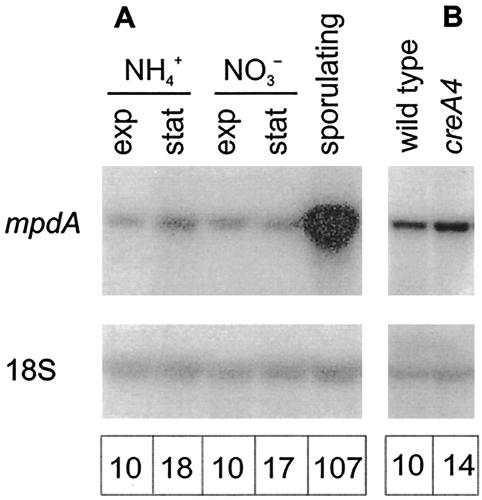
Hult and Gatenbeck (18) have proposed that the mannitol cycle (Fig. 1) plays a role in NADPH production in fungi. If this hypothesis is correct, then it would be expected that the expression of the mannitol cycle genes would be increased under conditions that require relatively large quantities of NADPH, such as fast growth and utilization of NO3− as a nitrogen source. However, in wild-type strain NW131, the levels of expression of mpdA were similar in mycelium growing exponentially and mycelium in the stationary growth phase after exhaustion of the nitrogen source (Fig. 3A). Likewise, mpdA expression levels were comparable on medium containing NH4+ and on medium containing NO3− as the nitrogen source (Fig. 3A). These results indicate that mpdA expression is not influenced by an NADPH requirement.
FIG. 3.
Northern analysis of mpdA transcription in A. niger. (A) Expression of mpdA in mycelium of wild-type strain NW131 growing on glucose and different nitrogen sources (NH4+ and NO3−) and in sporulating NW131. Exp, exponentially growing mycelium; stat, mycelium in stationary phase after exhaustion of the nitrogen source. (B) Comparison of mpdA expression in wild-type strain NW131 and creA mutant NW283 growing on glucose plus nitrate. Detailed culture conditions are described Materials and Methods. In all cases, the glucose concentration at the time of harvesting was at least 25 mM. 18S rRNA was used as a loading control. The numbers at the bottom indicate the relative levels of mpdA transcripts corrected for loading differences on the basis of the 18S signals. For panel A, the signal for NW131 growing exponentially on glucose plus ammonia was set at 10, whereas for panel B, the signal for NW131 growing on glucose plus nitrate was set at 10.
The level of expression of mpdA was high in conidiating mycelium growing on the surface of a liquid culture compared to mycelium growing submerged (Fig. 3A). Furthermore, MPD activity was increased more than sixfold in sporulating mycelium (Table 2). This strong increase in expression in sporulating mycelium is consistent with the presence of putative ABAA and BRLA binding sites in the promoter. Interestingly, MPP activity was also increased in sporulating mycelium (Table 2), although not to as great an extent as MPD activity. Another possible explanation for the increase in mpdA expression in sporulating mycelium is carbon catabolite derepression due to limited glucose availability. To investigate whether the putative CREA binding site in the mpdA promoter was functional, we tested the expression of mpdA in a creA mutant. Expression in an A. niger creA4 strain growing submerged was not increased compared to that in reference strain NW131 (Fig. 3B). These results agree with data reported by Singh et al. (34) indicating that MPD and MPP were not subject to carbon catabolite repression in A. nidulans.
TABLE 2.
Enzyme activities related to mannitol metabolism in vegetative and sporulating mycelia of wild-type strain NW131 and ΔmpdA strain NW306
| Enzyme | Sp act (nmol min−1 mg of protein−1) ina:
|
|||
|---|---|---|---|---|
| NW131
|
NW306 (ΔmpdA)
|
|||
| Vegetative | Sporulating | Vegetative | Sporulating | |
| MPD | 71 ± 35 | 457 ± 103 | NDb | NDb |
| MPP | 470 ± 46 | 1,190 ± 118 | 632 ± 136 | 1,574 ± 179 |
| MTD | 94 ± 26 | 174 ± 46 | 181 ± 40b | 164 ± 13 |
| Fructose 6-phosphate phosphatase | 17 ± 7 | 28 ± 6 | 9 ± 6 | 25 ± 1 |
Data are means and standard deviations from three to five independent determinations for vegetative mycelium and two or three independent determinations for sporulating mycelium. ND, not detected.
Significantly different from the corresponding value for wild-type strain NW131, as determined by Student's t test (95% probability).
Disruption of the mpdA gene.
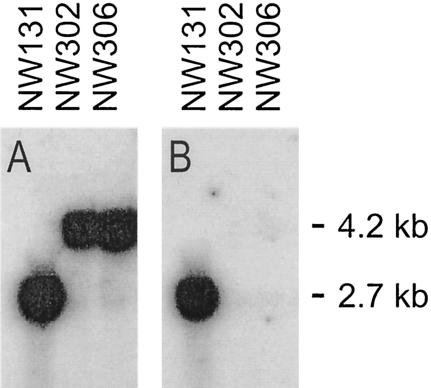
To investigate the role of MPD and mannitol production in A. niger physiology, we replaced a 0.7-kb EcoRV-SalI mpdA fragment, removing 0.2 kb of the promoter region and 0.5 kb of the coding region, with the A. niger argB gene. A. niger strain NW324 was transformed with linearized pIM4907. Of 50 arginine-prototrophic transformants tested, 19 lacked MPD activity. Gene replacement was confirmed by Southern analysis (data not shown). One ΔmpdA strain was selected and designated NW294. Since auxotrophies for unrelated functions (leucine, nicotinamide, and uridine requirements for NW294) might have interfered with the physiological characterization of the ΔmpdA strain, e.g., enhance growth differences, we performed a somatic recombination experiment to obtain a prototrophic strain. However, with somatic recombination NW294-N485, the mpdA disruption could not be separated from leuA1. This scenario positions mpdA on linkage group IV. One recombinant, NW302 (goxC17 leuA1 ΔmpdA), was used in a number of tests (see below). From somatic recombination NW294-N485, we also obtained strain NW305 (ΔargB goxC17), and this strain was transformed with the linearized mpdA disruption construct pIM4907 as well to obtain a prototrophic ΔmpdA strain. Of five transformants tested, two lacked MPD activity, and one of these strains was designated NW306. Gene replacement in NW306 and NW302 was confirmed by Southern analysis (Fig. 4). In the subsequent analyses, strain NW306 was compared to the isogenic reference strain NW131.
FIG. 4.
Southern analysis showing gene replacement at the mpdA locus. Chromosomal DNAs from the indicated strains were digested with NsiI and BglII. (A) Hybridization of the Southern blot with an EcoRI-BglII mpdA probe showing an increase in the size of the NsiI-BglII fragment from 2.7 to 4.2 kb due to gene replacement. (B) Hybridization with an EcoRV-SalI mpdA probe confirming the absence of this fragment in disruption strains NW302 and NW306.
Biochemical characterization of A. niger ΔmpdA strain NW306.
MPD activity was absent in mycelium of ΔmpdA strain NW306 growing exponentially in MM containing glucose as well as in sporulating mycelium growing on the surface of a glucose culture (Table 2). The level of mannitol 1-phosphate, one of the products of the reaction catalyzed by MPD, was 0.06 μmol g of dry weight−1 in mycelium of wild-type strain NW131 but was below the detection limit in ΔmpdA strain NW306 (Table 3). This result is consistent with the absence of MPD activity in NW306.
TABLE 3.
Metabolites accumulating in mycelia and conidiospores of wild-type strain NW131 and ΔmpdA strain NW306
| Metabolite | Concn (μmol g of dry wt−1) ina:
|
|||
|---|---|---|---|---|
| NW131
|
NW306 (ΔmpdA)
|
|||
| Mycelia | Spores | Mycelia | Spores | |
| Mannitol 1-phosphate | 0.06 ± 0.01 | — | 0.01 ± 0.01b | — |
| Glycerol | 213 ± 43 | 26 ± 9 | 221 ± 14 | 106 ± 66 |
| Erythritol | 371 ± 46 | 3 ± 2 | 312 ± 107 | 9 ± 6 |
| Mannitol | 140 ± 32 | 627 ± 63 | NDb | 220 ± 11b |
| Trehalose | 18 ± 3 | 105 ± 47 | 51 ± 27 | 307 ± 61b |
Mycelium samples were obtained from exponentially growing cultures with glucose as the carbon source. Data are means and standard deviations from three or four independent determinations. ND, not detected; —, not determined.
Significantly different from the corresponding value for wild-type strain NW131, as determined by Student's t test (95% probability).
Mannitol was not detected in mycelium of ΔmpdA strain NW306, but NW306 conidiospores still contained approximately 30% the mannitol level found in wild-type conidia (Table 3). Quantities of other polyols were similar in mycelia of NW131 and NW306 (Table 3). Conidia of ΔmpdA strain NW306 contained approximately threefold more trehalose than those of strain NW131 (Table 3). These observations, i.e., no mannitol in mycelium and lower mannitol and higher trehalose level in conidia, were confirmed by analysis of the other ΔmpdA strain, NW302 (data not shown).
Since ΔmpdA conidia still contained mannitol, while no MPD activity was detected, another metabolic pathway was apparently able to contribute to mannitol synthesis. One possibility is the operation of fructose 6-phosphate phosphatase activity and MTD activity, i.e., the reverse of the catabolic pathway for mannitol. Both activites were present in mycelia of NW131 and NW306 (Table 2).
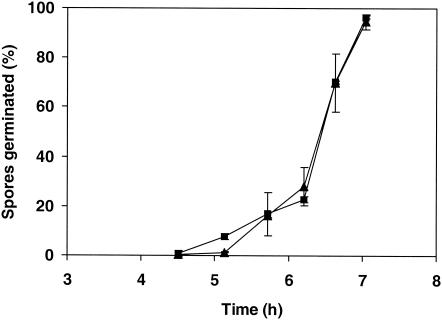
Spore germination and vegetative growth are not disturbed by disruption of mpdA.
The kinetics of germ tube formation of strain NW306 conidiospores on MM containing glucose could not be distinguished from that of conidiospores of the reference strain (Fig. 5). After 7 h, 95% ± 3% (mean and standard deviation) of NW131 spores and 97% ± 1% of NW306 spores had germinated. Biomass production levels during submerged growth on MM containing 2% d-glucose were very similar for the two strains. After exhaustion of the growth-limiting substrate (glucose), dry weights were 7.4 ± 0.2 g liter−1 (n = 3) for NW131 and 7.7 ± 0.5 g liter−1 (n = 3) for NW306. The levels of growth of wild-type and ΔmpdA strains on MM plates containing 1 or 10% d-glucose, 1% d-fructose, 1% d-mannitol, 1% d-xylose, 1% glycerol, or 1% acetate at 30, 37, or 42°C were comparable (data not shown). Although ΔmpdA strain NW306 was partially deficient in mannitol biosynthesis, sporulation was not delayed compared to that in the wild-type strain. One distinctive feature of the ΔmpdA strain was the accumulation of a yellow compound in colonies growing on plates. The yellow compound was produced on all carbon sources tested except for mannitol.
FIG. 5.
Kinetics of conidiospore germination for wild-type strain NW131 and ΔmpdA mutant NW306. Spores (2 × 104) were spot inoculated onto slides coated with MM containing 1% (wt/vol) glucose and solidified with agar. Germination was monitored by microscopic examination of the slides. Symbols: ▴, wild-type strain NW131; ▪, ΔmpdA strain NW306. Data are averages of two independent determinations and standard deviations.
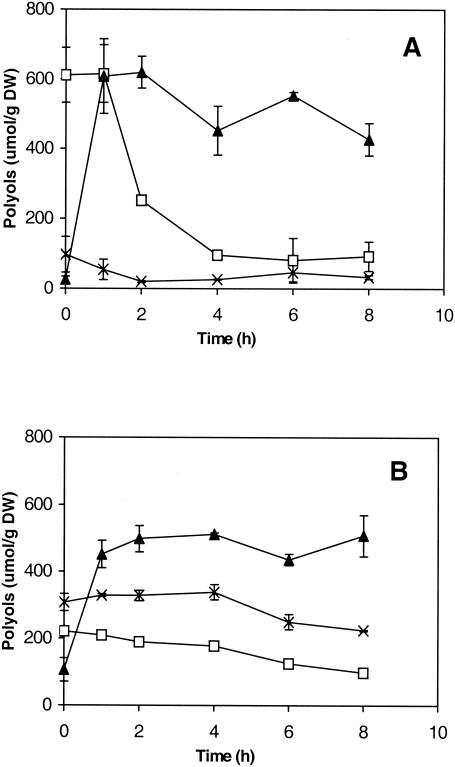
Glycerol is accumulated rapidly in germinating A. niger spores, whereas mannitol and trehalose are consumed (41). Mannitol and, to a lesser extent, trehalose may be used as sources of carbon to enable the rapid production of glycerol (11). However, glycerol accumulation during the germination of ΔmpdA spores was not different from that for the germination of wild-type spores (Fig. 6). In addition, the residual mannitol in ΔmpdA spores was only very slowly metabolized compared to the consumption rate in spores of reference strain NW131, indicating that mannitol is not required for glycerol production.
FIG. 6.
Intracellular polyol content of germinating conidiospores of A. niger wild-type strain NW131 (A) and ΔmpdA mutant NW306 (B) as a function of time after inoculation. DW, dry weight. Symbols: ▴, glycerol; □, mannitol; ×, trehalose. Data are averages of two independent determinations and standard deviations.
Inactivation of mpdA increases the sensitivity of spores to stress conditions.
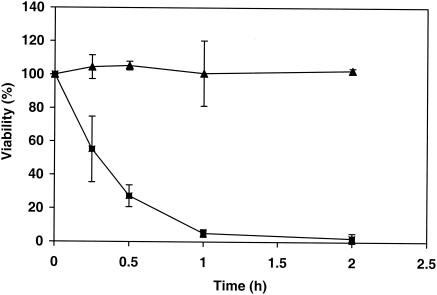
To investigate the effect of the inactivation of mpdA in A. niger on the survival of conidia during stress, we examined the viability of NW306 spores subjected to heat and oxidative treatments. Conidia from ΔmpdA strain NW306 were very sensitive to a high temperature. Following incubation for 1 h at 50°C, only 5% of ΔmpdA conidia were still viable, whereas the viability of wild-type conidia was 100% (Fig. 7). We first attempted to apply oxidative stress by exposure to H2O2, but wild-type A. niger conidia appeared to be resistant to 100 mM H2O2, presumably due to the presence of very active catalases. Treatment with 5 to 10 mM NaOCl, however, very effectively decreased the viability of A. niger conidia. NW306 conidia were much more sensitive to NaOCl than wild-type conidia, as no more than 0.5% of ΔmpdA conidia survived exposure for 1 h to 1 mM NaOCl, compared to 100% viability for NW131 conidia (Table 4). Since NaOCl is a basic compound and 1 mM NaOCl in water produces pH 9.8, the sensitivity of ΔmpdA conidia to NaOCl might have been due to an alkaline pH. Incubation of ΔmpdA conidia for 1 h in buffer at pH 10 however, did not affect the viability of NW306 spores, indicating that killing by NaOCl was in fact due to oxidative damage. Compared to wild-type conidia, ΔmpdA conidia were also very sensitive to other stress treatments, such as a freeze-thaw step and freeze-drying (Table 4), although the effect was less pronounced than that of heat and oxidative treatments. Mannitol is not required for long-term spore survival in A. niger, since storage of conidia in suspension for up to 24 days at either 4 or 25°C did not result in a loss of viability for NW306 (data not shown).
FIG. 7.
Viability of conidiospores of wild-type strain NW131 and ΔmpdA strain NW306 upon incubation at 50°C. Suspensions containing approximately 108 spores ml−1 in 0.05% (wt/vol) Tween 80- 0.9% (wt/vol) NaCl were incubated at 50°C for the indicated times. Spore viability was established by diluting aliquots, plating on complete medium containing 1% (wt/vol) glucose, and determining the number of CFU after 24 h of growth at 30°C. Symbols: ▴, wild-type strain NW131; ▪, ΔmpdA strain NW306. Data are averages of two independent determinations and standard deviations.
TABLE 4.
Viability of conidiospores of wild-type strain NW131 and ΔmpdA strain NW306 after different stress treatments
| Stress treatment | % Survival of spores of the indicated strain produced on MM containinga:
|
|||
|---|---|---|---|---|
| Glucose
|
Mannitol
|
|||
| NW131 | NW306 | NW131 | NW306 | |
| 50°C (1 h) | 101 ± 20 | 5 ± 2 | 129 ± 15 | 82 ± 8 |
| Freeze-thaw step | 76 ± 16 | 15 ± 3 | — | — |
| Lyophilization | 64 ± 16 | 23 ± 3 | — | — |
| NaOCl (1 mM, 1 h) | 107 ± 14 | 0.5 ± 0.4 | 107 ± 30 | 107 ± 30 |
Conidiospores were exposed to the indicated treatments. Viability was subsequently determined as the number of CFU after plating on complete medium and is given as a percentage of the value for the (untreated) control, which was set at 100%. Data are means and standard deviations from two or three independent determinations. —, not determined.
If the sensitivity of NW306 spores to heat treatment and oxidation was due to the lower level of mannitol in spores, then supplying mannitol during conidiogenesis might repair the defect. Indeed, spores of ΔmpdA strain NW306 produced on MM with a mixture of 50 mM mannitol and 1 mM glucose did contain mannitol (NW131, 621 ± 38 [mean and standard deviation] μmol g of dry weight−1; NW306, 1,150 ± 72 μmol g of dry weight−1) and were resistant to heat and oxidative stress (Table 4).
DISCUSSION
A. niger mpdA encodes MPD.
In order to obtain a better understanding of mannitol biosynthesis and its role in fungal physiology, we cloned the A. niger mpdA gene, which encodes MPD. The following lines of evidence indicate that the mpdA gene product is MPD: (i) the deduced MpdA protein sequence shows high homology to those of bacterial MPDs; (ii) A. niger mpdA multicopy transformants overproduce MPD up to 80-fold compared to the wild-type level; and (iii) A. niger transformants carrying a disruption in the mpdA locus lack MPD activity both during vegetative growth and during conidiation (Table 2). MPD purified from a multicopy transformant had an apparent molecular mass of 45 kDa (Fig. 2). Kiser and Niehaus (22) reported that A. niger MPD was a homodimer of 22-kDa subunits, a finding which may be explained by proteolytic degradation of their MPD preparation.
Inactivation of mpdA abolished mannitol biosynthesis in mycelium, but ΔmpdA conidiospores still contained approximately 30% the mannitol level found in wild-type conidia (Table 3). These findings indicate that MPD and MPP make up the major pathway for mannitol biosynthesis in A. niger but also that there is an alternative metabolic pathway which can partially take over the functions of MPD and MPP in mannitol biosynthesis. The residual mannitol in ΔmpdA conidia may be produced by an enzyme with fructose 6-phosphate phosphatase activity and MTD activity, i.e., the reverse of the catabolic pathway for mannitol. Both activities are present in A. niger (Table 2). It is unclear why such a pathway would not function in actively growing mycelium (mannitol is absent in ΔmpdA mycelium). A possible explanation is that fructose 6-phosphate phosphatase activity and/or MTD activity is regulated in vivo, i.e., inhibited in vegetative mycelium but not inhibited in sporulating mycelium.
A. niger MpdA shows no amino acid sequence similarity to two known eukaryotic MPDs, C. neoformans Mpd1 (38) and E. tenella MPD (GenBank accession no. AF055716). E. tenella MPD appears to be quite distinct, since it is a 67-kDa protein which can utilize both NAD+ and NADP+ as cofactors (32), whereas all other known MPDs have molecular masses in the range of 37 to 45 kDa and are specific for NAD+. It is rather surprising that A. niger MpdA is not homologous to C. neoformans Mpd1, the only other fungal MPD reported to date. However, the data reported by Suvarna et al. (38) do not establish that Mpd1 is the main MPD in C. neoformans. The kinetic data presented by Suvarna et al. (38) suggest that Mpd1 is more active with ethanol as a substrate than with mannitol 1-phosphate. C. neoformans Mpd1 might therefore be an alcohol dehydrogenase with some MPD activity, explaining why it is not homologous to A. niger MpdA.
Is the mannitol cycle involved in NADPH production?
The NADPH requirement of cells is high during active growth, i.e., for the biosynthesis of cell components and for the catabolism of certain nutrients, such as nitrate. In nonphotosynthetic microorganisms, the oxidation of glucose 6-phosphate to ribulose 5-phosphate is usually considered to be the main pathway for NADPH generation. Hult and Gatenbeck (18) have proposed the involvement of the mannitol cycle in NADPH generation in the fungus A. alternata and have suggested that this cyclic pathway is also important for NADPH production in other fungi (19). If the mannitol cycle is required for NADPH production, one might expect that the expression of the mannitol cycle genes would be increased under conditions that require relatively large quantities of NADPH. Moreover, interruption of the cycle might seriously compromise growth. We have no indications that this is the case in A. niger. First, the expression of mpdA and MPD activity were similar in nitrate- versus ammonia-cultured mycelia and in exponentially growing versus stationary-phase mycelia (i.e., high- versus low-NADPH-requiring conditions). Second, germination kinetics, growth, and final biomass levels in MM containing nitrate as the sole nitrogen source were identical in a ΔmpdA strain, in which the mannitol cycle is interrupted, and in an isogenic wild-type strain.
It is possible that the mannitol cycle is important for NADPH generation in A. alternata, but our data do not support the hypothesis that it operates in every fungus possessing all enzymes of this cycle. The mannitol cycle may contribute to NADPH production in A. niger when there is mannitol turnover during growth, but it is not essential. Singh et al. (34) came to a similar conclusion for A. nidulans.
Mannitol is essential for stress tolerance in conidiospores.
Conidiospores of an A. niger ΔmpdA strain were very sensitive to a high temperature, the presence of the strong oxidative compound NaOCl, a freeze-thaw step, and lyophilization (Table 4 and Fig. 7). Mannitol supply in the medium during sporulation repaired this deficiency (Table 4). These findings indicate that mannitol is essential for the protection of spores against cell damage which occurs under these stress conditions and that inactivation of mpdA results in conditional mannitol auxotrophy. Our results are in agreement with other reports describing fungal mannitol production to prevent oxidative damage, such as mannitol production during infection of the host by the human fungal pathogen C. neoformans (6) and excretion of mannitol by the phytopathogenic fungus A. alternata during plant infection (20). Interestingly, the plant induces MTD in response to fungal infection, presumably for the removal of mannitol to counteract the fungal protection mechanism (20). Since A. niger is a sapropytic fungus and is pathogenic only in immunocompromised individuals, it probably does not use mannitol production for defense during growth but rather uses mannitol to ensure maximal viability of spores to survive in unfavorable conditons.
Mannitol is known to scavenge ROS (35), but the mechanisms by which polyhydroxyalcohols could protect cells under conditions of high temperature, drying, or freezing are less well understood. Crowe et al. (9) have proposed a model in which the replacement of water by polyol molecules preserves the amorphous noncrystalline state of cells during drying. A similar mechanism could explain protection during freezing, i.e., prevention of ice crystal formation.
Our results suggest that mannitol biosynthesis is, to a certain extent, developmentally regulated in A. niger. The expression of mpdA strongly increased at the onset of sporulation (Fig. 3A) and, in particular, MPD activity but also MPP activity were higher in sporulating mycelium than in vegetative mycelium (Table 2). Furthermore, the mpdA promoter contains one putative BRLA response element (4) and two putative ABAA response elements (1) which are involved in development-specific transcription regulation in A. nidulans. Developmental control of mpdA and perhaps also of the gene encoding MPP is in agreement with the observation that A. niger conidiospores contain a high concentration of mannitol (41) (Table 3). Our data are consistent with a model in which mannitol biosynthesis is strongly increased during conidiogenesis to provide spores with a sufficient quantity of mannitol to ensure viability under adverse conditions.
Many fungal species accumulate trehalose and/or mannitol in their propagules, although the levels vary (13). For example, the trehalose level in A. nidulans spores is high compared to that in A. niger (14). Apparently, there is a species-specific preference for mannitol or trehalose accumulation in conidia. In A. nidulans conidia, trehalose is important for long-term spore survival and proper spore germination, suggesting a role as storage carbon (14). The high mannitol level in A. niger conidia is not required for the viability of conidiospores during prolonged storage or spore germination, since inactivation of mpdA had no adverse effects. However, the trehalose content in ΔmpdA conidia had increased about threefold to a level that would approximately match the decrease in the mannitol content on a carbon mass basis (Table 3), and this change might conceal any adverse effects of a decreased mannitol level. Thus, mannitol may play a role as storage carbon, but it is not essential.
Trehalose biosynthesis in A. niger has been investigated by Wolschek and Kubicek (43), who have cloned two genes encoding trehalose 6-phosphate synthase from A. niger. Disruption of one of these genes, tpsA, resulted in a 56% reduction in the conidial trehalose content and in a slightly reduced heat tolerance: 70% of ΔtpsA conidia survived incubation at 50°C, compared to 94% of wild-type conidia (43). These findings show that trehalose contributes to protection against a high temperature, but our data concerning the heat tolerance of the ΔmpdA strain demonstrate that mannitol is more significant as a stabilizing compound. The fact that, despite increased trehalose content, ΔmpdA conidia are heat sensitive indicates that trehalose cannot substitute for mannitol in protecting conidia against heat stress. It is possible that trehalose is localized in a compartment where it cannot contribute much to the protection of cells, but alternatively mannitol may somehow fulfill a special function. Shen et al. (33) reported a similar polyol preference in Saccharomyces cerevisiae during osmotic stress. Yeast cells normally accumulate glycerol in response to exposure to high salt concentrations. In a gpd1Δ gpd2Δ strain, lacking both glycerol 3-phosphate dehydrogenases, resulting in the inability to produce glycerol, mannitol or sorbitol accumulation could be achieved by the introduction of heterologous genes, but this effect only partially protected against osmotic stress. The authors suggested that glycerol, besides functioning as a solute to maintain cell turgor, has a role in redox balancing, i.e., the removal of excess NADH, during growth at reduced water activity. In A. niger conidiospores, mannitol may function as a pool of reducing power. It is possible that under stress conditions resulting in cell damage, NADPH produced during the catabolism of mannitol contributes significantly to repair mechanisms. The construction of an MTD mutant or an mpdA MTD double-disruption strain, which might be completely unable to produce mannitol, could resolve this matter.
In summary, the data presented in this study show that MPD and MPP form the major metabolic pathway for the biosynthesis of mannitol in A. niger. Moreover, increased expression of the gene encoding MPD, mpdA, in sporulating mycelium is consistent with the high mannitol content of conidiospores. We found no evidence for a role of mannitol as a reserve carbon source in conidia, but the data indicate that mannitol is essential for resistance to a variety of stress conditions.
Acknowledgments
The work described in this report was financially supported in part by EU grant QTRK3-199-00729 to J.V. and in part by the Ministry of Economic Affairs, the Ministry of Education, Culture and Science, and the Ministry of Agriculture, Nature Management and Fishery in the framework of an industrial relevant research program of The Netherlands Association of Biotechnology Centres.
We thank Henk Panneman and Jacques Benen for performing some initial experiments and J. W. Lengeler (University of Osnabrück) for providing E. coli JWL239.
REFERENCES
- 1.Adrianopoulos, A., and W. E. Timberlake. 1994. The Aspergillus nidulans abaA gene encodes a transcriptional activator that acts as a genetic switch to control development. Mol. Cell. Biol. 14:2503-2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allocco, J. J., H. Profous-Juchelka, R. W. Myers, B. Nare, and D. M. Schmatz. 1999. Biosynthesis and catabolism of mannitol is developmentally regulated in the protozoan parasite Eimeria tenella. J. Parasitol. 85:167-173. [PubMed] [Google Scholar]
- 3.Bos, C. J., A. J. M. Debets, K. Swart, A. Huybers, G. Kobus, and S. M. Slakhorst. 1988. Genetic analysis and the construction of master strains for assignment of genes to six linkage groups in Aspergillus niger. Curr. Genet. 14:437-443. [DOI] [PubMed] [Google Scholar]
- 4.Chang, Y. C., and W. E. Timberlake. 1992. Identification od Aspergillus brlA response elements (BREs) by genetic selection in yeast. Genetics 133:29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaturvedi, V., T. Flynn, W. G. Niehaus, and B. Wong. 1996. Stress tolerance and pathogenic potential of a mannitol mutant of Cryptococcus neoformans. Microbiology 142:937-943. [DOI] [PubMed] [Google Scholar]
- 6.Chaturvedi, V., B. Wong, and S. L. Newman. 1996. Oxidative killing of Cryptococcus neoformans by human neutrophils. Evidence that fungal mannitol protects by scavenging reactive oxygen intermediates. J. Immunol. 156:3836-3840. [PubMed] [Google Scholar]
- 7.Chaturvedi, V., A. Bartiss, and B. Wong. 1997. Expression of bacterial mtlD in Saccharomyces cerevisiae results in mannitol synthesis and protects a glycerol-defective mutant from high-salt and oxidative stress. J. Bacteriol. 179:157-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corina, D. L., and K. A. Munday. 1971. Studies on polyol function in Aspergillus clavatus: a role for mannitol and ribitol. J. Gen. Microbiol. 69:221-227. [DOI] [PubMed] [Google Scholar]
- 9.Crowe, J. H., F. A. Hoekstra, and L. M. Crowe. 1992. Anhydrobiosis. Annu. Rev. Physiol. 54:579-599. [DOI] [PubMed] [Google Scholar]
- 10.De Graaff, L., H. van den Broek, and J. Visser. 1988. Isolation and expression of the Aspergillus nidulans pyruvate kinase gene. Curr. Genet. 13:315-321. [DOI] [PubMed] [Google Scholar]
- 11.d'Enfert, C. 1997. Fungal spore germination: insights from the molecular genetics of Aspergillus nidulans and Neurospora crassa. Fungal Genet. Biol. 21:163-172. [Google Scholar]
- 12.Denning, D. W., M. J. Anderson, G. Turner, J. P. Latge, and J. W. Bennett. 2002. Sequencing the Aspergillus fumigatus genome. Lancet Infect. Dis. 2:251-253. [DOI] [PubMed] [Google Scholar]
- 13.Dijksterhuis, J., and R. A. Samson. 2002. Food and crop spoilage on storage, p. 39-52. In F. Kempken (ed.), The Mycota, vol. XI. Agricultural applications. Springer-Verlag KG, Berlin, Germany.
- 14.Fillinger, S., M.-K. Chaveroche, P. van Dijck, R. de Vries, G. Ruijter, J. Thevelein, and C. d'Enfert. 2001. Trehalose is required for the acquisition of tolerance to a variety of stresses in the filamentous fungus Aspergillus nidulans. Microbiology 147:1851-1862. [DOI] [PubMed] [Google Scholar]
- 15.Goosen, T., G. Bloemheuvel, C. Gysler, D. A. de Bie, H. W. J. van den Broek, and K. Swart. 1987. Transformation of Aspergillus niger using the homologous orotidine-5′-phosphate-decarboxylase gene. Curr. Genet. 11:499-503. [DOI] [PubMed] [Google Scholar]
- 16.Hammond, J. B. W., and R. Nichols. 1975. Changes in respiration and soluble carbohydrates during the post-harvest storage of mushrooms (Agaricus bisporus). J. Sci. Food Agric. 26:835-842. [Google Scholar]
- 17.Horikoshi, K., S. Iida, and Y. Ikeda. 1965. Mannitol and mannitol dehydrogenase in conidia of Aspergillus oryzae. J. Bacteriol. 89:326-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hult, K., and S. Gatenbeck. 1978. Production of NADPH in the mannitol cycle and its relation to polyketide formation in Alternaria alternata. Eur. J. Biochem. 88:607-612. [DOI] [PubMed] [Google Scholar]
- 19.Hult, K., A. Veide, and S. Gatenbeck. 1980. The distribution of the NADPH regenerating mannitol cycle among fungal species. Arch. Microbiol. 128:253-255. [DOI] [PubMed] [Google Scholar]
- 20.Jennings, D. B., M. Ehrenshaft, D. M. Pharr, and J. D. Williamson. 1998. Roles for mannitol and mannitol dehydrogenase in active oxygen-mediated plant defense. Proc. Natl. Acad. Sci. USA 95:15129-15133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jennings, D. H. 1984. Polyol metabolism in fungi. Adv. Microb. Physiol. 25:149-193. [DOI] [PubMed] [Google Scholar]
- 22.Kiser, R. C., and W. G. Niehaus. 1981. Purification and kinetic characterisation of mannitol 1-phosphate dehydrogenase from Aspergillus niger. Arch. Biochem. Biophys. 211:613-621. [DOI] [PubMed] [Google Scholar]
- 23.Kulmburg, P., M. Mathieu, C. Dowzer, J. Kelly, and B. Felenbok. 1993. Specific binding sites in the alcR and alcA promoters of the ethanol regulon for the CREA repressor mediating carbon catabolite repression in Aspergillus nidulans. Mol. Microbiol. 7:847-857. [DOI] [PubMed] [Google Scholar]
- 24.Kusters-van Someren, M. A., J. A. M. Harmsen, H. C. M. Kester, and J. Visser. 1991. Structure of the Aspergillus niger pelA gene and its expression in Aspergillus niger and Aspergillus nidulans. Curr. Genet. 20:293-299. [DOI] [PubMed] [Google Scholar]
- 25.Lenouvel, F., P. J. I. van de Vondervoort, and J. Visser. 2002. Disruption of the Aspergillus niger argB gene: a tool for transformation. Curr. Genet. 41:425-431. [DOI] [PubMed] [Google Scholar]
- 26.Melchers, W. J. G., P. E. Verweij, P. van den Hurk, A. van Belkum, B. E. de Pauw, A. A. Hoogkamp-Korstanje, and J. F. G. M. Meis. 1994. General primer-mediated PCR for detection of Aspergillus species. J. Clin. Microbiol. 32:1710-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pontecorvo, G., J. A. Roper, L. M. Hemmons, K. D. MacDonald, and A. W. J. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141-238. [DOI] [PubMed] [Google Scholar]
- 28.Ruijter, G. J. G., and J. Visser. 1996. Determination of intermediary metabolites in Aspergillus niger. J. Microbiol. Methods 25:295-302. [Google Scholar]
- 29.Ruijter, G. J. G., H. Panneman, and J. Visser. 1997. Overexpression of phosphofructokinase and pyruvate kinase in citric acid producing Aspergillus niger. Biochim. Biophys. Acta 1334:317-326. [DOI] [PubMed] [Google Scholar]
- 30.Ruijter, G. J. G., S. A. Vanhanen, M. M. C. Gielkens, P. J. I. van de Vondervoort, and J. Visser. 1997. Isolation of Aspergillus niger creA mutants. Effects on expression of arabinases and l-arabinose catabolic enzymes. Microbiology 143:2991-2998. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Schmatz, D. M. 1997. The mannitol cycle in Eimeria. Parasitology 114:S81-S89. [PubMed] [Google Scholar]
- 33.Shen, B., S. Hohmann, R. G. Jensen, and H. J. Bohnert. 1999. Roles of sugar alcohols in osmotic stress adaptation. Replacement of glycerol by mannitol and sorbitol in yeast. Plant Physiol. 121:45-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh, M., M. S. Scrutton, and M. C. Scrutton. 1988. NADPH generation in Aspergillus nidulans: is the mannitol cycle involved? J. Gen. Microbiol. 134:643-654. [DOI] [PubMed] [Google Scholar]
- 35.Smirnoff, N., and Q. J. Cumbes. 1989. Hydroxyl radical scavenging activity of compatible solutes. Phytochemistry 28:1057-1060. [Google Scholar]
- 36.Stoop, J. M. H., and H. Mooibroek. 1998. Cloning and characterization of NADP-mannitol dehydrogenase cDNA from the button mushroom, Agricus bisporus, and its expression in response to NaCl stress. Appl. Environ. Microbiol. 64:4689-4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoop, J. M. H., J. D. Williamson, and D. M. Pharr. 1996. Mannitol metabolism in plants: a method for coping with stress. Trends Plant Sci. 1:139-144. [Google Scholar]
- 38.Suvarna, K., A. Bartiss, and B. Wong. 2000. Mannitol-1-phosphate dehydrogenase from Cryptococcus neoformans is a zinc-containing long-chain alcohol/polyol dehydrogenase. Microbiology 146:2705-2713. [DOI] [PubMed] [Google Scholar]
- 39.Van Peij, N. N. M. E., J. Brinkmann, M. Vrsanska, J. Visser, and L. H. de Graaff. 1997. Beta-xylosidase activity, encoded by xlnD, is essential for complete hydrolysis of xylan by Aspergillus niger but not for induction of the xylanolytic enzyme spectrum. Eur. J. Biochem. 245:164-173. [DOI] [PubMed] [Google Scholar]
- 40.Vishniac, W., and M. Santer. 1957. The thiobacilli. Bacteriol. Rev. 21:195-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Witteveen, C. F. B., and J. Visser. 1995. Polyol pools in Aspergillus niger. FEMS Microbiol. Lett. 134:57-62. [DOI] [PubMed] [Google Scholar]
- 42.Witteveen, C. F. B., F. Weber, R. Busink, and J. Visser. 1994. Isolation and characterisation of two xylitol dehydrogenases from Aspergillus niger. Microbiology 140:1679-1685. [Google Scholar]
- 43.Wolschek, M. F., and C. P. Kubicek. 1997. The filamentous fungus Aspergillus niger contains two “differentially regulated” trehalose-6-phosphate synthase-encoding genes, tpsA and tpsB. J. Biol. Chem. 272:2729-2735. [DOI] [PubMed] [Google Scholar]