Abstract
The sheltering of chromosome ends from illegitimate DNA repair reactions and telomere length homeostasis are critical for preserving genomic integrity. Growing evidence implicates covalent protein modification by SUMO (small ubiquitin-like modifier) (sumoylation) in the regulation of numerous DNA transactions, including DNA repair and transcription, as well as heterochromatin formation and maintenance. We have recently shown that fission yeast Pli1p is a SUMO E3 ligase and that pli1 mutants, which are impaired for global sumoylation, are viable, but exhibit de-regulated homologous recombination and marked defects in chromosome segregation and centromeric silencing, as well as a consistent increase in telomere length. In this work, we explore the mechanisms underlying sumoylation-dependent telomere maintenance. We show that Pli1p, but not the related Nse2p, is the principal SUMO E3 ligase enzyme involved. Using both a pli1 mutation and a physiological “knockdown” of sumoylation, achieved by inducible expression of a dominant negative form of the conjugating enzyme Ubc9p, we further show that telomere lengthening induced by lack of sumoylation is not due to unscheduled telomere–telomere recombination. Instead, sumoylation increases telomerase activity, therefore suggesting that this modification controls the activity of a positive or negative regulator of telomerase.
Keywords: homologous recombination
The ends of linear chromosomes pose two special challenges for the cellular DNA replication and repair machinery. The first, also known as the end replication problem, is the progressive shortening of chromosome ends at each replication cycle because of the inability of lagging strand synthesis to fully replicate a linear template. The second, or end protection problem, is to shelter these chromosome ends from mechanisms that would ordinarily recognize these as double-strand breaks (DSBs) and initiate DNA repair processes with potentially disastrous consequences for genomic integrity (reviewed in ref. 1). Both these challenges are met by telomeres, which consist of an array of tandem DNA repeats with a characteristic asymmetry: a G-rich 5′-3′ strand that usually forms a 3′ single-stranded overhang and is replicated by leading strand synthesis, and the complementary C-rich 3′-5′ strand that is replicated by lagging strand synthesis. The overhang participates in chromosome end-protection both by recruiting capping proteins like Pot1 in fission yeast and higher eukaryotes (2) and by forming a specific DNA structure called the t-loop formed by the invasion of the 3′ single-stranded overhang into the duplex repeat array (3). Protected this way, telomeres prevent DNA repair activities such as homologous recombination (HR) and nonhomologous end joining (NHEJ) from acting at chromosome ends (reviewed in ref. 4). To overcome the end replication problem telomeres possess telomerase, a ribonucleoprotein enzyme complex composed of a template RNA molecule, the reverse transcriptase catalytic subunit and a number of protein subunits (reviewed in ref. 5). Telomerase extends the G-rich strand in the 5′- to -3′ direction by using its RNA component as a template to add new copies of the telomeric repeats (reviewed in ref. 6). These repeats are then directly bound by proteins such as TRF1/2 in mammals, or their homolog Taz1p in fission yeast, that are crucial for inhibition of recombination and for the maintenance of telomere length by both normal replication (7) and telomerase dependent synthesis (1). Moreover, in telomerase-positive immortal cell lines and in unicellular organisms telomere length is maintained within a narrow range that is species or cell type specific. The complexity of the telomere maintenance system is further underscored by the fact that mutations of telomerase or of proteins involved in end protection, telomeric silencing, DNA replication and repair or in cell cycle checkpoint functions lead to a modification of this homeostasis, senescence or cell death (6). Therefore, the study of the mechanisms governing telomere length regulation is critical to understanding the systems that protect genomic integrity.
A key observation came from phenotype analysis of fission yeast cells deleted for the pmt3 gene encoding SUMO (small ubiquitin-like modifier). Apart from very slow growth, aberrant chromosome segregation and enhanced sensitivity to DNA-damaging agents, pmt3Δ cells also display telomeres two to three times longer than wild type cells (8). SUMO is an evolutionarily conserved protein that resembles ubiquitin in its structure and its ability to be covalently attached to target proteins. Like ubiquitin, SUMO is conjugated via a conserved, ATP-dependent cascade of E1 activating (Aos1-Uba2), E2 conjugating (Ubc9) and E3 ligation steps to produce an isopeptide bond between the C-terminal glycine of SUMO and a specific lysine of the target protein (9). Unlike, for example, the conjugation to Lysine-48-linked polyubiquitin chains that tags proteins for proteasomal degradation, a unifying rationale for modification by SUMO (sumoylation) does not seem to exist. Rather, sumoylation affects each of its targets in specific ways by altering their conformation, stability or interaction and localization properties. Thus sumoylation has been implicated in a wide range of biological processes including transcriptional regulation, DNA replication and repair, nucleo-cytoplasmic transport and signal transduction (9–12).
A growing body of evidence implicates sumoylation in the control of homologous recombination (HR). For example, the mammalian Rad51 and Rad52 proteins, as well as the BLM and WRN helicases have been shown either to be modified by SUMO or to interact with SUMO or the Ubc9 E2 conjugating enzyme (13–16). Whereas the downstream targets of the modified proteins are still unknown in these cases, recent mechanistic insight into the role of SUMO in HR came from the study of proliferating cell nuclear antigen (PCNA) in budding yeast. PCNA is a trimeric sliding clamp and processivity factor for DNA polymerases. During S phase, both in the presence and absence of DNA-damage, PCNA is modified by SUMO and sumoylated PCNA recruits the helicase Srs2 that, in turn, disrupts Rad51 nucleoprotein filaments and thus inhibits unwanted recombination (17, 18). In this context, we have shown recently that sumoylation plays a critical role in the suppression of HR in specialized heterochromatin loci in fission yeast (19). In this system, mutation of the SUMO E3 ligase Pli1p was shown to drastically enhance the loss of an inserted reporter gene (ura4) by gene conversion between homologous sequences at the central core (cnt) and inner repeats (imr) of the Schizosaccharomyces pombe centromere. Furthermore, mutation of pli1 also leads to a significant increase in telomere length similar to that seen upon mutation of SUMO itself. Given that, by their tandemly repeated structure and their free 3′ overhangs, telomeres are potential but actively controlled HR substrates, it is conceivable that the increased telomere length in SUMO and pli1 mutants is due to a derepression of HR (8, 19). Indeed, a recombination-based mechanism underlying this effect might provide an attractive explanation, because in mammalian systems, some cancers, such as certain soft tissue sarcomas, maintain telomeres (and hence immortality) by a telomerase-independent pathway termed ALT (alternative lengthening of telomeres). This pathway requires the Mre11-Rad50-Nbs1 (MRN) complex involved in double-strand break repair (20, 21). Moreover, in ALT cells telomeres, MRN and the HR proteins Rad51 and Rad52 colocalize in the so-called ALT PML nuclear bodies (APBs; refs. 22 and 23), the principal components of which, PML and SP100, represent two well characterized SUMO substrates. Thus, whereas as yet poorly explored, there might exist an intriguing link between telomere length regulation, homologous recombination and sumoylation.
An alternative hypothesis is that the SUMO-related phenotypes at centromeres (“illegitimate” HR) and telomeres (aberrant elongation) arise by distinct mechanisms. In this work, we demonstrate that telomere elongation induced by lack (or reduction) of sumoylation requires telomerase activity and is not a consequence of unscheduled telomere–telomere recombination. Further, genetic analysis shows pli1 deletion to be dominant over most of the known mutations affecting telomere length control, with the exception of the telomere binding protein Taz1. These results suggest that SUMO acts in a novel pathway downstream of Taz1p to control telomere length, possibly by controlling the localization or activity of a regulator of telomerase.
Results and Discussion
Pli1p Is the Principal SUMO E3-Ligase Involved in Telomere Length Control.
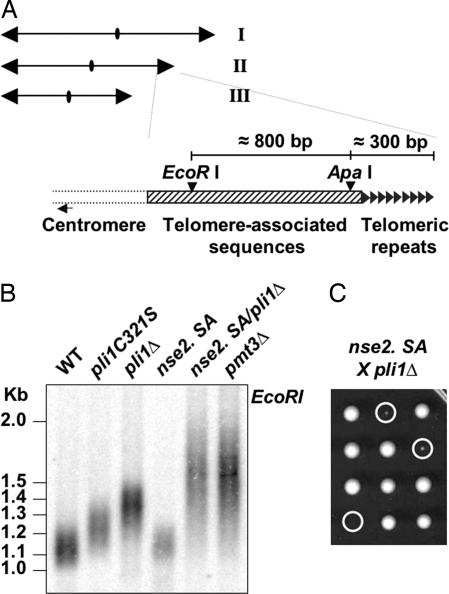
Two proteins possessing a RING domain associated with SUMO E3 ligase activity, Pli1p (19) and Nse2p (24), have so far been described in fission yeast. We have shown previously that pli1 mutants exhibit telomere elongation in inverse correlation with global sumoylation activity (ref. 19 and Fig. 1B). The role of Nse2p in telomere length control, however, has not been examined. As shown in Fig. 1B, the nse2.SA mutation, corresponding to a C195S,H197A conversion in the RING domain that abolishes the SUMO E3 ligase activity of Nse2p (24), does not lead to telomere elongation. A pli1Δ/nse2.SA double mutation, however, recapitulates both the telomere elongation (Fig. 1B) and the growth defects (Fig. 1C) associated with the SUMO encoding pmt3 gene deletion. These results suggest that Pli1p is the major SUMO E3 ligase implicated in telomere length control, and further, that Nse2p dependent sumoylation may account for the difference of telomere length between pli1Δ and pmt3Δ cells. It is noteworthy that a similar mechanism seems not to operate in budding yeast, because deletion of the Pli1-like E3 ligases Siz1 and -2 is not accompanied by telomere length increase (25), whereas mutation of the Nse2 homolog MMS21 (mms21-11) has been shown to lead to a very slight increase in telomere length (26).
Fig. 1.
Pli1p is the SUMO E3 Ligase involved in telomere length regulation in fission yeast. (A) The three chromosomes of haploid S. pombe as well as the EcoRI and ApaI sites in the telomere and telomere-associated sequences are shown. This telomeric structure is present on both arms of chromosome I and II whereas, on chromosome III, the telomeric repeats can be directly adjacent to the rDNA with no telomere associated sequences on one or both arms (P.B., unpublished observation). (B) Southern blot analysis of telomere length in single and double SUMO E3 ligase mutants pli1 and nse2, as indicated (EcoRI digestion and G-rich telomeric DNA probe). (C) Three representative tetrads from a pli1Δ × nse2.SA cross. Circles indicate double nse2.SA/pli1Δ segregants.
Deletion of pli1 Is Dominant over Most of the Known Mutations Affecting Telomere Length Control.
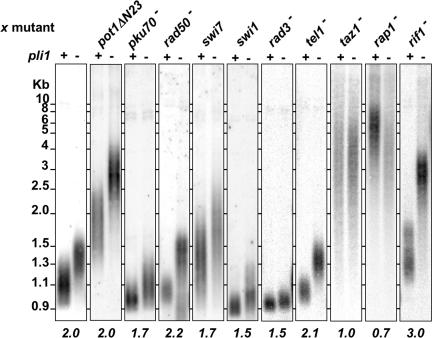
To investigate how low levels of sumoylation lead to telomere length increase, we deleted pli1+ in various mutant backgrounds affecting telomere metabolism. Comparison of telomere length of the double mutants with the corresponding single mutants shows that pli1 deletion has a dominant effect over the mutants of the telomere end protection proteins pot1ΔN23 (27), pku70Δ (2) and rad50Δ (28), the DNA replication proteins swi7 (an allele of pol1+, encoding the pol α catalytic subunit; refs. 29 and 30) and swi1 (a null allele of the Saccharomyces cerevisiae TOF1 homolog required for replication fork pausing, ref. 31), the checkpoint ATM and ATR homologs tel1Δ and rad3Δ (32) and the rif1Δ mutant (33) (Fig. 2). Indeed, the ratio between the median telomere length of double pli1Δ/x mutant over median telomere length of the corresponding single x mutant varies between 1.5 and 3. This indicates that Pli1p and these proteins act in separate pathways to control telomere length.
Fig. 2.
Genetic interactions of pli1 and genes involved in telomere metabolism. Southern blot analysis of telomere length in single and double mutants of genes involved in telomere metabolism and pli1 (EcoR I digestion and G-rich telomeric DNA probe). The numbers below the columns indicate the ratio of median telomere length of double pli1Δ/x mutant over median telomere length of the corresponding single x mutant.
In fission yeast, telomeric repeats are bound by the TRF1/2 homolog Taz1 (34). Recent work has suggested that this protein exerts both positive and negative roles in regulating telomere length, with the latter being mediated by two separate pathways: one involving Rap1 and the other involving Rif1 (33, 35). Whereas pli1+ deletion had no effect on the length of taz1Δ (x1) and a slight effect on rap1Δ telomeres (×0.72), it leads to a synergistic increase (×3) of telomere length in a rif1Δ background. This suggests that Pli1p works in a pathway downstream of Taz1p but parallel to Rif1p. To further assess whether Pli1p acts in the same pathway as Rap1p, we analyzed these mutants for cold sensitivity. Previous work has shown that taz1Δ cells are cold sensitive and that deletion of either rap1 or rif1 in a taz1Δ background exacerbates or suppresses, respectively, the taz1Δ cold sensitivity phenotype (35, 36). As pli1Δ/taz1Δ cells display similar cold sensitivity to taz1Δ cells (data not shown), we propose that Pli1p acts on telomere length downstream of Taz1p but independently of Rap1p and Rif1p.
Telomere Elongation Induced by Lack of Sumoylation Requires Telomerase Activity, Not Telomere–Telomere Recombination.
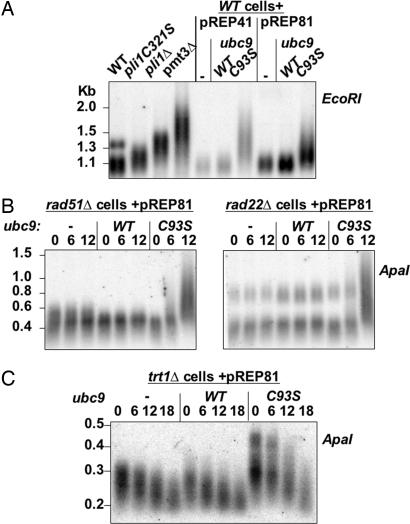
The aberrant lengthening of telomeres in pli1 mutant cells could be due to either telomerase or to telomore–telomere recombination (37). If Pli1p inhibits telomore–telomere recombination, telomere lengthening should not occur in a pli1Δ/rad22Δ (RAD52 homolog) double mutant, and a pli1Δ/trt1Δ (telomerase catalytic subunit) strain might not senesce or would senesce more slowly because of activation of the recombinational pathway for telomere maintenance. To test this hypothesis, however, we could use neither the pli1 deletion, as pli1Δ/rad22Δ and pli1Δ/rad51Δ double mutants are co-lethal (19), nor the pli1C321S hypomorphic mutant, because this mutation is leaky given the slow growth phenotype of pli1C321S/rad22Δ and pli1C321S/rad51Δ mutants (data not shown). Moreover, when we analyzed tetra-type tetrads from a trt1+/−, pli1+/− diploid, we noticed that pli1Δ/trt1Δ double mutants recurrently exhibited telomeres longer than the trt1Δ single mutants from the same tetra-type tetrad [see supporting information (SI) Fig. 5]. However, this cannot be a strict demonstration for the noninvolvement of Trt1p in the telomere elongation phenotype as pli1 deletion might just increase the activity of telomerase inherited from the parental diploid in the trt1Δ germinating spores. Further, given the larger size heterogeneity of trt1Δ telomeres, we sought to conduct this experiment under conditions of a fixed, given telomere length at the start. To this end, we established an inducible physiological “knockdown” of sumoylation by the controlled expression of Ubc9-C93S. The C93S mutation abolishes the capacity of the protein to form a thioester conjugate with SUMO, rendering this protein able to act as a dominant negative inhibitor of endogenous Ubc9p. In this system, cells are transformed with pREP41 or pREP81 plasmids containing no insert, or Ubc9 or Ubc9-C93S cDNAs under the control of two different attenuated forms of the thiamine repressible nmt1 promoter. Transformed cells are cultured under repressive conditions in medium containing thiamine (20 mg/liter) and transferring cells into medium without thiamine induces expression from the nmt1 promoter. For each case, samples for protein and DNA analysis were taken at precise generation times after induction. As shown in Fig. 3A, only WT cells expressing the dominant negative form of Ubc9p exhibit an increase of telomere length after twelve generations postinduction. Moreover, telomere length is again dependent on the degree of inhibition of sumoylation, because expression of Ubc9C93S from the stronger “41” form of the nmt1 promoter produces longer telomeres than when the weaker “81” form is used. We chose to continue with the very attenuated form of pREP81, as under these conditions, cell growth is only modestly affected. By using the pREP81 system, a significant elongation of telomeres was observed in rad51Δ, rad22Δ (Fig. 3B) and rad50Δ cells (data not shown), but not in fresh, nonsenescent trt1Δ cells (Fig. 3C). Thus, this system clearly establishes that telomere elongation induced by lack of sumoylation requires telomerase activity and is not a consequence of unscheduled telomere–telomere recombination.
Fig. 3.
Telomere elongation induced by lack of sumoylation requires telomerase activity, not telomore–telomere recombination. (A) Southern blot analysis of telomere length in cells of the indicated genotype (Left) and in WT cells transformed with pREP41 and pREP81 plasmids containing no, ubc9-WT, or ubc9-C93S insert under the control of the thiamine repressible nmt1 promoter at 12 generations after thiamine removal (EcoRI digestion and G-rich telomeric DNA probe). (B) Southern blot analysis of telomere length in rad51Δ and rad22Δ cells transformed with pREP81 plasmid containing no or ubc9-WT or ubc9-C93S at 0, 6 and 12 generations after nmt1 induction (ApaI digestion and G-rich telomeric DNA probe). (C) Southern blot analysis of telomere length in trt1Δ cells transformed with pREP81 plasmid containing no or ubc9-WT or ubc9-C93S at 0, 6, 12, and 18 generations after nmt1 induction (ApaI digestion and G-rich telomeric DNA probe). To start from fresh, nonsenescent trt1Δ cells, diploid cells heterozygous for trt1 transformed with the indicated plasmids were germinated under conditions selecting for the trt1 deletion and the presence of the plasmids.
Telomere Elongation Induced by Lack of Sumoylation Correlates with High Levels of G-Rich Single-Stranded Telomeric DNA.
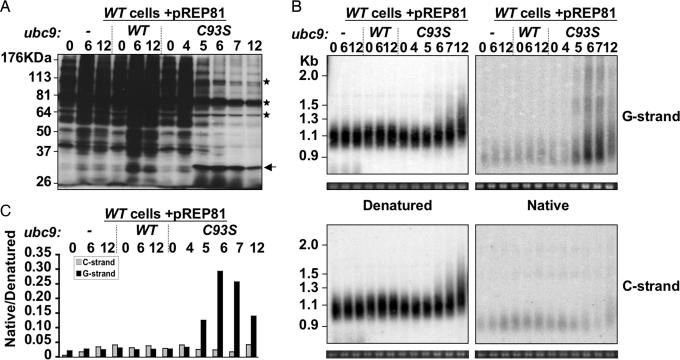
The inducible system also allows following the dynamics of telomere elongation in large volumes of cultures synchronous for loss of sumoylation. We took advantage of this to determine how fast telomere length increases after reduction of sumoylation. The Western Blot against SUMO (Pmt3p) in Fig. 4A shows that WT cells containing the pREP81-ubc9C93S plasmids start to reduce the overall levels of SUMO conjugates five generations after passage to medium without thiamine. This delay likely reflects the time needed for the cells to be depleted of thiamine and for the Ubc9C93S protein to be expressed and act as a dominant negative inhibitor. As shown by the regular telomeric Southern blots (Fig. 4B Left, “Denatured”), telomeres elongate concomitantly with loss of sumoylation, indicating that telomere length is very sensitive to levels of sumoylation in the cell. Production of single stranded G-rich DNA is a hallmark of telomerase activity. Therefore, in parallel with the regular telomeric Southern blot, we prepared “Native” blots in which the acid/base denaturation step before transfer onto nitrocellulose membrane has been omitted to leave preexisting double-stranded DNA intact (2). Surprisingly, we found low levels of both G-rich and C-rich single stranded DNA migrating at the level of 0.9 kb, corresponding to short telomeres of 100–150 bp (Fig. 4B Right, Native) in WT cells. However, the overall level of G-rich single-stranded DNA (Fig. 4B Right, Native) starts increasing at the 5th generation, reaches a peak at the 6th generation before decreasing to a steady state by the 12th generation only in the cells over-expressing Ubc9C93S, in parallel with the observed loss of sumoylation in these cells (Fig. 3C). Notably, this increase in signal intensity of up to 10-fold is specific to the G-rich strand and does not occur for the C-rich strand of the telomere. Moreover, the increase in G-strand signal is accompanied by an increase in telomere length. Therefore, loss of sumoylation likely induces a rapid elongation of the G-overhangs by telomerase, although is possible that some of the hybridization signal on the native gels is internal. In addition to low apparent molecular weight molecules with a G-overhang, that are in keeping with the preferential elongation of short telomeres by telomerase (38), other species of high apparent molecular weight were observed. These might originate from a subset of cells in which inhibition of sumoylation is very efficient, thus leading to very rapid elongation, or may result from long G-tails containing secondary structures, such as G quartets (39) formed in vivo or during the DNA manipulation steps. The presence of low levels of single stranded DNA (2–3%, Fig. 4C) at the level of short telomeres in WT cells is also of particular interest. Indeed, a recent report also found C-overhangs in human replicating cells (40) and S. pombe might provide an amenable genetic system for exploring their formation.
Fig. 4.
Telomere elongation occurs very rapidly after physiological knockdown of sumoylation and is associated with fast G-overhang production. WT cells were transformed with pREP81 plasmids containing no, ubc9-WT, or ubc9-C93S insert under the control of the nmt1 promoter, and cells were harvested 0, 4, 5, 6, 7, and 12 generations after nmt1 induction. (A) Whole-cell extracts were analyzed by Western blot by using an anti-pmt3 antibody. Asterisks indicate cross-reacting bands; the arrow indicates a possible Ubc9-SUMO conjugate. (B) Genomic DNA was digested with EcoRI and subjected to 1% agarose gel electrophoresis. The gels were cut in two and were either treated with HCl and NaOH to denature double-stranded DNA (Left, denatured) or with TBE (Right, native) before a SSC20X wash and transfer to the same nylon membrane. Samples were hybridized either to a C-rich probe (Upper) or to a G-rich probe (Lower). (C) Hybridization signals shown in B were quantified by using ImageQuantNT, and the relative intensities of native over denatured DNA for each sample and probe are plotted.
Collectively, our results implicate a sudden and rapid hyperactivation of telomerase in the telomere length increase, in only one or two cell divisions (i.e., between the 5th and 6th cell division in Fig. 4B) that is associated with low levels of cellular sumoylation. This is not due to an increase in the cellular levels of Trt1p (data not shown). Rather, it may result from more efficient recruitment to the telomere, increased initiation rate or processivity of elongation of the telomerase. One possible model would be that components of the SUMO conjugation pathway (e.g., SUMO, Pli1p or Ubc9p) interact directly with telomere proteins. The recent demonstration of a weak physical interaction between SUMO and Taz1 (41), for example, might suggest that longer telomeres, which contain more binding sites for Taz1, exhibit enhanced recruitment of the sumoylation machinery that, in turn, negatively regulates telomerase activity. SUMO might thus modify one of the components of the telomerase holoenzyme and affect the stability of the complex on the DNA-RNA hybrid. An analogous mechanism has been proposed for the sumoylation-dependent release of thymine DNA glycosylase (TDG) from apurinic sites on the DNA (42, 43). SUMO might also help to activate or localize proteins at the telomere that either compete with telomerase for G-overhang binding or actively remove telomerase from the telomere. An example for the latter has recently been described in S. cerevisiae, the helicase Pif1 (44). A further possibility is that hyposumoylation exerts a more general effect by, for example, opening an otherwise repressive chromatin structure, thus leading to the inappropriate activation of telomerase. Support for such a mechanism comes from recent studies in mice demonstrating that loss of retinoblastoma family proteins (Rb1, Rbl1 and Rbl2), DNA methyl transferase (DNMT1 or DNMT3a, -b) or histone methyl transferase (Suv39h1, -2) function is associated with abnormal telomere elongation (45–47).
Future work will be aimed identifying the specific SUMO substrates(s) as well as their downstream targets. One prediction from the present results is that such substrates should be responsive to Pli1 (i.e., PIAS-like) SUMO E3 ligase. Finally, it will be of great interest to know whether the findings obtained here in fission yeast are transposable to higher eukaryotic or mammalian model systems.
Materials and Methods
Fission Yeast Strains, Plasmids, Media, and Methods.
The S. pombe strains used in this study are listed in SI Table 1. Growth, maintenance and standard genetic methods for fission yeast strains were as described by (48). The density of liquid cultures was determined by Coulter counting. Double pli1Δ/x mutants were constructed by mating and tetrad analysis. For each mutant, two tetra-type tetrads (containing all possible types of segregants) were streaked three times before the telomere length was assessed. To ensure for telomere length stability it was verified that the single mutants and WT segregants had reached their known steady state telomere length. The trt1 deletion was kept in a heterozygous diploid, transformed with the plasmids and let to sporulate. Fresh trt1Δ haploids containing the plasmids were obtained by random spore analysis. Further details about strain and plasmid constructions are available upon request.
Preparation of S. pombe Whole-Cell Extracts and Western Blot Analysis.
Samples of 108 cells were harvested and washed with 1 ml of cold water, and the pellet was frozen at –80°C until Western blot analysis, as described (19).
Preparation of DNA and Southern Hybridization.
Samples of 4 × 108 cells were harvested, washed with 1 ml of cold water and the pellet was frozen at −80°C until gDNA was prepared by standard methods (49). For Southern blots, 1 μg of gDNA was digested 2 h with either EcoRI or ApaI. The digested DNA was separated by electrophoresis on a 1% agarose gel containing 0.0001% ethidium bromide. After electrophoresis, the gel was subjected to a 10 min. acid wash with 0.13 M HCl and two 10-min base washes with 0.5 M NaOH/1.5 M NaCl, except in the case of overhang analysis (Fig. 3B Right, Native) for which the gel was only subjected to a 10 min. wash with the 20×SSC transfer buffer (3 M NaCl, 0.3 M tri-sodium citrate, pH 7.0). DNA was transferred to a nitrocellulose membrane (Hybond-n + filters, Amersham) by capillary transfer and cross linked with UV Stratalinker (Stratagene). The membrane was prehybridized 1 h in hybridization buffer (5× SSC, 5× Denhardt's, 0.5% SDS, denatured sonicated salmon sperm DNA at 0.05 mg/ml) and hybridized overnight with hybridization buffer and the probe at 45°C. The probe was a telomeric repeat oligonucleotide (10 pmol), 5′-end labeled with T4 PNK and 40 μCi (1 Ci = 37 GBq) [γ-32P]ATP, and purified with a super fine G25 Sepharose column. After hybridization, the membrane was washed first in 2× SSC (low stringency) then in 0.2× SSC (high stringency) and exposed on a phosphor screen. The signal was detected by PhosphorImager 445SI and quantified by using ImagequantNT. The sequences of the telomeric probes used are: C-rich, tgtaaccgtgtgtaaccacgtaaccttgtaaccc; G-rich, gggttacaaggttacgtggttacacacggttaca
Supplementary Material
Acknowledgments
We thank Julie Cooper (Cancer Research UK, London, U.K.), Kazunori Tomita (Cancer Research UK, London, U.K.), Fuyuki Ishikawa (Department of Gene Mechanisms, Kyoto University, Kyoto, Japan), Junko Kanoh (Department of Gene Mechanisms, Kyoto University, Kyoto, Japan), Katsunori Tanaka (Department of Biophysics and Biochemistry, University of Tokyo, Mongo, Tokyo, Japan), Albert Pastink (Department of Toxicogenetics, Leiden University, Leiden, The Netherlands), Sang Dai Park (School of Biological Sciences, Seoul National University, Seoul, South Korea), Hiroshi Iwasaki (Division of Molecular and Cellular Biology, Yokohama City University, Yokohama, Japan), and Yufuko Akamatsu (Molecular Biology Program, Memorial Sloan–Kettering Cancer Center, New York, NY), and Amar Klar (National Cancer Institute, Frederick, MD) for providing us with strains; Miguel G. Ferreira for fruitful discussions; and Olivier Danot, Kyle M. Miller, and Andrew Bannister for useful comments on the manuscript. This work was supported by grants from the Human Frontiers in Science Program (HFSP), the Association pour la Recherche sur le Cancer (ARC), and the European Union. Fellowship support was from the Ministère de l'Education Nationale de la Recherche et de la Technologie (MENRT) and ARC (to B.X.), and from the Erasmus Program (to E.M.R.).
Abbreviations
- HR
homologous recombination
- SUMO
small ubiquitin-like modifier.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0605442104/DC1.
References
- 1.de Lange T. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 2.Baumann P, Cech TR. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 3.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 4.Ferreira MG, Miller KM, Cooper JP. Mol Cell. 2004;13:7–18. doi: 10.1016/s1097-2765(03)00531-8. [DOI] [PubMed] [Google Scholar]
- 5.Cech TR. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 6.Smogorzewska A, de Lange T. Annu Rev Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 7.Miller KM, Rog O, Cooper JP. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka K, Nishide J, Okazaki K, Kato H, Niwa O, Nakagawa T, Matsuda H, Kawamukai M, Murakami Y. Mol Cell Biol. 1999;19:8660–8672. doi: 10.1128/mcb.19.12.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson ES. Annu Rev Biochem. 2004;73:355–382. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 10.Müller S, Ledl A, Schmidt D. Oncogene. 2004;23:1998–2008. doi: 10.1038/sj.onc.1207415. [DOI] [PubMed] [Google Scholar]
- 11.Seeler JS, Dejean A. Nat Rev Mol Cell Biol. 2003;4:690–699. doi: 10.1038/nrm1200. [DOI] [PubMed] [Google Scholar]
- 12.Verger A, Perdomo J, Crossley M. EMBO Rep. 2003;4:137–142. doi: 10.1038/sj.embor.embor738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen Z, Pardington-Purtymun PE, Comeaux JC, Moyzis RK, Chen DJ. Genomics. 1996;36:271–279. doi: 10.1006/geno.1996.0462. [DOI] [PubMed] [Google Scholar]
- 14.Shen Z, Pardington-Purtymun PE, Comeaux JC, Moyzis RK, Chen DJ. Genomics. 1996;37:183–186. doi: 10.1006/geno.1996.0540. [DOI] [PubMed] [Google Scholar]
- 15.Eladad S, Ye TZ, Hu P, Leversha M, Beresten S, Matunis MJ, Ellis NA. Hum Mol Genet. 2005;14:1351–1365. doi: 10.1093/hmg/ddi145. [DOI] [PubMed] [Google Scholar]
- 16.Woods YL, Xirodimas DP, Prescott AR, Sparks A, Lane DP, Saville MK. J Biol Chem. 2004;279:50157–50166. doi: 10.1074/jbc.M405414200. [DOI] [PubMed] [Google Scholar]
- 17.Papouli E, Chen S, Davies AA, Huttner D, Krejci L, Sung P, Ulrich HD. Mol Cell. 2005;19:123–133. doi: 10.1016/j.molcel.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. Nature. 2005;436:428–433. doi: 10.1038/nature03665. [DOI] [PubMed] [Google Scholar]
- 19.Xhemalce B, Seeler JS, Thon G, Dejean A, Arcangioli B. EMBO J. 2004;23:3844–3853. doi: 10.1038/sj.emboj.7600394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krogh BO, Symington LS. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 21.Muntoni A, Reddel RR. Hum Mol Genet. 2005;14((Spec No 2)):R191–R196. doi: 10.1093/hmg/ddi266. [DOI] [PubMed] [Google Scholar]
- 22.Yeager TR, Neumann AA, Englezou A, Huschtscha LI, Noble JR, Reddel RR. Cancer Res. 1999;59:4175–4179. [PubMed] [Google Scholar]
- 23.Henson JD, Neumann AA, Yeager TR, Reddel RR. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- 24.Andrews EA, Palecek J, Sergeant J, Taylor E, Lehmann AR, Watts FZ. Mol Cell Biol. 2005;25:185–196. doi: 10.1128/MCB.25.1.185-196.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson ES, Gupta AA. Cell. 2001;106:735–744. doi: 10.1016/s0092-8674(01)00491-3. [DOI] [PubMed] [Google Scholar]
- 26.Zhao X, Blobel G. Proc Natl Acad Sci USA. 2005;102:4777–4782. doi: 10.1073/pnas.0500537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bunch JT, Bae NS, Leonardi J, Baumann P. Mol Cell Biol. 2005;25:5567–5578. doi: 10.1128/MCB.25.13.5567-5578.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartsuiker E, Vaessen E, Carr AM, Kohli J. EMBO J. 2001;20:6660–6671. doi: 10.1093/emboj/20.23.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh J, Klar AJ. Nature. 1993;361:271–273. doi: 10.1038/361271a0. [DOI] [PubMed] [Google Scholar]
- 30.Dahlen M, Sunnerhagen P, Wang TS. Mol Cell Biol. 2003;23:3031–3042. doi: 10.1128/MCB.23.9.3031-3042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Egel R, Beach DH, Klar AJ. Proc Natl Acad Sci USA. 1984;81:3481–3485. doi: 10.1073/pnas.81.11.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naito T, Matsuura A, Ishikawa F. Nat Genet. 1998;20:203–206. doi: 10.1038/2517. [DOI] [PubMed] [Google Scholar]
- 33.Kanoh J, Ishikawa F. Curr Biol. 2001;11:1624–1630. doi: 10.1016/s0960-9822(01)00503-6. [DOI] [PubMed] [Google Scholar]
- 34.Cooper JP, Nimmo ER, Allshire RC, Cech TR. Nature. 1997;385:744–747. doi: 10.1038/385744a0. [DOI] [PubMed] [Google Scholar]
- 35.Miller KM, Ferreira MG, Cooper JP. EMBO J. 2005;24:3128–3135. doi: 10.1038/sj.emboj.7600779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller KM, Cooper JP. Mol Cell. 2003;11:303–313. doi: 10.1016/s1097-2765(03)00041-8. [DOI] [PubMed] [Google Scholar]
- 37.Nakamura TM, Cooper JP, Cech TR. Science. 1998;282:493–496. doi: 10.1126/science.282.5388.493. [DOI] [PubMed] [Google Scholar]
- 38.Teixeira MT, Arneric M, Sperisen P, Lingner J. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 39.Keniry MA. Biopolymers. 2000;56:123–146. doi: 10.1002/1097-0282(2000/2001)56:3<123::AID-BIP10010>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 40.Cimino-Reale G, Pascale E, Alvino E, Starace G, D'Ambrosio E. J Biol Chem. 2003;278:2136–2140. doi: 10.1074/jbc.M208939200. [DOI] [PubMed] [Google Scholar]
- 41.Spink K, Ho JC, Tanaka K, Watts FZ, Chambers A. Biochem Genet. 2005;43:103–117. doi: 10.1007/s10528-005-1503-4. [DOI] [PubMed] [Google Scholar]
- 42.Baba D, Maita N, Jee JG, Uchimura Y, Saitoh H, Sugasawa K, Hanaoka F, Tochio H, Hiroaki H, Shirakawa M. Nature. 2005;435:979–982. doi: 10.1038/nature03634. [DOI] [PubMed] [Google Scholar]
- 43.Hardeland U, Steinacher R, Jiricny J, Schar P. EMBO J. 2002;21:1456–1464. doi: 10.1093/emboj/21.6.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boule JB, Vega LR, Zakian VA. Nature. 2005;438:57–61. doi: 10.1038/nature04091. [DOI] [PubMed] [Google Scholar]
- 45.Garcia-Cao M, Gonzalo S, Dean D, Blasco MA. Nat Genet. 2002;32:415–419. doi: 10.1038/ng1011. [DOI] [PubMed] [Google Scholar]
- 46.Gonzalo S, Jaco I, Fraga MF, Chen T, Li E, Esteller M, Blasco MA. Nat Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 47.Garcia-Cao M, O'Sullivan R, Peters AH, Jenuwein T, Blasco MA. Nat Genet. 2004;36:94–99. doi: 10.1038/ng1278. [DOI] [PubMed] [Google Scholar]
- 48.Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with Fission Yeast. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1993. [Google Scholar]
- 49.Moreno S, Klar A, Nurse P. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.