Abstract
Like many epithelial tumors, head and neck squamous cell carcinoma (HNSCC) contains a heterogeneous population of cancer cells. We developed an immunodeficient mouse model to test the tumorigenic potential of different populations of cancer cells derived from primary, unmanipulated human HNSCC samples. We show that a minority population of CD44+ cancer cells, which typically comprise <10% of the cells in a HNSCC tumor, but not the CD44− cancer cells, gave rise to new tumors in vivo. Immunohistochemistry revealed that the CD44+ cancer cells have a primitive cellular morphology and costain with the basal cell marker Cytokeratin 5/14, whereas the CD44− cancer cells resemble differentiated squamous epithelium and express the differentiation marker Involucrin. The tumors that arose from purified CD44+ cells reproduced the original tumor heterogeneity and could be serially passaged, thus demonstrating the two defining properties of stem cells: ability to self-renew and to differentiate. Furthermore, the tumorigenic CD44+ cells differentially express the BMI1 gene, at both the RNA and protein levels. By immunohistochemical analysis, the CD44+ cells in the tumor express high levels of nuclear BMI1, and are arrayed in characteristic tumor microdomains. BMI1 has been demonstrated to play a role in self-renewal in other stem cell types and to be involved in tumorigenesis. Taken together, these data demonstrate that cells within the CD44+ population of human HNSCC possess the unique properties of cancer stem cells in functional assays for cancer stem cell self-renewal and differentiation and form unique histological microdomains that may aid in cancer diagnosis.
Keywords: BMI1, CD44
Head and neck cancer is a common malignancy that affects ≈40,000 new patients in the United States each year (1). Despite advances in therapy, which have improved quality of life, survival rates have remained static for many years. Mortality from this disease remains high because of the development of distant metastases and the emergence of therapy-resistant local and regional recurrences. It is therefore essential that we develop a deeper understanding of the biology of this disease to develop more effective therapies.
Epithelial tumors, including head and neck squamous cell carcinoma (HNSCC), contain cellular heterogeneity, some of which is accounted for by ongoing mutations that occur because of genetic instability and environmental factors (2, 3). More recently, it has been hypothesized that functional heterogeneity may account for the fact that not all of the cancer cells in solid tumors have a similar ability to drive tumor formation (4, 5). This observation has led to the cancer stem cell (CSC) hypothesis, which suggests that a tumor can be viewed as an aberrant organ that is sustained, in a way similar to normal tissues, by a stem cell that drives tumorigenesis, as well as giving rise to a large population of differentiated progeny that make up the bulk of the tumor but that lack tumorigenic potential. In support of this hypothesis, recent studies have shown that, similar to leukemia and other hematologic malignancies (6–10), tumorigenic and nontumorigenic populations of breast cancer cells can be isolated based on their expression of cell-surface markers. In many cases of breast cancer, only a small subpopulation of cells had the ability to form new tumors (11, 12). This work strongly supports the existence of CSC in breast cancer. Further evidence for the existence of CSCs occurring in solid tumors has been found in CNS malignancies. By using culture techniques similar to those used to culture normal neuronal stem cells, it has been shown that CNS malignancies contain a small population of cancer cells that are clonogenic in vitro and initiate tumors in vivo, whereas the remaining cells in the tumor do not have these properties (13–16).
Using methods successfully used to identify CSCs in breast cancer, we studied HNSCC. In this study, we show that HNSCC contains a distinct population of cancer cells with the exclusive ability to produce tumors in mice and recreate the original tumor heterogeneity. We have identified a cell-surface marker that can enrich for this cell population and have provided evidence that this population possesses properties classically attributed to stem cells. We have also identified a gene previously implicated in self-renewal and tumorigenesis, BMI1 (17–21), which is differentially expressed at both the RNA and protein levels in the tumorigenic cell population and in tissue sections, defines microdomains of CSCs that are membrane CD44+ and nuclear BMI1+. This finding both provides insight into the possible molecular mechanisms mediating the self-renewal of these cells and demonstrating the value of identifying the CSC population in primary tumors to further characterize these cells at the molecular level and thus develop new treatment strategies targeted against this critical population of cancer cells.
Results
A mouse xenograft model of HNSCC was developed in which primary specimens obtained from patients undergoing surgical resection were implanted under the skin of immunocompromised mice, either nonobese diabetic/severe combined immunodeficient (NOD/SCID) (22) or Rag2/cytokine receptor common γ-chain double knockout (Rag2γDKO) (23), either as small (<2 mm) pieces of tumor or as cell suspensions in matrigel, ranging from 1–5 million total cells per injection. Of 25 samples of HNSCC tumors implanted in this way, 13 have given rise to tumors in the mice [9 of 16 at University of Michigan (UM), 4 of 9 at Stanford University (SU)]. Both the NOD/SCID (UM) and Rag2γDKO (SU) mouse model gave similar rates of tumor engraftment. These results indicate that either animal model is reliable. When solid tumor pieces were implanted into the mice, a small tumor nodule was evident in 6–10 weeks, on average, and reached a size of 1–1.4 cm in 4–6 months, on average. Single-cell suspensions produced small tumor nodules in 8–12 weeks, depending on the number of cells injected. For a comparison of the histology of tumors arising in mice with the original patient samples, see [supporting information (SI) Fig. 6.]
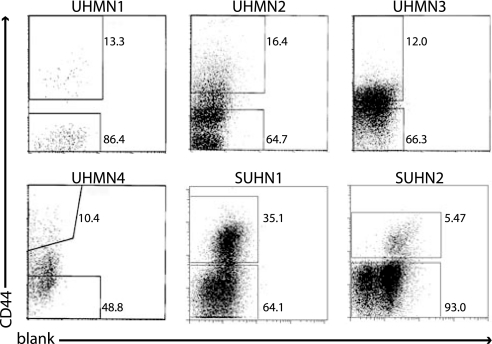
Of the tumor specimens that grew in mice, nine (seven from UM; UM 1, 2, 3, 4, 5, 6, and 7 and two from SU; SU1 and 2) were subjected to flow cytometry on cells obtained either immediately after removal from the patient (UM 3, 5, 6, and 7), or from tumors arising in the immunodeficient mice (UM1, 2, and 4 and SU1 and 2) to obtain purified populations of tumor cells for further transplants. It was not possible to use cells obtained directly from patient samples in all cases, because the specimens obtained from the clinic were frequently too small to obtain sufficient numbers of cells for these experiments. These nine subjects ranged in age from 22–72 years old. Three tumor specimens were harvested from the tongue, two each from the larynx and floor of mouth, and one each from the oropharynx and maxillary sinus. Three subjects had undergone previous treatment for their cancer >1 year before this study (UM3, 4, and 6). The degree of differentiation, evaluated by histologic architecture, varied from poorly to well differentiated (SI Table 2). Flow cytometry analysis revealed that the HNSCC specimens were heterogeneous with respect to the cell-surface marker CD44 (Fig. 1). Antigens associated with normal cell types (lineage markers CD2, CD3, CD10, CD18, CD31, CD64, and CD140b) were not expressed on the cancer cells. These lineage markers were used to eliminate “lineage (Lin)+ cells”, including normal leukocytes, fibroblasts, endothelial, and mesothelial cells (Lin+) from the tumor specimens during the cell-sorting experiments. In passaged tumors, mouse anti-H2K antibodies were used to eliminate contaminating mouse cells. In each tumor, a distinct population of CD44+ and CD44− cancer cells was identifiable. Importantly, similar results were obtained from tumors that had been passaged once through mice before sorting as from tumors analyzed directly from patients, indicating that a single passage did not significantly affect the expression of this marker. Single-cell suspensions of FACS-purified CD44+Lin− and CD44−Lin− cells at different doses were implanted into the mouse model to determine whether CD44 status could distinguish between tumorigenic and nontumorigenic cells (Table 1).
Fig. 1.
Isolation of tumorigenic cells. Flow cytometry was used to isolate subpopulations of cells based on their CD44 expression. Dead cells (7AAD+ or PI+) and Lin+ cells were eliminated from all analyses. The percentage of CD44+ vs. CD44− is shown.
Table 1.
Growth of HNSCC tumors in mice injected with CD44+Lin− or CD44−Lin− cells
| Sample (% CD44+) | Population | Cell count, in thousands |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| 500–650 | 200–300 | 100–150 | 40–50 | 20–25 | 10 | 5 | 2 | ||
| UMHN1* (13.4) | CD44+ | 1 of 1 | |||||||
| CD44− | 0 of 1 | ||||||||
| UMHN2* (16.4) | CD44+ | 2 of 2 | 1 of 2 | 1 of 1 | |||||
| CD44− | 0 of 2 | 0 of 1 | |||||||
| UMHN3† (12.0) | CD44+ | 1 of 1 | 3 of 3 | ||||||
| CD44− | 0 of 3 | 0 of 3 | 0 of 3 | ||||||
| UMHN4* (10.4) | CD44+ | 1 of 1 | |||||||
| CD44− | 0 of 1 | 0 of 1 | 0 of 1 | 0 of 1 | |||||
| UMHN5† (0.43) | CD44+ | 1 of 1 | 1 of 1 | 1 of 2 | |||||
| CD44− | 0 of 1 | 0 of 2 | |||||||
| UMHN6** (1.7) | CD44+ | 1 of 1 | 0 of 1 | ||||||
| CD44− | 0 of 2 | ||||||||
| UMHN7† (5.2) | CD44+ | 1 of 1 | 0 of 1 | ||||||
| CD44− | 0 of 1 | ||||||||
| SUHN1* (35.1) | CD44+ | 3 of 3 | 1 of 2 | 0 of 1 | 0 of 4 | 0 of 1 | |||
| CD44− | 1 of 6 | 0 of 2 | 0 of 2 | 0 of 2 | 0 of 1 | ||||
| SUHN2* (1.3) | CD44+ | 1 of 1 | |||||||
| CD44− | 0 of 1 | 0 of 2 | 0 of 1 | ||||||
UMHN samples were all grown in NOD/SCID mice. SUHN samples were all grown in RagγDKO mice.
*Samples that were passaged once through mice before sorting for CD44+ and CD44− populations.
†Samples that were directly sorted from patient samples for CD44+ and CD44− populations.
Initial data from UM using large numbers of implanted cells indicated that CD44+ cells could form tumors, whereas CD44− cells could not. Similarly, initial data from SU indicated that, at equivalent doses of cells, CD44+ cells initiated tumor growth much more efficiently than CD44− cells. In subsequent experiments, larger numbers of CD44− cells were injected than CD44+ cells, on the assumption that this would increase the likelihood of detecting any tumor-initiating cells that may be present, but rare, within the CD44− population. In all cases, the number of cells injected, the range of cell doses, and the number of mice injected with each dose was severely limited by the number of cells that could be sorted from primary and first-passage tumors.
In the tumors analyzed in this study, a total of 20 of 31 injections of CD44+ cells formed tumors, whereas only 1 of 40 injections of CD44− cells did so (P < 6 × 10−9, Fisher's exact test). As few as 5 × 103 CD44+Lin− cells obtained directly from a patient's tumor or from early-passage xenograft tumors gave rise to new tumors (Table 1). In contrast, up to 5 × 105 CD44− cells failed to form tumors. When >4 × 104 CD44+Lin− HNSCC cells were injected, tumors formed within 10–16 weeks (7 of 7). In experiments where 5–25 × 103 CD44+Lin− HNSCC cells were injected, tumors formed in 10 of 17 injections. No detectable tumors developed at any dose of CD44−Lin− cells, with one exception (1 of 40). The latter case occurred early in the study, in an experiment where cells were FACS-sorted only once, and the tumor could have arisen as a result of contamination of the CD44− population with a small number of CD44+ cells, further indicated by the fact that the tumor in question was ≈3-fold smaller than the tumor initiated by an equivalent dose of CD44+ cells. In all other experiments, cells were double-sorted, yielding a population purity of >95% (data not shown). Even after >24–48 weeks, CD44−Lin− injection sites revealed no detectable tumor growth. Implanted Lin+ cells did not grow any tumors in the mouse model.
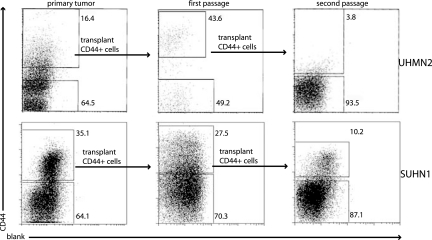
Tumors resulting from implanted CD44+ cells showed the original tumor morphology upon histologic examination (Fig. 2). Furthermore, analysis of tumors arising from implantation of CD44+Lin− cells revealed that these cells gave rise to new tumors that contained cells that were, again, phenotypically diverse for CD44 expression. This result indicates that the CD44+ cells can give rise to more CD44+ cells as well as CD44− cells. Upon resorting and passaging of the CD44+Lin− and CD44−Lin− populations, again, only CD44+Lin− cells initiated new tumors. CD44+Lin− cells from UM4 and SU2 have been successfully serially passaged through two rounds of tumor formation. CD44+Lin− cells from UM2 and SU1 (Fig. 3) have been passaged through three rounds of tumor formation in mice. CD44−Lin− cells selected from passaged tumors never resulted in new tumor formation. Analysis of the passaged tumors by flow cytometry, after each successive sort and implantation, confirmed that the resulting tumors again contained both CD44+ and CD44− cells.
Fig. 2.
Tumor morphology. (A) Representative tumor in a mouse at the CD44+ injection site. (B) Histology of the tumor resulting from a CD44+ injection site. (C) Histology from the corresponding primary tumor.
Fig. 3.
Phenotypic diversity in tumors arising from CD44+ Lin− cells. CD44 staining pattern of live cancer cells from primary unpassaged tumor, tumor resulting from the implantation of CD44+Lin− cells from the primary tumor (first passage), and tumor resulting from the implantation of CD44+Lin− cells from the first passage tumor (second passage).
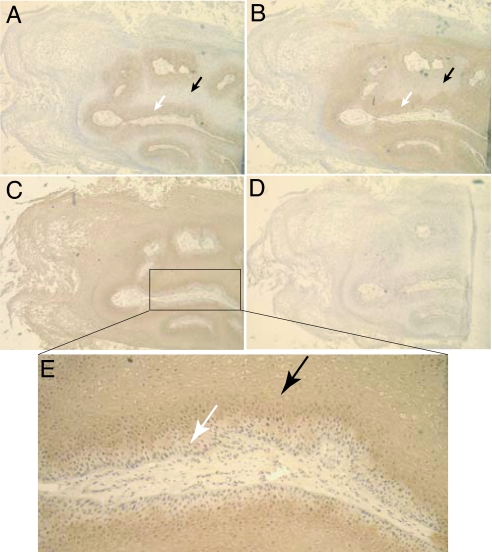
In addition to the nine tumor samples described above, additional tumors (those that did not take in the mouse model and those that were too small to provide sufficient cell numbers for the xenograft experiments) were analyzed by flow cytometry and immunohistochemistry (IHC) for CD44 expression (SI Table 3 and Fig. 4). The majority of HNSCC contain a subpopulation of CD44+ cells, with the percentage of CD44+Lin− cells in the tumors varying from 0.1–41.72%, n = 33 (SI Table 3). Nine of 33 samples analyzed had <1% of the cells expressing CD44. IHC for CD44 was performed to determine the physical location of the tumorigenic population of cells within the tumor. In the case of moderately to well differentiated HNSCC samples, where a clear hierarchy of cell differentiation is present within the tumor (i.e., primitive basal layer cells and cells with the phenotype of differentiating keratinocytes), CD44 expression was detected in the basal layer but not in the differentiated cells (Fig. 4). To confirm that the basal cell layer represented undifferentiated cells, serial sections were stained with an antibody to Cytokeratin 5/14 (CK5/14), a marker of normal squamous epithelial stem and progenitor cells (24), or with an antibody to Involucrin, a differentiated keratinocyte marker (25). It was found that CD44 costained with CK5/14, whereas CD44 and Involucrin staining were mutually exclusive. It was also clear on these sections that the Involucrin+CD44− cells in these tumors retained the differentiated phenotype of mature or maturing squamous epithelium, whereas the Involucrin−CD44+ cells have a basaloid phenotype. The latter data were confirmed on four moderately to well differentiated primary patient tumors (see SI Fig. 7).
Fig. 4.
Immunohistochemical analysis of a well differentiated HNSCC. Serial sections were stained with antibodies to CD44 (A), Cytokeratin 5/14 (B), Involucrin (C and E), or an isotype control (D). A horseradish peroxidase-conjugated secondary antibody was used for detection. CD44+ regions were CK5/14+ and Involucrin− (white arrows), whereas CD44− regions were CK5/14− and Involucrin+ (black arrows).
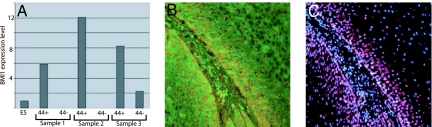
We then decided to investigate the properties of the CD44+ versus CD44− populations of cells at the level of gene expression. CD44+Lin− and CD44−Lin− cells were sorted for RNA purification, and quantitative RT-PCR was performed for a variety of stem cell-related genes. In three primary patient samples, it was found that BMI1 was expressed at high levels compared with human ES cells in the CD44+ population but was undetectable in the CD44− population in two of the three samples and 4-fold lower in the third sample (Fig. 5). These samples represented one well differentiated, one moderately to well differentiated, and one poorly differentiated sample, respectively. Expression of β-actin was used as a control for RNA quality, and the data are shown relative to β-actin expression. We then performed immunofluorescent costaining for CD44 and BMI1 on tissue sections and found that the BMI1 protein was present in the nuclei of the CD44+ cells of the tumors, with staining appearing strongest in the most basal regions, decreasing with distance from the basal region, and absent or rarely seen in the CD44− regions of the tumors. This result was obtained for four different well or moderately to well differentiated primary patient samples that were analyzed in this way (Fig. 5). In poorly differentiated tumors, costaining between CD44 and BMI1 was also observed, even in the absence of the distinct tissue architecture that is seen in well differentiated tumors (see SI Fig. 8).
Fig. 5.
Quantitative RT-PCR and immunofluorescence for BMI1. (A) CD44+Lin− and CD44−Lin− cells from three HNSCC samples were double-sorted, and purified RNA was assayed for the presence of BMI1 transcripts by quantitative RT-PCR. Human embryonic stem cells (ES) were used as a positive control for PCR, and transcript levels were normalized to expression of β-actin. All three samples were primary patient samples. BMI1 was expressed in the CD44+Lin− population but was undetectable in the CD44−Lin− population in samples 1 and 2 and 4-fold lower in the CD44−Lin− population in sample 3. (B and C) A well differentiated HNSCC was costained for CD44 (green) and Bmi1 (red). (B) The CD44/BMI1 overlay. (C) The BMI1/Hoechst 33342 overlay, to highlight nuclei in blue. BMI1 protein was seen in the nuclei of the majority of CD44+ cells and was rare in the CD44− region of the tumor.
Discussion
The CSC hypothesis suggests that only the CSC within the tumor can self-renew and proliferate extensively to form new tumors. It has been demonstrated for several different types of cancer that a distinct subset of cells initiates tumors in vivo, whereas the remaining cells do not (7, 11, 16). Further evidence for the CSC hypothesis comes from clinical observations, where tumors often respond to chemotherapy initially, but frequently recur, suggesting that residual stem cells remaining after therapy are responsible for tumor regeneration. In HNSCC, as in other cancers, characterization of CSCs will allow for the development of new treatments that are specifically targeted against this critical population of cells, particularly their ability to self-renew, resulting in more effective therapies.
In this study, we have provided evidence for the existence of a developmental hierarchy within human HNSCC; that is, HNSCC contain a subpopulation of tumorigenic cancer cells with both self-renewal and differentiation capacity.
Our initial finding was that all tumorigenic potential was contained within the CD44+Lin− population of HNSCC. The ability to initiate tumor growth with cell doses ranging from 5,000 to 20,000 of these cells, combined with the need to inject at least 1 × 106 cells to obtain tumors from whole tumor-cell suspensions, and the lack of tumor formation by CD44−Lin− cells at much higher cell doses, provided the strongest evidence that the CD44+Lin− population contains the HNSCC stem cell population. Although the CD44+ population is clearly significantly enriched for the CSC in these tumors, the cell doses required to initiate tumors indicate that it may not be pure. We were unable to precisely estimate the CSC frequency within this population because of the difficulties imposed by limited sample size and the robustness of the mouse model. It still remains to further fractionate the CD44+ population to further purify the CSC.
Upon reanalysis of tumors arising from the transplantation of purified CD44+ cells, both CD44+ and CD44− cells were again present, and serial transplants demonstrate that, with each tumor passage, only CD44+Lin− and not the CD44−Lin− cells can initiate a new tumor. It was observed in the small number of samples for which serial transplants were performed that there was a decrease in the proportion of CD44+ cells in the secondary mice compared with the initial patient sample and the primary mice. This difference may be accounted for by the different time points at which the animals were killed; the secondary tumor was allowed to grow larger, over a longer period, than the primary. Alternatively, the decrease in the percentage of CD44 over serial transplant may suggest a loss of self-renewal activity of the putative stem cell population. However, the tertiary mouse from SU1 contained 13.8% CD44+ cells (data not shown), and thus a further decrease between secondary (10.2% CD44+) and tertiary recipients did not occur. Furthermore, calculations demonstrate that, even when the %CD44+ cells is decreased from one tumor to the next, an overall expansion of the CD44+ cells occurs in these mice. For example, the SU1 secondary mouse was injected with 25,000 CD44+ cells from the primary tumor, and, based on the total number of cells recovered from the tumor, 400,000 CD44+Lin− cells were present in the secondary tumor, indicating at least a 16-fold expansion. Thus, over multiple transplants, even with the variation in the proportion of CD44+ cells present, a large expansion of the CD44+ population occurs. The cause of the variation in the percentage of CD44+ cells over serial transplant remains to be determined. Additional variables, such as differences in the degree of tumor dissociation from one day to the next, may be involved. Although we have not obtained direct evidence for self-renewal by clonal analysis, the exclusive ability of the CD44+ population to serially transplant tumors provides strong evidence that self-renewal is a property of at least some fraction of the CD44+ population.
Further evidence for a developmental hierarchy in HNSCC comes from the histology and IHC studies done on moderately to well differentiated tumors. First, the tumors demonstrate cytologic and architectural features similar to normal squamous epithelium, including differentiation from a basal layer toward an apical layer containing cells with mature squamous morphology and the formation of keratin (keratin pearls were often present). Second, CD44 clearly stains regions of the tumors that have basal cell morphology and that costain with the basal layer marker CK5/14. Finally, the differentiation marker Involucrin stains the regions of the tumor that are negative for CD44, and vice versa. One caveat to this is that such an analysis is not possible to perform in poorly differentiated tumors in which there is not a distinct tissue architecture corresponding to a basal layer giving rise to differentiated cells. In these tumors, IHC demonstrates a more random distribution of CD44+ cells. However, the consistent restriction of tumorigenicity to the CD44+ population and the reappearance of both CD44+ and CD44− cells in tumors derived from purified CD44+ cells indicates that a hierarchy remains, but that differentiation in these tumors has likely been blocked at an earlier stage.
Finally, quantitative RT-PCR combined with immunofluorescent staining of tumor sections indicates that the tumorigenic population of cells differentially expresses the gene BMI1. BMI1 has been shown to play a role in the self-renewal of hematopoietic and neuronal stem cells (20, 26) and is considered to be a stem cell-related gene. BMI1 has also been implicated in tumorigenesis, primarily in leukemias (17), but also in several human cancers, including colorectal carcinoma, liver carcinomas, and non-small-cell lung cancer (21). The finding that BMI1 is differentially expressed in the tumorigenic population of HNSCC suggests a potential functional role for BMI1 in this tumor, a possibility that remains to be investigated. The combination of CD44+ staining at the cell membrane and BMI1 staining in the nucleus allows the definition of HNSCC CSC microdomains in the primary tumor and may be useful in the diagnosis of HNSCC in primary sites as well as cells in lymph nodes or distant metastases.
In summary, we have demonstrated that, in HNSCC, there exists a developmental hierarchy, including a population of cells that possess the properties of CSCs. This population can be enriched by selecting for cells that express the cell-surface marker CD44. It is not yet known whether these cells are CSCs at the developmental stage of normal oral squamous epithelium stem cells or progenitors, because the developmental lineages in these tissues are not yet defined.
In addition to the significance of this work to the study of HNSCC, our findings have implications for the field of cancer biology in general. First, this is the second epithelial tumor in which CD44 has been identified as a key cell-surface marker of CSC (breast cancer being the first), suggesting that CD44 is likely to be an important marker for the identification of CSCs in other tumors of epithelial origin. This is perhaps not surprising because, although the differentiation of most stem cells is likely restricted to the organ in which they reside (27–29), similarities in the organization of epithelial tissues suggest that similarities in their respective stem cells may exist. Second, we demonstrate that genes with potentially important biological activities can be identified that are differentially expressed between subpopulations of tumor cells, thus emphasizing the importance of identifying and isolating the appropriate subpopulations of tumor cells before performing large-scale gene-expression and proteomic analyses. Because the CSCs typically make up a small fraction (10% or less) of the tumor cells, and the genes of interest may be expressed at low levels (e.g., transcription factors), analyses of whole tumors may not detect the most important molecular players in the etiology and pathology of cancer.
Materials and Methods
After obtaining informed consent, tumors were obtained from subjects at either the UM or SU hospitals. Animal care and experimental protocols were performed in accordance with procedures and guidelines established by the UM and SU Administrative Panels for Lab Animal Care. Experiments at UM used NOD/SCID mice, and experiments at SU used Rag2γDKO mice.
Primary Tumor Implantation.
NOD/SCID (UM) or Rag2γDKO (SU) mice were anesthetized with ketamine/xylazine, and, once the mice were asleep, an ≈3-mm incision was made, and small pieces (<2 mm) of fresh tumor were implanted on both sides of the flank. Tumors were pinched into their final position, and the incision was sealed with a liquid adhesive suture or a surgical staple.
Tumor Digestion.
Tumors were cut into small fragments, further minced with a sterile scalpel, and then placed in a solution of Media 199 and 200 units/ml Collagenase III. The mixture was incubated at 37°C for up to 3 h to allow complete digestion. Every 15 min, the solution was mixed through a 10-ml pipette to encourage dissociation. Cells were filtered through 40-μm nylon mesh and washed twice with HBSS/2% Heat Inactivated Calf Serum (HICS), then stained for flow cytometry or injected into mice as whole-tumor single-cell suspensions.
Single-Cell Suspension Injections.
Up to two million dissociated tumor cells or varying numbers of FACS-sorted cells were suspended in a volume of 100 μl of RPMI medium 1640 per injection. One hundred microliters of Matrigel (BD Pharmingen, Franklin Lakes, NJ) was added and mixed to form a final volume of 200 μl per injection. The mice were then injected s.c. on the flank with the suspension.
Flow Cytometry.
The single-cell suspensions were washed in HBSS/2% HICS and counted and then resuspended in 100 μl per 106 cells of HBSS and incubated with 1 mg/ml Sandoglobin for 10 min. The suspensions were then washed with HBSS/2% HICS, resuspended in 100 μl per 106 cells of HBSS, and stained with antibodies. Anti-CD44 (Pharred, phycoerythrin (PE)-, or allophycocyanin (APC)-conjugated, clone G44–26; BD Pharmingen) was used at a 1:50 dilution; lineage markers anti-CD2, CD3, CD10, CD16, CD18, CD31, CD64, and CD140b (all diluted 1:50; BD Pharmingen) were used to allow identification of contaminating nontumor cells from patient samples. Tumors that had been passaged in the mouse were incubated with anti-H2kd or H2kb (diluted 1:100; BD Pharmingen). Antibodies were directly conjugated to various fluorochromes, or unconjugated and detected with secondary antibodies, depending on the experiment. Stained cells were washed and resuspended at 0.5 ml per 106 cells with 7-aminoactinomycin (7-AAD; BD Pharmingen) or propidium iodide to allow exclusion of nonviable cells. Cells were sorted with a BD FACSVantage flow cytometer.
IHC and Immunofluorescence.
Upon receipt of a tumor specimen, a small piece was kept aside and frozen in optimal cutting temperature (OCT) embedding media. Seven-micron sections were cut, fixed in ice-cold acetone for 5 min, and air-dried. Slides were then rinsed in PBS/0.1% Tween 20, and blocked in PBS with 0.5% BSA and, in the case of IHC, 0.3% hydrogen peroxide, and 0.1% sodium azide. For CD44 staining, 20 μg/ml of mouse IgG was added to the blocking solution. For CK5/14, Involucrin, and BMI1, 5% goat serum was added to the blocking solution. Sections were incubated in blocking solution at room temperature (RT) for 30 min. Sections were then incubated with the primary antibody diluted in PBS with 0.5% BSA for 30 min at RT. For BMI1, sections were incubated overnight at 4°C. For CD44 staining, biotinylated clone G44–26 (Pharmingen) was used at a dilution of 1:50. For CK5/14, unconjugated clone LH8 (Abcam, Cambridge, U.K.) was used at a dilution of 1:100. For Involucrin, unconjugated clone SY5 (Abcam) was used at a dilution of 1:200. For BMI1, unconjugated clone 1.T.21 (Abcam) was used at a dilution of 1:100. The slides were then washed twice in PBS with 0.1% Tween 20 for 5 min each. A secondary incubation of 30 min at RT was then performed with 1:1,000 Avidin–HRP (Pharmingen) or 1:400 Avidin–Alexa Fluor 488 (Molecular Probes, Eugene, OR), 1:200 biotinylated goat anti-mouse IgM (Jackson ImmunoResearch, West Grove, PA), or biotinylated 1:1,000 goat anti-mouse IgG (Jackson ImmunoResearch), or 1:200 goat anti-mouse IgG–Alexa Fluor 594, for CD44, CK5/14, Involucrin, and BMI1, respectively. Slides were again washed, and for CK5/14 and Involucrin, a tertiary incubation with Avidin–HRP was performed. The sections were then incubated with 3,3′-diaminobenzidine (DAB), as directed by the manufacturer (peroxidase substrate kit; Vector Laboratories, Burlingame, CA), counterstained with hematoxylin, dehydrated, and coverslipped with histomount for IHC. For immunofluorescence, double staining was done sequentially as follows: blocking, unconjugated BMI1, goat anti-mouse Alexa Fluor 594, CD44-biotin, and Avidin–Alexa Fluor 488. Slides were then immersed in 1:1,000 Hoechst 33342 (Invitrogen, Carlsbad, CA) for 1 min, rinsed in PBS/0.1% Tween 20, and coverslipped with Fluoromount G (Southern Biotech, Birmingham, AL).
Quantitative RT-PCR.
Sorted cells were pelleted, resuspended in 1 ml of TRIzol (Invitrogen), and incubated on ice for 10 min. Two microliters of linear acrylamide (Ambion, Austin. TX) were added and mixed, and RNA was purified by standard techniques. cDNA was then synthesized by using the SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen), as directed by the manufacturer. cDNA was diluted to give 200 cell equivalents per microliter. For quantitative PCR, TaqMan gene expression assays (Applied Biosystems, Foster City, CA) were used, which contain prevalidated primers and TaqMan probes for the individual genes in question. Specifically, for human BMI1 and the β-actin control, Applied Biosystems product numbers Hs00180411_m1 and 4333762, respectively, were used. The sequence of these commercially available TaqMan primers and probes is not provided by the manufacturer. PCR was performed as instructed by the manufacturer, in triplicate, on an ABI 7500 real-time PCR system (Applied Biosystems). cDNA isolated from undifferentiated human embryonic stem cells was used as a positive control, and gene expression was normalized to β-actin expression.
Supplementary Material
Acknowledgments
We thank Drs. D. B. Chepeha and T. N. Teknos for their assistance in obtaining tumor samples at UM and Libuse Jerabek and Manja Muijtjens for laboratory and animal support at SU. This work was supported by the National Institutes of Health through UM's Special Program of Research Excellence (SPORE) in Head and Neck Cancer Grant 5 P50 CA097248 (to M.E.P.) and an anonymous gift fund for CSC research at SU (I.L.W. and L.E.A.).
Abbreviations
- CSC
cancer stem cell
- HNSCC
head and neck squamous cell carcinoma
- IHC
immunohistochemistry
- Lin
lineage
- NOD/SCID
nonobese diabetic/severe combined immunodeficient
- Rag2γDKO
Rag2 common cytokine receptor γ-chain double knockout
- UM
University of Michigan
- SU
Stanford University.
Footnotes
Conflict of interest statement: I.L.W. was a member of the scientific advisory board of Amgen and owns significant Amgen stock; and I.L.W. cofounded and consulted for Systemix, is a cofounder and director of Stem Cells, Inc., and recently cofounded Cellerant, Inc.
This article contains supporting information online at www.pnas.org/cgi/content/full/0610117104/DC1.
References
- 1.Jain S, Khuri FR, Shin DM. Curr Probl Cancer. 2004;28:265–286. doi: 10.1016/j.currproblcancer.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Aubele M, Werner M. Anal Cell Pathol. 1999;19:53–58. doi: 10.1155/1999/960923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Golub TR. N Engl J Med. 2001;344:601–602. doi: 10.1056/NEJM200102223440809. [DOI] [PubMed] [Google Scholar]
- 4.Pardal R, Clarke MF, Morrison SJ. Nat Rev Cancer. 2003;3:895–902. doi: 10.1038/nrc1232. [DOI] [PubMed] [Google Scholar]
- 5.Reya T, Morrison SJ, Clarke MF, Weissman IL. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 6.Blair A, Hogge DE, Ailles LE, Lansdorp PM, Sutherland HJ. Blood. 1997;89:3104–3112. [PubMed] [Google Scholar]
- 7.Bonnet D, Dick JE. Nat Med. 1997;3:730–737. doi: 10.1038/nm0797-730. [DOI] [PubMed] [Google Scholar]
- 8.Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, Gotlib J, Li K, Manz MG, Keating A, et al. N Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 9.Lapidot T, Sirard C, Vormoor J, Murdoch B, Hoang T, Caceres-Cortes J, Minden M, Paterson B, Caligiuri MA, Dick JE. Nature. 1994;367:645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 10.Miyamoto T, Weissman IL, Akashi K. Proc Natl Acad Sci USA. 2000;97:7521–7526. doi: 10.1073/pnas.97.13.7521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dontu G, Jackson KW, McNicholas E, Kawamura MJ, Abdallah WM, Wicha MS. Breast Cancer Res. 2004;6:R605–R615. doi: 10.1186/bcr920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hemmati HD, Nakano I, Lazareff JA, Masterman-Smith M, Geschwind DH, Bronner-Fraser M, Kornblum HI. Proc Natl Acad Sci USA. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ignatova TN, Kukekov VG, Laywell ED, Suslov ON, Vrionis FD, Steindler DA. Glia. 2002;39:193–206. doi: 10.1002/glia.10094. [DOI] [PubMed] [Google Scholar]
- 15.Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 16.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 17.Lessard J, Sauvageau G. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 18.Molofsky AV, Pardal R, Iwashita T, Park IK, Clarke MF, Morrison SJ. Nature. 2003;425:962–967. doi: 10.1038/nature02060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park IK, Morrison SJ, Clarke MF. J Clin Invest. 2004;113:175–179. doi: 10.1172/JCI20800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park IK, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. Nature. 2003;423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 21.Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Cell. 2004;118:409–418. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Prochazka M, Gaskins HR, Shultz LD, Leiter EH. Proc Natl Acad Sci USA. 1992;89:3290–3294. doi: 10.1073/pnas.89.8.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldman JP, Blundell MP, Lopes L, Kinnon C, Di Santo JP, Thrasher AJ. Br J Haematol. 1998;103:335–342. doi: 10.1046/j.1365-2141.1998.00980.x. [DOI] [PubMed] [Google Scholar]
- 24.Chu PG, Weiss LM. Histopathology. 2002;40:403–439. doi: 10.1046/j.1365-2559.2002.01387.x. [DOI] [PubMed] [Google Scholar]
- 25.Walts AE, Said JW, Siegel MB, Banks-Schlegel S. J Pathol. 1985;145:329–340. doi: 10.1002/path.1711450406. [DOI] [PubMed] [Google Scholar]
- 26.Molofsky AV, He S, Bydon M, Morrison SJ, Pardal R. Genes Dev. 2005;19:1432–1437. doi: 10.1101/gad.1299505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Massengale M, Wagers AJ, Vogel H, Weissman IL. J Exp Med. 2005;201:1579–1589. doi: 10.1084/jem.20050030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wagers AJ, Christensen JL, Weissman IL. Gene Ther. 2002;9:606–612. doi: 10.1038/sj.gt.3301717. [DOI] [PubMed] [Google Scholar]
- 29.Wagers AJ, Sherwood RI, Christensen JL, Weissman IL. Science. 2002;297:2256–2259. doi: 10.1126/science.1074807. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.