Abstract
Activation of the transcription factor NF-κB after stimulation through antigen receptors is important for lymphocyte differentiation, activation, proliferation, and protection against apoptosis. Much progress has been made in understanding the molecular events leading to NF-κB activation, but how this activation is eventually down-regulated is less well understood. Recent studies have indicated that Bcl10 functions downstream of lymphocyte antigen receptors to promote the activation of the IκB kinase complex leading to the phosphorylation and degradation of the IκB inhibitors of NF-κB. Bcl10 has also been implicated in the pathogenesis of mucosa-associated lymphoid tissue lymphoma, possibly in association with its nuclear localization. Here, we provide evidence that the IκB kinase complex phosphorylates Bcl10 after T cell antigen receptor stimulation and causes its proteolysis via the β-TrCP ubiquitin ligase/proteasome pathway. These findings document a negative regulatory activity of the IKK complex and suggest that Bcl10 degradation is part of the regulatory mechanisms that precisely control the response to antigens. Mutants of Bcl10 in the IKK phosphorylation site are resistant to degradation, accumulate in the nucleus, and lead to an increase in IL-2 production after T cell antigen receptor stimulation.
Keywords: NF-kB, ubiquitination, signal transduction
The NF-κB signaling cascade is critically involved in the regulation of the immune, inflammatory, and antiapoptotic responses (1). The NF-κB proteins are sequestered in the cytoplasm through physical interaction with inhibitors of the IκB family. After a number of extracellular signals, IKK, a cytoplasmic kinase complex, becomes activated and phosphorylates the IκBs, leading to their degradation through the ubiquitin-proteasome pathway. NF-κB dimers then translocate to the nucleus and activate their target genes. T cell activation by the antigen receptor leads to the activation of multiple signaling pathways, including NF-κB. Research over the past decade has uncovered more than a dozen proteins that link the T cell antigen receptor (TCR) to IKK (1). Among those, Bcl10 plays a pivotal role in regulating T cell functions, together with two interacting partners: CARMA1 (CARD11/Bimp3), a member of the family of membrane-associated guanylate kinases (MAGUK), which was identified by several groups as a Bcl10 interacting protein (2–5), and MALT1, a member of the paracaspase family (6, 7). Thus, Bcl10, CARMA1, and MALT1 (also called the CBM complex) represent three essential players of NF-κB activation in antigen-stimulated lymphocytes, and their deficiency results in profound defects in proliferation, cytokine production, and NF-κB activation in response to antigen stimulation (8–10).
Recently, it has been shown that two kinases, PDK1 and its downstream target PKCθ, are involved in the recruitment of the CBM and the IKK complexes to the lipid rafts of the plasma membrane (11). This in turn leads to the activation of the ubiquitin ligase activity of TRAF6 (12) and the polyubiquitination of NEMO through Lys-63-linked chains (13). Autoubiquitination of TRAF6 allows recruitment and activation of a complex that includes the kinase TAK1, which acts as an IKK kinase (14).
While the events that lead to NF-κB activation are reasonably well understood, how this activation is terminated is less clear. Resynthesis of the IκB inhibitors as well as deubiquitination of certain critical components of the cascade by the CylD and A20 deubiquitinases have been identified as negative regulators of the pathway. Here, we demonstrate that, in T cells, Bcl10 is phosphorylated on multiple sites by IKK in response to TCR activation and subsequently degraded in the nucleus through the ubiquitin/proteasome machinery. These results suggest that negative regulation of Bcl10 by IKK represents a negative-feedback loop required to selectively terminate NF-κB signaling in response to TCR activation. Consequently, expression of a nondegradable form of Bcl10 in a mouse T cell hybridoma gives rise to an enhancement of IL-2 production in response to TCR stimulation. Interestingly, expression of this mutant also results in its nuclear localization in response to TCR activation, and to its accumulation in speckles similar to the ones observed in certain types of MALT lymphoma.
Results
Bcl10 Degradation Depends on NEMO and CARMA1 and Is Blocked by Proteasome Inhibitors.
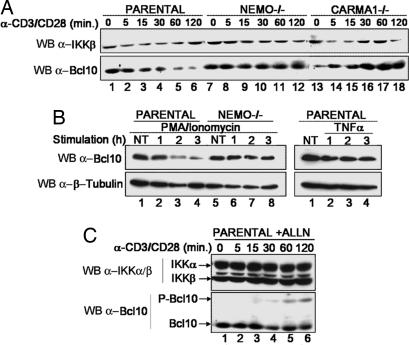
It has been recently shown that Bcl10 undergoes degradation upon TCR stimulation of various T cell types (15). To test whether Bcl10 degradation depends on NEMO and CARMA1, we performed TCR ligation using antibodies against CD3 and CD28 in WT Jurkat cells and in NEMO- or CARMA1-deficient Jurkat cell lines (16, 17). As reported previously by Scharschmidt and collaborators (15), we observed that Bcl10 was degraded within 2 h in WT Jurkat cells after TCR ligation (Fig. 1A, lanes 1–6) or phorbol 12-myristate 13-acetate (PMA)/ionomycin treatment (Fig. 1B Left, lanes 1–4). Interestingly, we observed that Bcl10 was not degraded in NEMO-deficient (Fig. 1 A, lanes 7–12, and B, lanes 5–8) or CARMA1-deficient cells (lanes 13–18).
Fig. 1.
NEMO-, CARMA1-, and proteasome-dependent degradation of Bcl10 in T cells in response to PMA and CD3/CD28 costimulation. (A) Kinetics of Bcl10 degradation in response to CD3/CD28 costimulation. Jurkat parental cells (lanes 1–6) as well as NEMO-deficient (lanes 7–12) or CARMA1-deficient (lanes 13–18) cells were treated with CD3/CD28 antibodies for the indicated period, and the amount of Bcl10 was measured by immunoblotting. The level of expression of IKKβ is shown as a loading control. (B) PMA/ionomycin but not TNF-α induces Bcl10 degradation. Jurkat cells (Left, lanes 1–4, and Right) or NEMO-deficient cells (lanes 5–8) were stimulated for the indicated period (hours) with PMA or TNF-α. Levels of Bcl10 and β-tubulin as a loading control were determined by Western blotting. (C) Bcl10 is degraded by the proteasome. Jurkat cells were pretreated with ALLN for 30 min and treated with ALLN and CD3/CD28 antibodies for the indicated period, and the levels of Bcl10, IKKα, and IKKβ were determined by Western blotting.
To test whether Bcl10 degradation is specific for TCR ligation, Jurkat cells were treated with TNF-α (Fig. 1B Right). No Bcl10 degradation was observed, whereas IκBα was efficiently degraded (data not shown).
Taken together, these data demonstrate that Bcl10 is specifically degraded after TCR or PMA stimulation and that this degradation process requires NEMO and CARMA1. We also demonstrated that a proteasome inhibitor, ALLN, efficiently blocked Bcl10 degradation (Fig. 1C) and induced the appearance of a slower migrating form of Bcl10 after 30 min of CD3/CD28 costimulation (lanes 4–6), which likely represents a hyperphosphorylated isoform of Bcl10 (see below).
Inhibition of IKK Prevents Bcl10 Degradation.
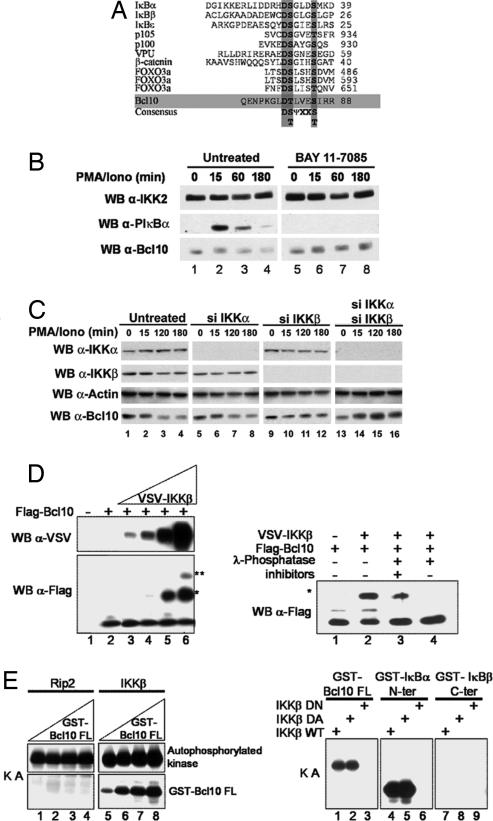
Because we observed that TCR-induced Bcl10 degradation depends on NEMO and the proteasome pathway, we hypothesized that Bcl10 could be phosphorylated by the NEMO/IKK complex. Analysis of the Bcl10 protein sequence revealed a Threonine and a Serine (Thr-81 and Ser-85) in an environment similar to the consensus phosphorylation site/E3 ligase recognition sequence found within IκBs, p105, p100, and Foxo3A (Fig. 2A).
Fig. 2.
IKKβ-dependent Bcl10 phosphorylation is required for its degradation. (A) Sequence alignment of proteins that are polyubiquitinated by the β-TrCP E3 ligase. The ubiquitination of these proteins is caused by phosphorylation by IKK, except for VPU and β-catenin. The phosphoacceptor sites are the S and T indicated in bold. In the consensus sequence, ψ represents an hydrophobic amino acid, and X represents any amino acid. (B) Pharmacological inhibition of IKK inhibits Bcl10 degradation. Jurkat cells were pretreated with the IKK inhibitor Bay 11-7085 for 30 min. Time course analysis of Bcl10 expression (bottom gel) and IκBα phosphorylation [using a phosphospecific anti-IκBα antibody (middle gel)] reveals that Bay 11-7085 abrogates PMA/ionomycin-induced IκBα phosphorylation and Bcl10 degradation. IKK2 was used as a loading control. (C) siRNA-mediated depletion of IKKβ, and to a lesser extent IKKα, inhibits Bcl10 degradation. Jurkat cells were either left untransfected or transfected with IKKβ siRNA, IKKβ siRNA, or both, as mentioned in the figure. Cells were then treated with PMA/ionomycin for the indicated time. The level of expression of IKKα, IKKβ, actin (loading control), and Bcl10 were determined by Western blotting. (D) (Left) Ex vivo phosphorylation of Bcl10 by IKKβ. (Left) Flag-tagged Bcl10 was expressed in HEK-293T cells in the absence (−) or presence (+) of increasing amounts of VSV-tagged IKKβ. Bcl10 was analyzed by Western blotting with anti-Flag antibody and reveals the IKK-dependent accumulation of higher-molecular-weight products (∗ and ∗∗). Expression of transfected IKKβ was also monitored by immunoblotting with anti-VSV. (Right) Lysates of cells cotransfected with Flag-Bcl10 and VSV-IKKβ (lane 2) were treated with λ-phosphatase alone (lane 4) or with λ-phosphatase and phosphatase inhibitors (lane 3), and analyzed by Western blotting with anti-Bcl10 anti-serum. (E) In vitro phosphorylation of Bcl10 by IKKβ. (Left) VSV-Rip2 and VSV-IKKβ were transiently transfected in HEK293T cells, and kinase activity was assayed (KA) by immune complex reaction in the presence of increasing amounts of GST-Bcl10 (1, 2.5, 5, and 10 μg). (Right) VSV-IKKβ WT, Flag-IKKβ DA (a constitutively active mutant), and VSV-IKKβ DN (a kinase-dead mutant) were transiently transfected into HEK-293T cells and immunoprecipitated. In vitro kinase assays (KA) were performed by using 2.5 μg of GST-Bcl10, GST-IκBα N-ter (as a positive control), or GST-IκBβ C-ter (as a negative control).
To determine whether Bcl10 degradation could be blocked by pharmacological inhibition of IKK, we tested the effect of the IKK inhibitor Bay 11-7085 on PMA/ionomycin-induced degradation of Bcl10 (Fig. 2B) and observed a complete inhibition of Bcl10 as well as IκBα phosphorylation (compare lanes 6–8 and 2–4). Because Wegener et al. (18), using the same type of experiments, did not observe any effect on signal-induced Bcl10 degradation, we decided to inactivate IKK by RNA interference (RNAi) (Fig. 2C). The results confirmed and extended the data obtained in our BAY 11-7085 experiment: down-regulation of IKKβ (Fig. 2C, bottom panel, lanes 9–12) completely inhibited PMA/ionomycin-induced Bcl10 degradation, whereas that of IKKα was less efficient (lanes 5–8).
Bcl10 Is Phosphorylated by the NEMO/IKK Complex.
To test whether IKKβ could induce Bcl10 phosphorylation, we transfected HEK-293T cells with Flag-tagged Bcl10 alone or together with increasing amounts of VSV-tagged IKKβ (Fig. 2D Left). Under these conditions, we observed that IKKβ overexpression led to the accumulation of slower migrating forms of Bcl10 (lanes 4–6). Treatment with λ phosphatase (Fig. 2D Right) indicates that these slowly migrating bands represent hyperphosphorylated forms of Bcl10.
To evaluate whether IKK directly phosphorylates Bcl10 in vitro, we generated a recombinant GST-Bcl10 fusion protein. We then performed in vitro kinase assays using GST-Bcl10 as a substrate and immunoprecipitated IKKβ or RIP2 derived from transfected HEK-293T cells as a source of kinase activity (Fig. 2E). Under these conditions, GST-Bcl10 was phosphorylated by IKKβ (Fig. 2E Left, lanes 5–8) but not by Rip2 (lanes 1–4). This result demonstrates that IKKβ, but not RIP2, directly phosphorylates Bcl10. RIP2 may, however, activate an intermediate kinase, because it has been shown to be involved in TCR-induced Bcl10 phosphorylation (19).
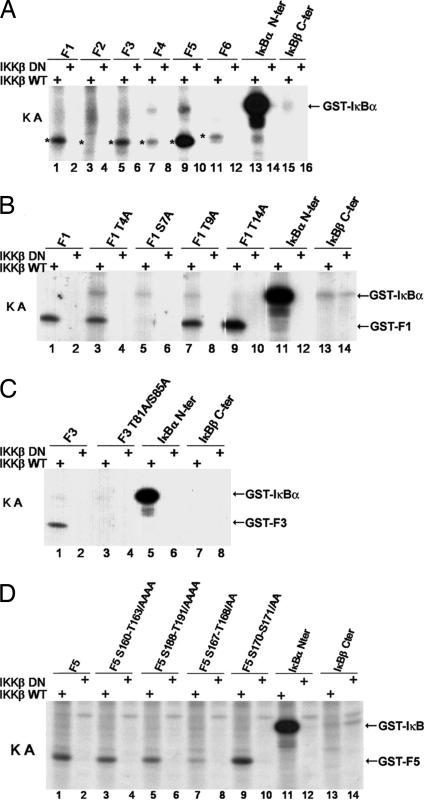
Further experiments were performed to determine whether the putative consensus sequence of Bcl10 is the actual target of IKK phosphorylation. To this end, we produced several nonoverlapping fragments of Bcl10 fused to GST and used them as substrates in an IKKβ-immunocomplex kinase assay. Three of these fragments were phosphorylated by IKKβ, F1 (amino acids 1–38), F3 (74–112), and F5 (155–196) (Fig. 3A, lanes 1, 5, and 9), whereas the other fragments, F2 (39–73), F4 (113–154), and F6 (197–232) were not (Fig. 3A, lanes 3, 7, and 11). To narrow down the potential phosphorylation site(s), we mutated several individual Ser or Thr residues in fragments F1, F3, and F5 and used them as substrates in the IKKβ kinase assay. In F1, mutation of Ser-7 abolished phosphorylation, whereas mutation of Thr-4, -9, or -14 did not (Fig. 3B, compare lane 5 with lanes 1, 3, 7, and 9). As mentioned above, F3 contained the consensus recognition site for IKK phosphorylation/β-TrCP ubiquitination. Therefore, we mutated Thr-81 and Ser-85 to Ala and observed that the phosphorylation of the mutant peptide was totally abrogated (Fig. 3C, compare lane 3 with lane 1), confirming that the DTLVES sequence is a bona fide IKKβ phosphorylation site. Finally, various mutations (Ser or Thr clusters) were introduced into F5 as indicated in Fig. 3D, and in vitro phosphorylation experiments were performed. Although mutation of three clusters (amino acids 160–164, 170–171, and 188–191) did not result in a decrease of F5 phosphorylation (compare lanes 3, 5, 9 to lane 1), phosphorylation was attenuated by substitution of Ser-167 and Thr-168 to Ala (compare lane 7 to lane 1). Incidentally, we observed that the appearance of the major slowly migrating band observed in Fig. 2 was strongly affected by the substitution of these phosphorylation sites [see supporting information (SI) Fig. 7].
Fig. 3.
Mapping of IKKβ-induced Bcl10 phosphorylation sites. (A) In vitro phosphorylation of Bcl10 fragments (F1–F6) by IKKβ. VSV-tagged IKKβ, either WT or dominant negative (DN), were expressed in HEK-293T cells, and immunoprecipitates were used for in vitro kinase assays (KA) with fragments of Bcl10 fused to GST, as indicated above the lanes (the relevant bands are indicated by asterisks). (B–D) Mutation analysis of IKK-mediated Bcl10 phosphorylation. GST fused to fragment F1 [amino acids 11–38 (B)], F3 [74–112 (C)], or F5 [155–196 (D)] of Bcl10 and mutants of the indicated Ser (S) or Thr (T) residues were subjected to in vitro kinase assays as described in A.
We finally examined whether F1, F3, and F5 could be phosphorylated by IKKα in vitro. Our results demonstrated that IKKα and IKKβ phosphorylate Bcl10 in vitro on the same phosphorylation sites (data not shown).
Bcl10 Interacts with and Is Ubiquitinated by β-TrCP.
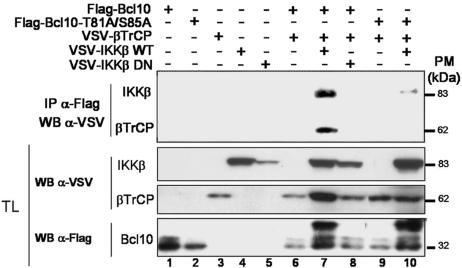
Because the sequence surrounding Thr-81 and Ser-85 shows a strong homology to the consensus recognition site for the E3 ubiquitin ligase β-TrCP, its phosphorylation by IKK is expected to recruit β-TrCP to Bcl10. To assess whether Bcl10/β-TrCP interaction can take place ex vivo, the two proteins were expressed in HEK-293T cells either alone, together, or in association with active or inactive forms of IKKβ, as indicated in Fig. 4. Bcl10/β-TrCP association was demonstrated by coimmunoprecipitation and was found to depend on the cotransfection of IKKβ (Fig. 4, compare lane 7 with lane 6). Mutation of the two phosphoacceptor residues (T81A/S85A) abolished Bcl10–β-TrCP interaction (Fig. 4, lanes 9 and 10). In addition, we could also detect an interaction between Bcl10 and IKKβ (Fig. 4, lane 7), which was reduced by the mutation of the two phosphoacceptor residues (lane 10). In addition, Bcl10/β-TrCP association requires the presence of IKKβ and of the two conserved phosphoacceptor residues of Bcl10 and results in Bcl10 ubiquitination (SI Fig. 8).
Fig. 4.
The presence of active IKKβ is necessary for the association between Bcl10 and β-TrCP. HEK-293T cells were transfected with the indicated constructs. Cell extracts were immunoprecipitated with anti-Flag and immunoblotted with anti-VSV (Upper), and total lysates (TL) were blotted with the indicated antibodies (Lower).
Thr-81 and Ser-85 of Bcl10 Contribute to TCR-Induced Bcl10 Degradation.
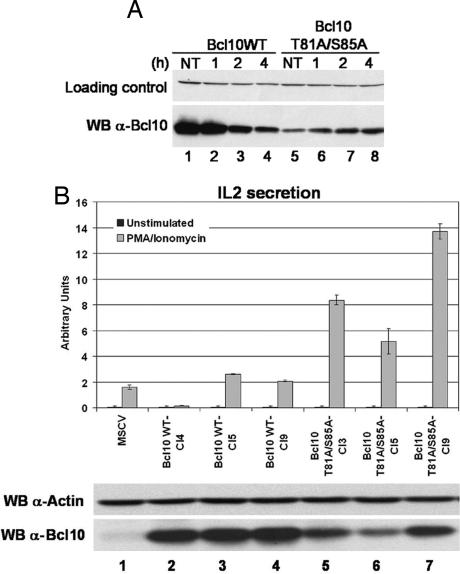
Experiments in Figs. 2 and 4 were performed by overexpression in nonlymphoid cells. To further our understanding of the regulation of Bcl10 in T cells, we tested the impact of TCR stimulation on the degradation of Bcl10 in E29.1 cells (an antigen-specific murine T cell hybridoma) stably expressing the WT form of human Bcl10 or the T81A/S85A mutant. After incubation with anti-TCR mAb H57 in the presence of cycloheximide (to avoid Bcl10 resynthesis) for the indicated period, E29.1 cells were lysed, and Bcl10 degradation was measured by immunoblotting with an antibody that only recognizes its human form. This experiment showed that TCR stimulation induced a strong reduction of WT Bcl10 within 2 h after stimulation (Fig. 5A, lanes 1–4), whereas the T81A/S85A mutant was resistant to this degradation (lanes 5–8).
Fig. 5.
The Bcl10 T81A/S85A mutation prevents degradation and increases IL-2 production. (A) Bcl10 degradation in response to TCR stimulation is prevented by mutation of the T81/S85 phosphorylation site. E29.1 murine T cells transduced with WT human Bcl10 (lanes 1–4) or the T81A/S85A mutant (lanes 5–8) were treated with anti-TCR mAb for the indicated period in the presence of cycloheximide, and the expression of Bcl10 was determined by Western blotting. (B) Increased IL-2 production in cells expressing Bcl10 T81A/S85A. (B Lower) Levels of Bcl10 were measured in E29.1 cells expressing the empty vector MSCV (lane 1), in three individual clones expressing WT Bcl10 (lanes 2–4) and in three individual clones expressing Bcl10 T81A/S85A (lanes 5–7). (B Upper) IL-2 production after 14 h of PMA/ionomycin treatment was assayed in the E29.1 clones.
Enhancement of IL-2 Production in Bcl10 T81A/S85A-Expressing Cells.
Because Bcl10 degradation results in down-regulation of NF-κB activity, it is expected that its stabilization will lead to an increased production of IL-2 in activated T cells. To confirm this prediction, individual E29.1 clones expressing either Bcl10 WT or Bcl10 T81A/S85A were isolated by limiting dilution. These clones were first assayed for expression of Bcl10 by Western blot analysis. Three cell lines expressing comparable levels of either WT or mutant Bcl10 were selected for further studies (Fig. 5B Lower). E29.1 cell lines were stimulated with PMA/ionomycin, and IL-2 production was assayed by ELISA (Fig. 5B Upper). When compared with E29.1 clones expressing the empty vector MSCV or Bcl10 WT, cells expressing mutant Bcl10 demonstrated a marked enhancement of IL-2 production.
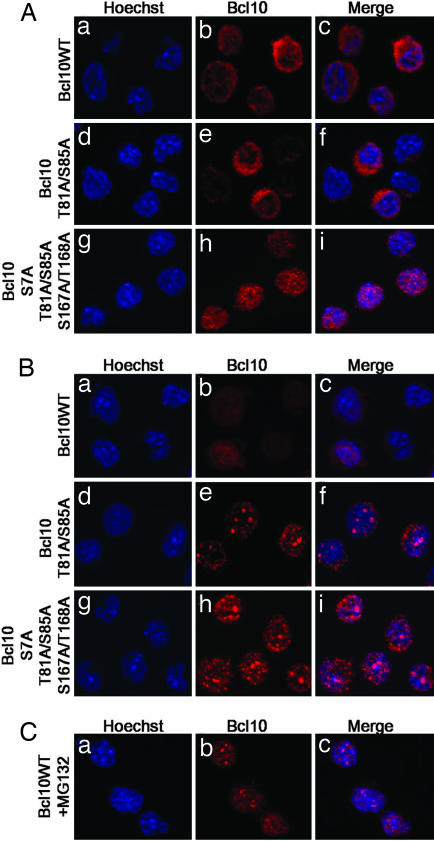
Mutation of the T81/S85 Bcl10 Phosphorylation Sites Results in Its Nuclear Localization upon TCR Stimulation.
Because nuclear localization of Bcl10 has been reported in a number of cases of MALT lymphoma, we determined the subcellular distribution of Bcl10 and its mutants before and after TCR stimulation (Fig. 6). In resting E29.1 cells, WT and T81A/S85A Bcl10 showed a diffuse staining that was detected in the cytoplasm (Fig. 6 Aa–Af). However, substitution of the in vitro IKK phosphorylation sites (Bcl10 S7A/T81A/S85A/S167A/T168A) unexpectedly resulted in a slightly granular nuclear staining (Fig. 6 Ag–Ai). After 5 h of TCR stimulation, whereas WT Bcl10 was still localized in the cytoplasm (Fig. 6 Ba–Bc; most of Bcl10 is degraded at this time, but the rest of it still shows cytoplasmic staining), Bcl10 T81A/S85A and Bcl10 S7A/T81A/85A/S167A/T168A were mainly localized in punctate structures in the nucleus (Fig. 6 Bd–Bi). Interestingly, after 2 h of TCR stimulation in the presence of the proteasome inhibitor MG132, WT Bcl10 was also localized in punctate structures in the nucleus (Fig. 6C).
Fig. 6.
Subcellular localization of WT and mutant (T81A/S85A and S7A/T81A/S85A/S167A/T168A) Bcl10 in untreated (A) or TCR-activated (B and C) E29.1 cells. Mutation of the T81/S85 phosphorylation site is sufficient to induce nuclear localization of Bcl10 upon TCR stimulation. A similar localization can be observed for the WT molecule in the presence of proteasome inhibitors [MG132 (C)]. In each panel, E29.1 cells were stained with Hoechst to reveal DNA (Left) and with anti-Bcl10 antiserum (Center).
Thus, these experiments suggest that in response to TCR signaling, Bcl10 translocates to the nucleus before being degraded.
Discussion
The data presented in this study demonstrate that Bcl10 is the target of a negative regulatory event, which is part of the shut-off response that usually follows activation of the NF-κB pathway. We provide evidence that TCR stimulation induces Bcl10 degradation through the ubiquitin/proteasome machinery, after phosphorylation by the IKK complex and ubiquitination by the β-TrCP E3 ligase. Transfection and in vitro kinase assays reveal that both IKKα and IKKβ are able to phosphorylate Bcl10 on three distinct sites, although we observed that Bcl10 is preferentially phosphorylated by IKKβ (data not shown), in accordance with the fact that IKKβ siRNA is more efficient than IKKα siRNA at blocking Bcl10 degradation after PMA/ionomycin treatment (Fig. 2C). Interestingly, we observed that Bcl10 is not degraded in response to TNF-α, another inducer of NF-κB. The molecular mechanism by which Bcl10 is degraded appears to be similar to the one that affects the members of the IκB family, with regard to their phosphorylation, ubiquitination, and proteolysis, although the efficiency of phosphorylation as well as the kinetics of degradation appear to be different. This molecular event is indeed a negative regulatory mechanism of T cell activation because expression of a nondegradable form of Bcl10 leads to a significant increase in IL-2 production (Fig. 5).
It has been shown by Daniel Krappmann's group (15) that Bcl10 is degraded through the lysosomal pathway in a NEMO-independent manner. Although we cannot completely exclude the existence of such a pathway under certain conditions (the NEMO-independent degradation has been demonstrated only in pre-B cells by Krappmann et al., and the involvement of lysosomes has only been shown in the case of PMA-stimulated T cells), our data clearly demonstrate that Bcl10 degradation is NEMO-dependent and completely prevented by proteasome inhibitors in TCR-activated T cells (Fig. 1). In addition, Krappmann et al. have reported recently that IKKβ, independently of NEMO, phosphorylates the C-terminal region of Bcl10 (corresponding to fragment 4 in Fig. 3) upon TCR stimulation and thereby interferes with Bcl10/MALT1 association and Bcl10-mediated NEMO ubiquitination (18). The reason why we have not been able to observe these IKKβ-mediated phosphorylation events is currently unclear, but the possibility exists that under different conditions, IKK might phosphorylate different regions of Bcl10, thus inducing different outcomes.
Several groups have investigated the subcellular localization of MALT1 and Bcl10. Nakagawa and colleagues (20) have demonstrated that MALT1 contains two nuclear export signals (NES) at its C terminus that are responsible for its cytoplasmic localization, and suggest that MALT1 is responsible for the cytoplasmic retention of Bcl10. Another study by Yeh and colleagues (21) has shown that after TNF-α treatment, Bcl10 is phosphorylated by Akt on its last C-terminal Ser, allowing its interaction with Bcl3 and its subsequently nuclear translocation. Here, we show that Bcl10 translocates to the nucleus, where it is degraded after TCR stimulation (Fig. 6). This nuclear localization can be visualized when degradation is prevented by incubation with proteasome inhibitors or by mutation of the IKK phosphorylation sites (Fig. 6). One known site of nuclear degradation is represented by the promyelocytic leukemia (PML) protein nuclear bodies (22), but we could not demonstrate colocalization of nuclear Bcl10 with these structures (data not shown).
The mechanisms controlling Bcl10 subcellular localization are not fully understood. Several immunohistological studies have observed nuclear localization of Bcl10 in MALT lymphomas (23–25). On the basis of these findings, it has been suggested that nuclear Bcl10 expression may be a determinant in the pathogenesis of these lymphoma. According to those observations, our finding that the T81A/S85A mutant of Bcl10 accumulates in the nucleus of T cells after TCR stimulation suggests that mutations of the consensus sequence DS/TψXXS/T might be associated with some lymphoma. This could take place through at least two distinct but not exclusive mechanisms: Bcl10 stabilization could increase NF-κB activation, and the subsequent expression of pro survival genes and/or Bcl10 stabilization and nuclear translocation could have a role in the transcriptional regulation of a particular subset of genes. Interestingly, two studies have revealed mutations that affect the conserved aspartic acid (D) of the consensus sequence (DS/TψXXS/T): one in a diffuse large B cell lymphoma (26), and the other in a mesothelioma (27). Interestingly, it has been observed in the case of β-catenin (see Fig. 2A) that this aspartic acid, which is specifically involved in the interaction with β-TrCP (28, 29), is subjected to mutations in a large number of cancers (30–32).
Finally, the revelation that Bcl10 is a physiological target of the NEMO/IKK complex adds to the complexity of T cell activation and places Bcl10 at the center of the regulation of TCR-induced NF-κB activation.
Materials and Methods
Cell Culture.
HEK293T cells were maintained in DMEM (Invitrogen) supplemented with 10% FCS, penicillin (500 units/ml), and streptomycin (500 μg/ml) (Invitrogen). Plat E cells were cultivated in DMEM 10% FCS, penicillin, streptomycin, supplemented with blasticidine (10 μg/ml) and puromycin (1 μg/ml). WT Jurkat cells, 8321 cells (Nemo-deficient cells; a gift from A. Ting, Immunobiology Center, New York, NY), JPM50.6 (CARMA1-deficient cells; a gift from X. Lin, Harvard Medical School, Boston, MA), and E29.1 mouse T cell hybridoma were maintained in standard growth medium (RPMI Glutamax I) supplemented with 10% FCS, penicillin, streptomycin. E29.1 cell line derivatives expressing WT or mutant Bcl10 were generated by retroviral infection (using the MSCV backbone) and maintained in standard growth medium supplemented with 1 μg/ml puromycin.
Antisera.
The antisera used for immunoprecipitation or Western blot were as follows: mAb anti-VSV P5D4 (Upstate), mAb anti-Bcl10 (Upstate), mAb anti-flag M2 (Sigma-Aldrich), mAb anti-β-tubulin (Sigma-Aldrich), mAb anti-Actin (Sigma), mAb anti-IKKα (Imgenex), mAb anti-IKKβ (Abgent), rabbit polyclonal anti-Flag (Sigma-Aldrich), rabbit polyclonal anti-IKKα/β H-470 (Santa Cruz Biotechnology), and rabbit polyclonal anti-pIκBα (Cell Signaling).
Plasmids and Constructs.
The pCR3-VSV-CARMA1, pCR3-VSV-RIP2, and pCR3-Flag-Bcl10 WT expression vectors were obtained from M. Thome (Biomedical Research Center, Epalinges, Switzerland). The pcDNA3-VSV-βTRCP construct was a gift from R. Benarous (Institut Cochin de Génétique Moléculaire, Paris, France), and the retroviral expression vector MSCV was kindly provided by T. W. Mak (Princess Margaret Hospital, Toronto, ON, Canada). The pcDNA3-VSV-IKKβ was generated in our laboratory. The WT ubiquitin expression vector encoding an octameric tandem fusion of HA-ubiquitin was a gift from M. Treier (European Molecular Biology Laboratory, Heidelberg, Germany).
All PCR-generated DNA fragments used to construct the GST fusions were cloned into the BamHI–EcoRI restriction site of the pGEX-KT vector (Amersham Pharmacia). The GST-IκBα N-ter and the GST-IκBβ C-ter has been described (33). The Flag-Bcl10 mutants were generated by PCR mutagenesis.
MSCV-Bcl10 WT or mutants were created by PCR and cloned into HpaI–XhoI restriction sites of the MSCV retroviral vector (Clontech).
Activation, Immunoprecipitations, and Immunoblots.
TCR activation.
E29.1 cells were activated with H57 mAb (R & D Systems) coated overnight at 4°C on six-well plates. Human T cells were activated as described (34).
Treatment with chemicals or siRNA.
The following reagents were purchased and used as indicated: 100 ng/ml PMA (Sigma-Aldrich) and 1 μM calcium ionophore (Sigma-Aldrich), Bay 11-7085 (5 μg/ml; Calbiochem), 10 μg/ml human TNF-α (PeproTech) or mouse TNF-α (Apotech), 100 μM N-acetyl-Leu-Leu-norleucinal (calpain inhibitor I; Sigma-Aldrich), 25 μM MG 132 (Sigma-Aldrich) or 50 μg/ml cycloheximide (Sigma-Aldrich). For transient expression of siRNA, we used smart pool IKKα and smart pool IKKβ (Dharmacon).
After treatment, cells were lysed in 100 μl of 1× TNE buffer (50 mM Tris, pH 8.0/0.5% Nonidet P-40/200 mM NaCl/0.1 mM EDTA, supplemented with protease and phosphatase inhibitors) and subjected either to direct immunoblotting or to immunoprecipitation followed by immunoblotting as described in the figure legends (34).
Immunofluorescence Microscopy.
E29.1 stable clones were activated as described above and then harvested on circular glass slides coated with polylysine (Sigma-Aldrich) in a 24-well dish. The cells were then fixed in 4% formaldehyde in PBS for 20 min at room temperature followed by treatment with 50 mM ammonium chloride. Cells were then permeabilized in 0.2% Triton in PBS and washed in PBS supplemented with 1 mg/ml BSA. The primary antibody (anti-Bcl10) was applied for 1 h at room temperature. Cells were then washed and incubated for 45 min with the anti-mouse CY3-labeled secondary antibody, while the nucleus was stained with DAPI. Cell preparations were mounted in Mowiol, and the specimens were examined by using an ApoTome imaging system (Imager Z1; Zeiss) with a ×60/1.4 OIL DIC objective (Zeiss). Images were analyzed by using AxioVision software (Zeiss).
Immune Complex Kinase Assays.
HEK 293T cells were transfected with an IKKβ-expressing vector, lysed with 1× TNE buffer after 48 h, and immunoprecipitated. Production of fusion proteins and kinase assays were performed as described (34) by using the kinase buffer 20 mM Hepes/10 mM MgCl2/50 mM NaCl (pH 7.5).
IL-2 Assays.
To evaluate PMA/ionomycin-induced IL-2 production by the different E29.1 clones, wells of 24-well Falcon tissue culture plates (2 × 105 cells per well) were activated in duplicate with 5 ng/ml PMA and 1 mg/ml ionomycin for 14 h at 37°C. Serial dilutions of supernatant were then tested in triplicate for IL-2 content. These assays were performed by measuring IL-2 production with an ELISA kit (R & D Systems).
Supplementary Material
Acknowledgments
We thank Margot Thome for critical reading of the manuscript and for the gift of Bcl10 and CARMA1 expression vectors, Adrian Ting for the gift of the 8321 NEMO-deficient cell line, and Xin Lin for the gift of the CARMA1-deficient cell line. This work was supported by Ligue Contre le Cancer Contract R05/75-22 (R.W.), Association pour la Recherche sur le Cancer Contract 3119 (A.I.), and Cancéropôle Axe 2 (A.I.). C.L. is the recipient of a Ministère de la Recherche et Technologie fellowship.
Abbreviations
- PMA
phorbol 12-myristate 13-acetate
- TCR
T cell antigen receptor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0606982104/DC1.
References
- 1.Weil R, Israël A. Cell Death Differ. 2006;13:826–833. doi: 10.1038/sj.cdd.4401856. [DOI] [PubMed] [Google Scholar]
- 2.Bertin J, Wang L, Guo Y, Jacobson MD, Poyet JL, Srinivasula SM, Merriam S, DiStefano PS, Alnemri ES. J Biol Chem. 2001;276:11877–11882. doi: 10.1074/jbc.M010512200. [DOI] [PubMed] [Google Scholar]
- 3.Gaide O, Martinon F, Micheau O, Bonnet D, Thome M, Tschopp J. FEBS Lett. 2001;496:121–127. doi: 10.1016/s0014-5793(01)02414-0. [DOI] [PubMed] [Google Scholar]
- 4.McAllister-Lucas LM, Inohara N, Lucas PC, Ruland J, Benito A, Li Q, Chen S, Chen FF, Yamaoka S, Verma IM, et al. J Biol Chem. 2001;276:30589–30597. doi: 10.1074/jbc.M103824200. [DOI] [PubMed] [Google Scholar]
- 5.Wang L, Guo Y, Huang WJ, Ke X, Poyet JL, Manji GA, Merriam S, Glucksmann MA, DiStefano PS, Alnemri ES, Bertin J. J Biol Chem. 2001;276:21405–21409. doi: 10.1074/jbc.M102488200. [DOI] [PubMed] [Google Scholar]
- 6.Lucas PC, Yonezumi M, Inohara N, McAllister-Lucas LM, Abazeed ME, Chen FF, Yamaoka S, Seto M, Nunez G. J Biol Chem. 2001;276:19012–19019. doi: 10.1074/jbc.M009984200. [DOI] [PubMed] [Google Scholar]
- 7.Uren AG, O'Rourke K, Aravind LA, Pisabarro MT, Seshagiri S, Koonin EV, Dixit VM. Mol Cell. 2000;6:961–967. doi: 10.1016/s1097-2765(00)00094-0. [DOI] [PubMed] [Google Scholar]
- 8.Egawa T, Albrecht B, Favier B, Sunshine MJ, Mirchandani K, O'Brien W, Thome M, Littman DR. Curr Biol. 2003;13:1252–1258. doi: 10.1016/s0960-9822(03)00491-3. [DOI] [PubMed] [Google Scholar]
- 9.Ruefli-Brasse AA, French DM, Dixit VM. Science. 2003;302:1581–1584. doi: 10.1126/science.1090769. [DOI] [PubMed] [Google Scholar]
- 10.Ruland J, Duncan GS, Elia A, del Barco Barrantes I, Nguyen L, Plyte S, Millar DG, Bouchard D, Wakeham A, Ohashi PS, Mak TW. Cell. 2001;104:33–42. doi: 10.1016/s0092-8674(01)00189-1. [DOI] [PubMed] [Google Scholar]
- 11.Lee KY, D'Acquisto F, Hayden MS, Shim JH, Ghosh S. Science. 2005;308:114–118. doi: 10.1126/science.1107107. [DOI] [PubMed] [Google Scholar]
- 12.Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ. Mol Cell. 2004;14:289–301. doi: 10.1016/s1097-2765(04)00236-9. [DOI] [PubMed] [Google Scholar]
- 13.Zhou H, Wertz I, O'Rourke K, Ultsch M, Xiao W, Dixit VM. Nature. 2004;427:167–171. doi: 10.1038/nature02273. [DOI] [PubMed] [Google Scholar]
- 14.Chen ZJ. Nat Cell Biol. 2005;7:758–765. doi: 10.1038/ncb0805-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scharschmidt E, Wegener E, Heissmeyer V, Rao A, Krappmann D. Mol Cell Biol. 2004;24:3860–3873. doi: 10.1128/MCB.24.9.3860-3873.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.He KL, Ting AT. Mol Cell Biol. 2002;22:6034–6045. doi: 10.1128/MCB.22.17.6034-6045.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang D, Matsumoto R, You Y, Che T, Xue-Yan L, Gaffen SL, Lin X. Mol Cell Biol. 2004;24:164–171. doi: 10.1128/MCB.24.1.164-171.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wegener E, Oeckinghaus A, Papadopoulou N, Lavitas L, Schmidt-Supprian M, Ferch U, Mak TW, Ruland J, Heissmeyer V, Krappmann D. Mol Cell Biol. 2006;23:13–23. doi: 10.1016/j.molcel.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 19.Ruefli-Brasse AA, Lee WP, Hurst S, Dixit VM. J Biol Chem. 2003;279:1570–1574. doi: 10.1074/jbc.C300460200. [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa M, Hosakawa Y, Yonezumi M, Izumiyama K, Susuki R, Tsuzuki S, Asaka M, Seto M. Blood. 2005;106:4210–4216. doi: 10.1182/blood-2004-12-4785. [DOI] [PubMed] [Google Scholar]
- 21.Yeh PY, Kuo SH, Yeh KH, Chuang SE, Hsu CH, Chang WC, Lin HI, Gao M, Cheng AL. J Biol Chem. 2005;281:167–175. doi: 10.1074/jbc.M511014200. [DOI] [PubMed] [Google Scholar]
- 22.Bailey D, O'Hare P. Biochem J. 2005;329:271–281. doi: 10.1042/BJ20050873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu H, Ye H, Dogan A, Ranaldi R, Hamoudi RA, Bearzi I, Isaacson PG, Du MQ. Blood. 2001;98:1182–1187. doi: 10.1182/blood.v98.4.1182. [DOI] [PubMed] [Google Scholar]
- 24.Maes B, Demunter A, Peeters B, De Wolf-Peeters C. Blood. 2002;99:1398–1404. doi: 10.1182/blood.v99.4.1398. [DOI] [PubMed] [Google Scholar]
- 25.Ye H, Dogan A, Karran L, Willis TG, Chen L, Wlodarska I, Dyer MJ, Isaacson PG, Du MQ. Am J Pathol. 2000;157:1147–1154. doi: 10.1016/S0002-9440(10)64630-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SH, Shin MS, Kim HS, Park WS, Kim SY, Lee HK, Park JY, Oh RR, Jang JJ, Park KM, et al. Cancer Res. 1999;59:5674–5677. [PubMed] [Google Scholar]
- 27.Willis TG, Jadayel DM, Du MQ, Peng H, Perry AR, Abdul-Rauf M, Price H, Karran L, Majekodunmi O, Wlodarska I, et al. Cell. 1999;96:35–45. doi: 10.1016/s0092-8674(00)80957-5. [DOI] [PubMed] [Google Scholar]
- 28.Megy S, Bertho G, Gharbi-Benarous J, Evrard-Todeschi N, Coadou G, Ségéral E, Iehle C, Quéméneur E, Benarous R, Girault J-P. J Biol Chem. 2005;280:29107–29116. doi: 10.1074/jbc.M501628200. [DOI] [PubMed] [Google Scholar]
- 29.Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich N. Mol Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 30.Chan EF, Gat U, McNiff JM, Fuchs E. Nat Genet. 1999;21:410–413. doi: 10.1038/7747. [DOI] [PubMed] [Google Scholar]
- 31.Legoix P, Bluteau O, Bayer J, Perret C, Balabaud C, Belghiti J, Franco D, Thomas G, Laurent-Puig P, Zucman-Rossi J. Oncogene. 1999;18:4044–4046. doi: 10.1038/sj.onc.1202800. [DOI] [PubMed] [Google Scholar]
- 32.Machin P, Catasus L, Pons C, Munos J, Matias-Guiu X, Prat J. Hum Pathol. 2002;33:206–212. doi: 10.1053/hupa.2002.30723. [DOI] [PubMed] [Google Scholar]
- 33.Weil R, Laurent-Winter C, Israël A. J Biol Chem. 1997;272:9942–9949. doi: 10.1074/jbc.272.15.9942. [DOI] [PubMed] [Google Scholar]
- 34.Weil R, Schwamborn K, Alcover A, Bessia C, Di Bartolo V, Israël A. Immunity. 2003;18:13–26. doi: 10.1016/s1074-7613(02)00506-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.